ABSTRACT
Carbonic anhydrase IX (CAIX) is a pH-regulating enzyme that plays a key role in maintaining an alkaline intracellular pH under hypoxic conditions. It is overexpressed in a variety of solid cancers, including breast cancer (BC), and has been implicated in the migration, invasion and stemness of breast cancer cells. Therefore, CAIX recently emerged as a novel therapeutic target for the treatment of BC. To gain an insight into the mechanism of action of CAIX inhibitors, we investigated the impact of CAIX knock-down on the transcriptional response to hypoxia in 2 BC cell lines – MCF7 and MDA-MB-231, by performing a global gene expression analysis. This showed that CAIX knock-down had a relatively minor effect on the global transcriptional response to hypoxia, however it blocked hypoxia-induced upregulation of stanniocalcin-1 (STC1), a secreted glycoprotein that has been shown to promote tumor progression and metastasis in BC. Kaplan-Meier survival analysis showed that high STC1 expression is significantly associated with poor survival in patients with basal-type breast cancer but not luminal A and HER2+ subtypes. Moreover, the association was particularly high in a subgroup of basal-type BC patients with TP53 mutations thus revealing a putative cooperation of STC1 with mutated TP53 in generating highly aggressive BC subgroup. Taken together, these findings show that CAIX inhibitors at least partially act through blocking STC1 induction in BC cells and reveal a subgroup of BC patients, who potentially would benefit most from the treatment with CAIX inhibitors.
| Abbreviations | ||
| AZM | = | acetazolamide |
| BC | = | breast cancer |
| CAIX | = | carbonic anhydrase IX |
| DEG | = | differentially expressed gene |
| HIF-1α | = | hypoxia inducible factor 1 α subunit |
| NF-κB | = | nuclear factor kappa B |
| PI3K | = | phosphatidylinositol-4,5-bisphosphate 3-kinase |
| STC1 | = | stanniocalcin-1 |
| TNBC | = | triple negative breast cancer |
Introduction
Carbonic anhydrase IX (CAIX) is a transmembrane zinc metalloenzyme that catalyzes the reversible hydration of carbon dioxide to bicarbonate and protons (CO2 + H2O ↔ HCO3− + H+).Citation1 The expression of CAIX in normal tissues is restricted to stomach, duodenum, small intestine and gallbladder (http://www.proteinatlas.org/), where it had been implicated in the pH regulation and cell adhesion.Citation2-5 CAIX knockout mice showed only mild phenotypes mostly related to hyperplasia of the gastric mucosa and increased pit cell to chief cell ratio, which was not associated with increased tumorigeneicity.Citation4,6
On the other hand, CAIX is overexpressed in a variety of solid cancers, including breast cancer (BC).Citation5,7,8 In the majority of BC cell lines, its expression level is relatively low under well-oxygenated conditions, while it is strongly induced in hypoxic conditions.Citation9-11 Hypoxic signaling is mediated by hypoxia-inducible transcription factors HIF-1 and HIF-2 that activate multiple gene expression programs required for the adaptation of tumor cells to hypoxia, including reprogramming of glucose metabolism and pH regulation.Citation12 Accelerated rate of glycolysis in combination with poor vasculature perfusion leads to the accumulation of a large amount of lactic acid resulting in acidification of the tumor microenvironment. Extracellular pH in tumor tissues can be as low as 6.7.Citation13 In normal cells, this could lead to a lowering of the intracellular pH, which in turn can result in the disruption of a variety of biologic functions and trigger apoptosis,Citation14-16 whereas cancer cells have evolved a dynamic pH regulatory system that allows them to adapt to acidic microenvironment. Furthermore, several lines of evidence suggest that acidosis-induced signaling not only helps the cancer cells to survive but also promotes invasiveness, growth and acquisition of stem cell phenotype.Citation17,18 CAIX is a direct transcriptional target of HIF-1 and was shown to be one of the most strongly upregulated genes in response to hypoxia and a robust biomarker of tumor hypoxia.Citation19,20 It is an essential component of the pH regulatory system in tumors - the bicarbonate produced at the extracellular surface by CAIX is transported into the cytoplasm by monocarboxylate transporters and Cl−/HCO3− exchangers, which helps to maintain an alkaline intracellular pH, while the proton remains in the extracellular space thus further contributing to its acidification.Citation21,22
Several studies have demonstrated that increased CAIX expression in tumor tissues is associated with distant metastasis and poor survival in BC patients.Citation8,9,23 Our recent study showed that high CAIX mRNA expression is significantly associated with poor survival in patients with basal-like and triple negative breast cancer (TNBC), but not in the luminal A and HER2+ subtypes.Citation11 At least partially, this could be due to the fact that CAIX is overexpressed more frequently in TNBC than in other BC subtypes via hypoxia-independent mechanisms of HIF-1α expression.Citation9,24 It is also possible that CAIX mediates TNBC-specific signaling events leading to metastasis and resistance to chemotherapy. In fact, in a previous study we found that silencing of CAIX reduced invasiveness and self-renewal capacity under hypoxic conditions and had a synergistic effect with doxorubicin on decreasing the spheroid-forming efficiency in TNBC cells.Citation11
Taken together, these studies have established CAIX as a therapeutic target for the treatment of TNBC.Citation9,11,25 However, the molecular mechanisms underlying the tumor-promoting effects of CAIX and signaling pathways affected by the inhibition of CAIX remained unknown.
In the current study, we investigated the impact of CAIX knock-down on the transcriptional response to hypoxia in 2 BC cell lines – MCF7 and MDA-MB-231, representing luminal A and TNBC subtypes, respectively, by performing a global gene expression analysis. Results revealed that the genetic silencing of CAIX and/or pharmacological inhibition of CAs by acetazolamide abrogated hypoxia-induced upregulation of stanniocalcin-1 (STC1), which is significantly associated with poor survival in patients with basal-type BC and has been previously shown to stimulate the invasiveness and tumor progression in TNBC.Citation26,27
Results
Genetic silencing of CAIX in BC cell lines
The generation of CAIX-depleted BC cell lines and the biologic effects of CAIX silencing were reported before.Citation11 Briefly, MDA-MB-231 and MCF7 cell lines representing the TNBC and luminal A subtypes, respectively, were stably transfected with a pool of 3 plasmids encoding CAIX-specific shRNAs or scrambled shRNA. CAIX depletion did not have an impact the cell proliferation or death rates in any of the cell lines, while it inhibited the clonal spheroid-forming ability, which is a surrogate marker for the self-renewal capacity, and the invasiveness of the MDA-MB-231 cells under hypoxia.Citation11
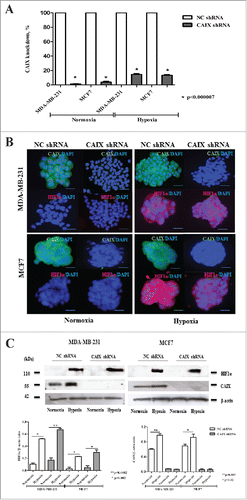
In the current study, we exploited the same CAIX-depleted and control cell lines. qRT-PCR analysis showed that CAIX mRNA expression was decreased by 98 and 85% in MDA-MB-231 cells and by 95 and 88% in MCF7 cells under normoxia and hypoxia (). Immunofluorescence (IF) and Western blotting analysis confirmed that CAIX mRNA and protein levels were significantly decreased in the CAIX-depleted cells as compared with the control cells both under hypoxic and normoxic conditions ( and ). The hypoxic conditions were verified by IF and Western blotting with anti-CAIX and anti-HIF1α antibodies, which showed a strong induction of HIF1α under hypoxic conditions both in the CAIX-depleted and the control cells, while the CAIX expression was induced by hypoxia in the control cells only ( and ).
Figure 1. Verification of CAIX knock-down and exposure to hypoxia in MDA-MB-231 and MCF7 cells. (A) The percent knockdown of CAIX mRNA expression measured by qRT-PCR in MDA-MD-231-shCAIX, MCF7-shCAIX and the respective negative control cells grown at normoxic or hypoxic conditions. (B) Immunofluoresnce and (C) Western blot analysis of CAIX and HIF1α expression in MD-231-shCAIX, MCF7-shCAIX and the respective negative control cells grown as multicellular spheroids in serum-free medium at normoxic or hypoxic conditions. Quantitative analysis of the band intensity normalized to β-actin is shown in the lower panel.
Characterization of the transcriptional response to hypoxia
To interrogate the hypoxia response pathways affected by the depletion of CAIX in BC cells, we performed gene expression profiling using SurePrint G3 Human Gene Expression 60K microarrays in MDA-MB-231-shCAIX, MCF7-shCAIX and the respective control cells grown as multicellular spheroids under hypoxic or normoxic conditions. The Pearson correlation coefficients between the biologic replicates ranged from 0.93 to 0.95 in the hypoxic cells and from 0.93 to 0.96 in the normoxic cells, thus showing a relatively good reproducibility of the hypoxia experiments.
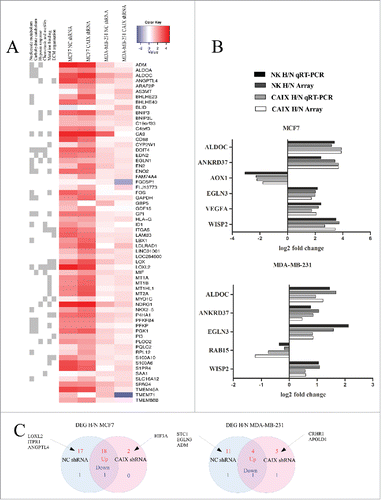
A total of 32 and 85 genes were differentially expressed (log FC >1; adj p-value <0.1) in MDA-MB-231-shCAIX cells and MDA-MB-231-shNC cells, and 239 and 165 genes in MCF7-shCAIX and MCF-shNC cells, respectively, upon the exposure to hypoxia. The top 20 differentially expressed genes (DEGs) in each of the cell lines are shown in the . Eleven DEGs were selected for validation by qRT-PCR and all of them showed a good concordance between the microarray and qRT-PCR results (). Our results indicate that MCF7 cells have stronger transcriptional response to hypoxia than MDA-MB-231 cells. The majority of DEGs are upregulated in response to hypoxia in all 4 cell lines and overall the transcriptional response to hypoxia is similar in the CAIX-depleted and control cells. However, a subset of genes, which were upregulated by hypoxia in the control cells, were unaffected or even downregulated by exposure to hypoxia in the CAIX depleted cells. These included GBP5, FGD5P1, TMEM71 and ID1, and some genes that were not included in the list of top 20 DEGs. Apparently, CAIX is required for the induction of their expression in hypoxic conditions. Though, the functional role of these genes in the cellular response to hypoxia is currently unknown.
Figure 2. Transcriptional response to hypoxia in the CAIX-depleted and control cell lines. (A) A merged list of top 20 DEGs altered by hypoxia in the MDA-MB-231-shCAIX and MCF7-shCAIX and the respective negative control cells. In the heatmap, hypoxia vs normoxia log fold change is shown. Similar GO terms associated with the DEGs were grouped and the GO term groups are shown on the left. (B) Validation of microarray results by qRT-PCR in the same RNA samples that were used for gene expression profiling. The figure shows the log2-fold change in gene expression in hypoxia (H) vs normoxia (N) in the CAIX-depleted and control cells. (C) Venn diagram showing the number of hypoxia response genes (genes that are associated with the GO term “response to oxygen levels”) upregulated (red) and downregulated (blue) by hypoxia in the CAIX-depleted and control cells.
The majority of DEGs were associated with GO terms involved in hypoxia response (“response to hypoxia,” “response to oxygen levels” etc.), carbohydrate metabolism (“canonical glycolysis,” “carbohydrate catabolic process,” fructose metabolic process” etc.), nucleoside and RNA metabolism (“purine nucleotide metabolic process,” “ribonucleotide metabolic process,” “mRNA catabolic process” etc.), chemotaxis and motility (“neutrophil chemotaxis,” “regulation of cell migration” etc.), metal ion binding (“metal ion binding,” “zinc ion binding” etc.) and ECM organization and adhesion (“extracellular structure organization,” “cell junction assembly” etc.) (). In MCF7 cells, the majority of enriched GO terms overlapped between the CAIX-depleted and control cells, whereas in MDA-MB-231 cells, several GO terms related to response to hypoxia and metabolism were significantly enriched in the control cells but not the CAIX-depleted cells.
To focus specifically on the genes that are involved in the cellular response to hypoxia, we selected a set of 272 genes associated with the GO term “response to oxygen levels” and performed a detailed DEG analysis. This analysis revealed 19 genes that were significantly altered by hypoxia in both MCF7-shCAIX and the respective control cells, 18 genes that were altered in the controls cells only and 2 genes were significantly upregulated in the CAIX-depleted cells only (). In MDA-MB-231 cells, 5 genes were altered both in the CAIX-depleted and control cells, 12 – in the control cells and 6 in the CAIX –depleted cells only (). Hypoxia-regulated genes that most significantly differ between the CAIX-depleted and control cells are listed in the .
Table 1. Hypoxia-regulated genes that significantlyTable Footnote* differ between the CAIX-depleted and control cells.
CAIX is required for the induction of STC1 expression under hypoxia
Among the hypoxia response genes that were upregulated in MDA-MB-231 control cells but not in the CAIX-depleted cells was STC1 – a hypoxia inducible gene that has been shown to play an oncogenic role in breast cancer.Citation27,28 Its expression pattern was validated by qRT-PCR in an independent hypoxia treatment experiment. STC1 mRNA expression was induced 1.8-fold by the exposure to hypoxia in MDA-MB-231-shNC cells, whereas in MDA-MB-231-shCAIX cells it was not significantly altered thus suggesting that CAIX is required for STC1 induction in hypoxia (). In MCF7 cells, STC1 expression increased 3.7-fold upon exposure to hypoxia, yet the induction was only partially abolished by the depletion of CAIX. These results were confirmed at protein level by Western blot analysis ().
Figure 3. Levels of STC1 and CAXII expression in the CAIX-depleted and control cell lines. (A) qRT-PCR analysis and (B) Western blot analysis of STC1 expression in MDA-MD-231-shCAIX, MCF7-shCAIX and the respective negative control cells grown at normoxic or hypoxic conditions with and without treatment with 1 µM acetazolamide (AZM). Quantitative analysis of the STC1 band intensity normalized to β-actin is shown in the lower panel. (C) qRT-PCR analysis of CAXII mRNA expression in MDA-MD-231-shCAIX, MCF7-shCAIX and the respective negative control cells grown at normoxic or hypoxic conditions.
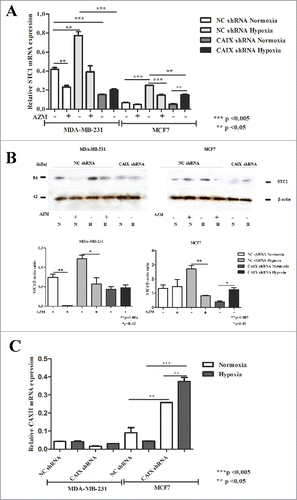
Given that luminal BC cells T47D and MCF10A cells have been shown to express high levels of CAXII,Citation29 we hypothesized that MCF7 cells may also express CAXII in response the loss of CAIX. Therefore, next we analyzed CAXII mRNA expression levels in the same set of cell lines by qRT-PCR. The results confirmed that MCF7-shCAIX cells overexpressed CAXII mRNA both at normoxia and hypoxia, while no induction of CAXII was detected in MDA-MB-231 cells (). These data suggest that CAXII expression compensates for the CAIX loss in MCF7 but not in MDA-MB-231 cells.
To investigate whether CAIX enzymatic activity is required for mediating STC1 induction, the cells were treated with a non-selective CA inhibitor acetazolamide (AZM) – a sulfonamide derivative that inhibits mammalian CAII, VI, VII, IX and XII isoforms.Citation30 The treatment with 1μM AZM reduced STC1 mRNA and protein expression level in normoxia and fully abolished STC1 induction by hypoxia in MDA-MB-231-shNC cells ( and ). Similar effect of AZM on STC1 induction by hypoxia was observed in MCF7 cells. These data show that the induction of STC1 expression under hypoxia is mediated by CA enzymatic activity and support the hypothesis that CAXII expression functionally compensates the knock-down of CAIX in MCF7 cells.
High STC1 mRNA expression is associated with poor survival in basal breast cancer
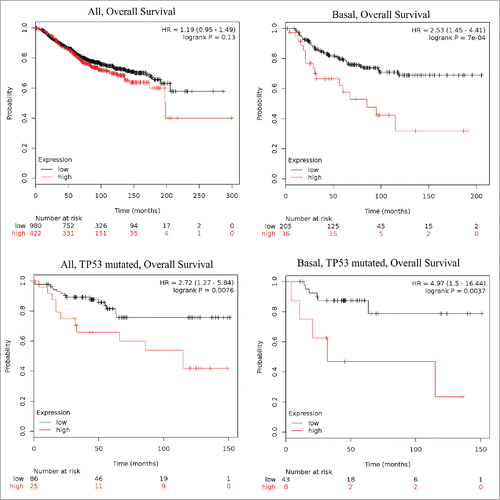
To investigate a possible link between STC1 expression level and the clinical outcome in BC patients, we exploited Kaplan-Meier plotter – an online tool for drawing survival plots based on gene expression data and survival information in a cohort of 5143 BC patients with a mean follow-up of 69 months.Citation31 The patients were split by upper tertile of STC1 expression into groups with high vs low STC1 mRNA expression. When all patients were analyzed together, no significant association between STC1 level and relapse free survival (RFS), overall survival (OS) or distant metastasis free survival (DMFS) was observed. However, when the intrinsic molecular subtypes were analyzed separately, Kaplan-Meier survival analysis revealed a significant association between high STC1 expression and worse OS and RFS in patients with basal subtype (defined as ESR1-/HER2- BC), while no such correlations were found for luminal A and HER2+ subtypes (, ). In luminal B subtype, high STC1 expression was associated with shorter DMFS but not with RFS and OS. Furthermore, the association between high STC1 expression and shorter OS and RFS was even stronger, when a subgroup of basal-type BC cases with TP53 mutations were analyzed separately. Taken together these data show that increased expression of STC1 is associated with poor prognosis in basal-type BC but not in other subtypes and reveal a novel link between STC1 and mutated TP53 in driving progression of basal BC.
Table 2. Association between STC1 mRNA expression and survival in various subtypes of breast cancer.
Figure 4. Kaplan-Meier plots showing the association between STC1 mRNA expression level and overall survival in various subgroups of BC patients. Red lines – patients with high STC1 expression, black lines – patients with low STC1 expression, vertical lines – censored events, cut-off – upper tertile of STC1 expression over entire BC data set.
Discussion
Due to the restricted expression pattern in normal tissues, overexpression in tumor tissues and the functional significance in cancer progression, CAIX was proposed as a target for the treatment of renal cell carcinoma (RCC) more than 16 y ago.Citation32 Since then, several sulfonamide and coumarin-based small molecule inhibitors and CAIX-binding monoclonal antibodies have been developed.Citation21 Clinical trials with cG250 - a monoclonal antibody against CAIX, have demonstrated a clinically meaningful disease stabilization in a subgroup of metastatic RCC patients with high CAIX expression,Citation33 while no clinical benefit was found for patients with localized RCC.Citation34 This antibody, however, binds to the PG-like domain, not the catalytic domain of CAIX, hence its mechanism of action most likely is related to antibody-mediated cell cytotoxicity, not the inhibition of CAIX enzymatic activity. Several clinical trials with small molecule inhibitors have been initiated (for example, NCT02215850), however the results have not been reported so far, hence their therapeutic potential is not known yet. More recently, CAIX has been proposed as a drug target for the treatment of other cancers, including BC.Citation21,35 Preclinical studies have shown that CAIX has a significant effect on the survival, migration and self-renewal capacity of BC cells and its inhibition reduces the growth rate of primary breast tumors and metastasis.Citation9,11,25,36 Molecular pathways affected by the CAIX inhibition, however, are largely unknown. Gaining an insight into the molecular mechanisms of CAIX inhibitors could help to select rational drug combinations and to define patient subgroups for further clinical trials.
To the best of our knowledge, 2 studies have reported on alterations in gene expression profiles elicited by up- or downregulation of CAIX before. A study by Shin et al. (2011) showed that forced overexpression of CAIX in a cervical carcinoma cell line altered Rho-GTPase signaling leading to weakened cell-cell adhesion, and increased cell migration and invasion.Citation37 Radvak et al. (2012) studied gene expression profiles in hypoxic fibrosarcoma cells and showed that CAIX knockdown affected the expression levels of several genes involved in focal adhesion leading to the reduced adhesion and spreading of CAIX-depleted cells.Citation38
In the current study, we investigated the impact of CAIX knock-down on the transcriptional response to hypoxia in 2 BC cell lines. One of the hypoxia-response genes whose induction was completely abrogated in the CAIX-depleted MDA-MB-231 cells was STC1 encoding stanniocalcin-1 – a secreted glycoprotein that has been implicated in multiple physiologic processes, such as maintenance of calcium homeostasis, reducing oxidative stress, cytoprotection and wound healingCitation39-41 and is overexpressed in a variety of cancers, including but not limited to glioma, colorectal, lung and breast cancer.Citation26,27,42-44 STC1 expression was found to be induced by hypoxia in various normal tissues and it is likely to act in a paracrine and/or autocrine manner.Citation45-47 It was shown to inhibit hypoxia-induced cell apoptosis and the production of reactive oxygen species in cardiomyocytes and to improve poststroke functionality.Citation46,48 On the other hand, elevated expression of STC1 in tumor tissues appears to promote the progression of cancer and metastasis and is associated with shorter survival and poor response to therapy in the majority of cancers.Citation28,44,49,50 STC1 knock-down in murine and human BC cell lines had no effect on cell proliferation, while it reduced cell invasiveness and metastasis in mice models.Citation27 In line with this, a study by Han et al. (2016) showed that the treatment with recombinant human STC1 significantly increased the invasiveness of TNBC cells and this effect was mediated by the phosphorylation of JNK/c-Jun leading to the upregulation of MMP9.Citation51 Moreover, the level of STC1 expression was found to be higher in TNBC than in other BC subtypes and it was associated with shorter survival in patients with basal-type BC.Citation51
Our results show that the hypoxia-induced upregulation of STC1 is abrogated by the depletion of CAIX in MDA-MB-231 cells. This suggests that at least partially the biologic effects caused by the inhibition of CAIX – the reduced invasiveness and self-renewal capacityCitation11 are mediated by blocking of STC1 induction. We observed that STC1 expression was induced by hypoxia in MCF7 cells too, however the knock-down of CAIX only partially reduced its induction. At the same time, the expression of CAXII was markedly increased in the CAIX-depleted MCF7 cells but not MDA-MB-231 cells. CAXII is another cancer-associated, cell surface carbonic anhydrase isoenzyeme that has been previously shown to be expressed in luminal BC cell lines but not in MDA-MB-231 cells.Citation29,52 Moreover, gene expression data available at Kaplan Meier plotter suggest that CAXII is expressed at higher levels in luminal BC than in basal BC tissues (for example, the expression range for probe 215867_x_at (CA12) is 129–32243, median 6270 for luminal A subtype, while it is only 60–7411, median 517 for basal subtype BC). We hypothesized that CAXII may functionally compensate the loss of CAIX in MCF7 cells. This idea was supported by the fact that AZM – a non-selective CA inhibitor that acts on both CAIX and CAXII fully abolished the induction of STC1 in MCF7 cells. Therefore, effective blocking of hypoxia-induced upregulation of STC1 in breast cancer would require dual CAIX and CAXII inhibitors.
In our previous study, Kaplan-Meier survival analysis revealed that high CAIX expression is associated with poor survival specifically in patients with basal-type BC.Citation11 Likewise, here we found a strong association between high STC1 expression and shorter OS and RFS in patients with the basal-type BC but not with the other BC subtypes. The association was even stronger, when a subgroup of TP53-mutated basal-type BC cases was analyzed – the prognosis was significantly worse in patients with TP53 mutations and high STC1 level than in patients with TP53 mutations and low STC1 expression level. This finding suggests that the activation of STC1 signaling cooperates with the loss of TP53 in generating highly aggressive BC phenotype. Although the molecular basis of this interplay is unknown, it suggests that this particular subgroup of patients may benefit most from CAIX/CAXII inhibitors.
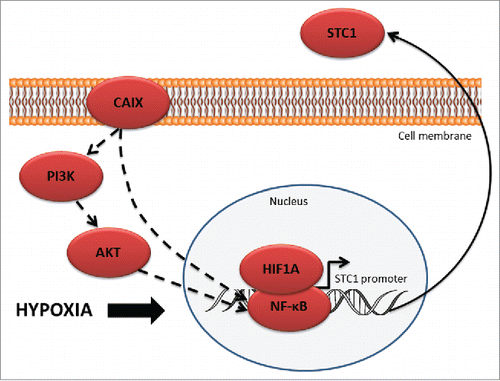
The mechanism by which CAIX regulates STC1 expression remains to be elucidated. The biologic data mining suggested that the regulatory mechanism may involve NF-κB (). Both, NF-κB and HIF-1α have been shown to bind to the STC1 promoter and regulate its transcription.Citation53 NF-κB pathway is known to be induced by hypoxia, while CAIX depletion has been shown to block the hypoxic induction of NF-κB activity by inhibiting its translocation to the nucleus.Citation54 Hence, we hypothesize that the hypoxia-induced upregulation of STC1 requires interaction between NF-κB and HIF-1α. Given that CAIX overexpression has been shown to activate PI3K/AKT signaling pathway,Citation55 and AKT is involved in the activation of NF-κB by phosphorylating I-κB and RELA,Citation56 one of the possibilities is that CAIX-dependent NF-κB activation may happen through PI3K/AKT pathway.
Due to the ubiquitous expression in normal tissues and cytoprotective physiologic functions, STC1 as such is unlikely to serve as a drug target. Our study revealed that the inhibition of CAIX blocks the hypoxia-induced upregulation of STC1 in BC cells, thus providing a new insight into the mechanism of action of CAIX/XII inhibitors and suggesting that CAIX/XII inhibition may serve as a novel strategy for tumor-specific inhibition of STC1. Furthermore, it showed that a subgroup of basal-type BC patients with high STC1 expression and TP53 mutations may benefit most from this treatment.
Materials and methods
Cell culture and exposure to hypoxia
The human breast cancer cell lines MDA-MB-231 and MCF7 were purchased from the European Collection of Cell Cultures (ECACC, UK) and maintained as recommended by the manufacturer. The cell cultures were monitored for mycoplasma infections at least once per month using PCR Mycoplasma Test Kit I/C (PromoKine, #PK-CA91–1096). The generation of stable CAIX knockdown cells using shRNAs was described previously.Citation11 The obtained cell lines with stable knockdown of CAIX were designated as MDA-MB-231-shCAIX and MCF7-shCAIX, and the respective negative controls as MDA-MB-231-shNC and MCF7-shNC. The efficiency of CAIX silencing was assessed by qRT-PCR and immunofluorescence before the experiments.
The transfected cells were plated at density 2 × 103 cells per ml of serum-free DMEM/F12 medium supplemented with EGF (20 ng/ml, R&D Systems, #236-EG-200), bFGF (10 ng/ml, SantaCruz, #sc-4573), hydrocortisone (50 ng/ml, Sigma-Aldrich, #H0135–1MG) and 1 × B27 (Invitrogen, #17504001) and grown as multicellular spheroids for 5 d. To establish hypoxic conditions, the cells were cultured at 1% oxygen, 94% N2 and 5% CO2 at 37°C using humidified multi-gas incubator (Sanyo, Sanyo Electric Co.,Ltd.) for 48 hours. The normoxic control cells were incubated at 37°C with 5% CO2 in a humidified incubator (Panasonic, Panasonic Healthcare Co., Ltd.).
To evaluate the effects of pharmacological CA inhibition on the expression of STC1, the spheroids were treated with 1 µM acetazolamide (AZM) (Thermo Fisher Scientific, #L07562) and cultured for 48 h under hypoxia or normoxia. 10% DMSO was used as the solvent control.
Immunofluorescence
Spheroids were attached onto poly-L-lysine-coated slides by centrifuging at 800 rpm for 5 min using a Shandon cytospin and then fixed in ice cold methanol/acetone (1:1) for 20 min. The slides were washed in PBS, blocked with 3% BSA for 30 min and then incubated with rabbit anti-CAIX antibody (abcam, #ab15086) (dilution 1:50) or mouse monoclonal anti-HIF1α antibody (R&D Systems, #MAB1935) (dilution 1:50) for 12 h at +4°C. After washing the slides were incubated with goat anti-rabbit IgG secondary antibody (Jackson ImmunoResearch, #111–095–006) conjugated with fluorescein (FITC) or goat anti-mouse IgG secondary antibody (Jackson ImmunoResearch, #115–165–0) conjugated with cyanine (Cy3), respectively for 1 hour at room temperature. The stained slides were mounted with Prolong gold antifade reagent with DAPI (Invitrogen, Life Technologies, USA) and the images were acquired using Leica DM3000 microscope (Leica Microsystems, Germany).
RNA isolation and gene expression profiling
Total RNA was isolated using mirVana PARIS Kit (Thermo Fisher Scientific, #AM1556) according to manufacturer's protocol. RNA was treated with DNAse I (Ambion, #AM1906) and the quantity and quality was assessed using Agilent 2100 Bioanalyzer and RNA 6000 Nano kit (Agilent technologies, #5067–1511). Gene expression profiling was performed using SurePrint G3 Human Gene Expression 8 × 60K microarrays (Agilent, #G4851A) containing 60 000 probes according to manufacturer's instructions. Briefly, 50 ng of total RNA were converted to cRNA, labeled with Cy3 and hybridized on SurePrint arrays. The arrays were scanned at 4 μm resolution in PowerScanner (Tecan) and the data were extracted using GenePix 6.0.1.22 software (Molecular Devices). The analysis was performed in 2 biologic replicates.
Quantitative RT-PCR
For validation of microarray data, 7 genes were selected for qRT-PCR analysis. Two µg of total RNA were converted to cDNA using random hexamer primers and Revert Aid M-MuLV Reverse Transcriptase (Thermo Fisher Scientific, #K1622). qRT-PCR was performed using 2 µl of 1:10 diluted cDNA reaction mixtures, 6 µl SYBR green (Thermo Fisher Scientific, #4367659), 3,8 µl dH2O, 0,2 µl 10pM forward and reverse primer mixture and ViiA 7 real-time PCR system (Thermo Fisher Scientific). The primer sequences and concentrations are available upon request. All qRT-PCR experiments were performed in duplicates. To normalize the expression data, the normalization factor was calculated as the geometric mean of the 3 most stable reference genes (ACTB, PGK1, YWHAZ; the primer sequences are available upon request) selected from 7 frequently used housekeeping genes using geNorm software
Western blot analysis
Protein and total RNA were isolated simultaneously from the same cell samples using mirVana PARIS Kit (Thermo Fisher Scientific, #AM1556). Ten micrograms of protein were applied per lane and separated by 10% SDS-PAGE, and electroblotted onto nitrocellulose membranes. The membranes were blocked with 10% (w/v) fat-free milk and then incubated with the following primary antibodies: CAIX (1:1000) (abcam, #ab15086), HIF1α (1:2000) (abcam, #ab51608), STC1 (1:5000) (R&D Systems #AF2958-SP), β-actin (1:4000) (abcam, #ab8224). After washing, the blots were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG F(ab')2-HRP (1:2000) (Santa Cruz, #sc-3837), donkey anti-goat IgG F(ab')2-HRP (1:2000) (Santa Cruz, #sc-3851) or chicken anti-mouse IgG-HRP (1:2000) (Santa Cruz, #sc-2962) secondary antibodies, respectively. The blots were developed with ECL Select Western Blotting Detection Reagents (GE Healthcare, #RPN2235) according to manufacturer's instructions. For quantification of protein band intensity, the data was analyzed using software ImageJ, averaged between duplicates and represented in the graphs as ratio of protein of interest against β-actin.
Meta-analysis by Kaplan-Meier Plotter
The correlation between STC1 mRNA expression level and survival was assessed using the Kaplan-Meier Plotter (http://kmplot.com/analysis/) - an online tool that integrates gene expression and clinical data on 5143 BC patients. Relapse-free survival (RFS), overall survival (OS) and distant metastasis free survival (DMFS) were selected as survival endpoints. For STC1 expression, Affymetrix ID 204595_s_at probe set was used. The upper tertile of STC1 expression (computed over entire data set) was used to split patients into the groups with high or low STC1 expression. This cutoff was selected based on the distribution of gene expression values given in the Beeswarm graphs. The patient groups were compared by Kaplan-Meier survival plots and the hazard ratio (HR; 95% confidence intervals) and log-rank P values were calculated. Biased arrays (i.e. those with 2 or more parameters out of the 95% range of all arrays) and redundant samples were excluded from the analysis.
Statistical analysis
The microarray data were background subtracted, log2- transformed and normalized using quantile method between replicates and adjustment to 70% quantile for all samples as recommended by Agilent. The data have been deposited to the ArrayExpress (ArrayExpress accession E-MTAB-5512 http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-5512/). Limma package was applied to identify differentially expressed genes between the cell cultures grown in hypoxia and normoxia. FDR method was used for the multiple testing correction and adjusted (adj) P-value of <0.1 was considered to be significant. Hierarchical cluster analysis was performed using ad hoc composed R-language script (R Development Core Team). Gene ontology (GO) analysis was performed with GOstats package. Pathway Studio (Elsevier) was used for the biologic data mining.
For comparing with microarray data, the obtained qRT-PCR data were log2-transformed and represented in the graphs as log2 fold-change. For STC1, CAIX and CAXII expression analysis the data are represented in graphs as means ± SD. Statistical significance was determined by the nonparametric Mann-Whitney U test and differences were considered to be significant at P < 0.05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by the Latvian National Research Program BIOMEDICINE 2014–2017.
References
- Li Y, Tu C, Wang H, Silverman DN, Frost SC. Catalysis and pH control by membrane-associated carbonic anhydrase IX in MDA-MB-231 breast cancer cells. J Biol Chem 2011; 286:15789-96; PMID:21454639; https://doi.org/10.1074/jbc.M110.188524
- Svastova E, Zilka N, Zat'ovicova M, Gibadulinova A, Ciampor F, Pastorek J, Pastorekova S. Carbonic anhydrase IX reduces E-cadherin-mediated adhesion of MDCK cells via interaction with beta-catenin. Exp Cell Res 2003; 290:332-45; PMID:14567991; https://doi.org/10.1016/S0014-4827(03)00351-3
- Csaderova L, Debreova M, Radvak P, Stano M, Vrestiakova M, Kopacek J, Pastorekova S, Svastova E. The effect of carbonic anhydrase IX on focal contacts during cell spreading and migration. Front Physiol 2013; 4:271; PMID:24101905; https://doi.org/10.3389/fphys.2013.00271
- Leppilampi M, Karttunen TJ, Kivela J, Gut MO, Pastorekova S, Pastorek J, Parkkila S. Gastric pit cell hyperplasia and glandular atrophy in carbonic anhydrase IX knockout mice: studies on two strains C57/BL6 and BALB/C. Transgenic Res 2005; 14:655-63; PMID:16245156; https://doi.org/10.1007/s11248-005-7215-z
- Ondriskova E, Debreova M, Pastorekova S. Chapter 10 - tumor-associated carbonic anhydrases IX and XII A2 - Supuran, Claudiu T. In: Simone GD, ed. Carbonic anhydrases as biocatalysts. Amsterdam: Elsevier, 2015:169-205; https://doi.org/10.1016/B978-0-444-63258-6.00010-X
- Gut MO, Parkkila S, Vernerova Z, Rohde E, Zavada J, Hocker M, Pastorek J, Karttunen T, Gibadulinova A, Zavadova Z, et al. Gastric hyperplasia in mice with targeted disruption of the carbonic anhydrase gene Car9. Gastroenterology 2002; 123:1889-903; PMID:12454846; https://doi.org/10.1053/gast.2002.37052
- Trastour C, Benizri E, Ettore F, Ramaioli A, Chamorey E, Pouyssegur J, Berra E. HIF-1alpha and CA IX staining in invasive breast carcinomas: Prognosis and treatment outcome. Int J Cancer 2007; 120:1451-8; PMID:17245699; https://doi.org/10.1002/ijc.22436
- Tan EY, Yan M, Campo L, Han C, Takano E, Turley H, Candiloro I, Pezzella F, Gatter KC, Millar EK, et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer 2009; 100:405-11; PMID:19165203; https://doi.org/10.1038/sj.bjc.6604844
- Lou Y, McDonald PC, Oloumi A, Chia S, Ostlund C, Ahmadi A, Kyle A, Auf dem KU, Leung S, Huntsman D, et al. Targeting tumor hypoxia: Suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res 2011; 71:3364-76; PMID:21415165; https://doi.org/10.1158/0008-5472.CAN-10-4261
- Chen CL, Chu JS, Su WC, Huang SC, Lee WY. Hypoxia and metabolic phenotypes during breast carcinogenesis: Expression of HIF-1alpha, GLUT1, and CAIX. Virchows Arch 2010; 457:53-61; PMID:20526721; https://doi.org/10.1007/s00428-010-0938-0
- Ivanova L, Zandberga E, Silina K, Kalnina Z, Abols A, Endzelins E, Vendina I, Romanchikova N, Hegmane A, Trapencieris P, et al. Prognostic relevance of carbonic anhydrase IX expression is distinct in various subtypes of breast cancer and its silencing suppresses self-renewal capacity of breast cancer cells. Cancer Chemother Pharmacol 2015; 75:235-46; PMID:25422154; https://doi.org/10.1007/s00280-014-2635-1
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell 2012; 148:399-408; PMID:22304911; https://doi.org/10.1016/j.cell.2012.01.021
- Mazzio EA, Smith B, Soliman KF. Evaluation of endogenous acidic metabolic products associated with carbohydrate metabolism in tumor cells. Cell Biol Toxicol 2010; 26:177-88; PMID:19784859; https://doi.org/10.1007/s10565-009-9138-6
- Chiche J, Ilc K, Laferriere J, Trottier E, Dayan F, Mazure NM, Brahimi-Horn MC, Pouyssegur J. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res 2009; 69:358-68; PMID:19118021; https://doi.org/10.1158/0008-5472.CAN-08-2470
- Adams DJ, Wahl ML, Flowers JL, Sen B, Colvin M, Dewhirst MW, Manikumar G, Wani MC. Camptothecin analogs with enhanced activity against human breast cancer cells. II. Impact of the tumor pH gradient. Cancer Chemother Pharmacol 2006; 57:145-54; PMID:16001167; https://doi.org/10.1007/s00280-005-0008-5
- Webster KA, Discher DJ, Kaiser S, Hernandez O, Sato B, Bishopric NH. Hypoxia-activated apoptosis of cardiac myocytes requires reoxygenation or a pH shift and is independent of p53. J Clin Invest 1999; 104:239-52; PMID:10430605; https://doi.org/10.1172/JCI5871
- Gupta SC, Singh R, Pochampally R, Watabe K, Mo YY. Acidosis promotes invasiveness of breast cancer cells through ROS-AKT-NF-kappaB pathway. Oncotarget 2014; 5:12070-82; PMID:25504433; https://doi.org/10.18632/oncotarget.2514
- Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, McLendon R, Lindner D, Sloan A, Rich JN. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ 2011; 18:829-40; PMID:21127501; https://doi.org/10.1038/cdd.2010.150
- Kaluz S, Kaluzova M, Liao SY, Lerman M, Stanbridge EJ. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: A one transcription factor (HIF-1) show? Biochim Biophys Acta 2009; 1795:162-72; PMID:19344680; https://doi.org/10.1016/j.bbcan.2009.01.001
- Tafreshi NK, Lloyd MC, Proemsey JB, Bui MM, Kim J, Gillies RJ, Morse DL. Evaluation of CAIX and CAXII expression in breast cancer at varied O2 Levels: CAIX is the superior surrogate imaging biomarker of tumor hypoxia. Mol Imaging Biol 2016; 18:219-31; PMID:26276155; https://doi.org/10.1007/s11307-015-0885-x
- McDonald PC, Winum JY, Supuran CT, Dedhar S. Recent developments in targeting carbonic anhydrase IX for cancer therapeutics. Oncotarget 2012; 3:84-97; PMID:22289741; https://doi.org/10.18632/oncotarget.422
- Chiche J, Ilc K, Brahimi-Horn MC, Pouyssegur J. Membrane-bound carbonic anhydrases are key pH regulators controlling tumor growth and cell migration. Adv Enzyme Regul 2010; 50:20-33; PMID:19895836; https://doi.org/10.1016/j.advenzreg.2009.10.005
- Hussain SA, Ganesan R, Reynolds G, Gross L, Stevens A, Pastorek J, Murray PG, Perunovic B, Anwar MS, Billingham L, et al. Hypoxia-regulated carbonic anhydrase IX expression is associated with poor survival in patients with invasive breast cancer. Br J Cancer 2007; 96:104-9; PMID:17213826; https://doi.org/10.1038/sj.bjc.6603530
- Bernardi R, Gianni L. Hallmarks of triple negative breast cancer are emerging at last? Cell Res 2014; 24:904-5; PMID:24810303; https://doi.org/10.1038/cr.2014.61
- Ward C, Meehan J, Mullen P, Supuran C, Dixon JM, Thomas JS, Winum JY, Lambin P, Dubois L, Pavathaneni NK, et al. Evaluation of carbonic anhydrase IX as a therapeutic target for inhibition of breast cancer invasion and metastasis using a series of in vitro breast cancer models. Oncotarget 2015; 6:24856-70; PMID:26259239; https://doi.org/10.18632/oncotarget.4498
- Jeon M, Han J, Nam SJ, Lee JE, Kim S. STC-1 expression is upregulated through an Akt/NF-kappaB-dependent pathway in triple-negative breast cancer cells. Oncol Rep 2016; 36:1717-22; PMID:27461417; https://doi.org/10.3892/or.2016.4972
- Chang AC, Doherty J, Huschtscha LI, Redvers R, Restall C, Reddel RR, Anderson RL. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clin Exp Metastasis 2015; 32:15-27; PMID:25391215; https://doi.org/10.1007/s10585-014-9687-9
- Ma X, Gu L, Li H, Gao Y, Li X, Shen D, Gong H, Li S, Niu S, Zhang Y, et al. Hypoxia-induced overexpression of stanniocalcin-1 is associated with the metastasis of early stage clear cell renal cell carcinoma. J Transl Med 2015; 13:56; PMID:25740019; https://doi.org/10.1186/s12967-015-0421-4
- Li Y, Wang H, Oosterwijk E, Tu C, Shiverick KT, Silverman DN, Frost SC. Expression and activity of carbonic anhydrase IX is associated with metabolic dysfunction in MDA-MB-231 breast cancer cells. Cancer Invest 2009; 27:613-23; PMID:19367501; https://doi.org/10.1080/07357900802653464
- Supuran CT. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008; 7:168-81; PMID:18167490; https://doi.org/10.1038/nrd2467
- Mihaly Z, Kormos M, Lanczky A, Dank M, Budczies J, Szasz MA, Gyorffy B. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res Treat 2013; 140:219-32; PMID:23836010; https://doi.org/10.1007/s10549-013-2622-y
- Grabmaier K, Vissers JL, De Weijert MC, Oosterwijk-Wakka JC, Van Bokhoven A, Brakenhoff RH, Noessner E, Mulders PA, Merkx G, Figdor CG, et al. Molecular cloning and immunogenicity of renal cell carcinoma-associated antigen G250. Int J Cancer 2000; 85:865-70; PMID:10709109; https://doi.org/10.1002/(SICI)1097-0215(20000315)85:6%3c865::AID-IJC21%3e3.0.CO;2-Q
- Siebels M, Rohrmann K, Oberneder R, Stahler M, Haseke N, Beck J, Hofmann R, Kindler M, Kloepfer P, Stief C. A clinical phase I/II trial with the monoclonal antibody cG250 (RENCAREX(R)) and interferon-alpha-2a in metastatic renal cell carcinoma patients. World J Urol 2011; 29:121-6; PMID:20512580; https://doi.org/10.1007/s00345-010-0570-2
- Chamie K, Donin NM, Klopfer P, Bevan P, Fall B, Wilhelm O, Storkel S, Said J, Gambla M, Hawkins RE, et al. Adjuvant weekly girentuximab following nephrectomy for high-risk renal cell carcinoma: The ARISER randomized clinical trial. JAMA Oncol 2016; 3:913-920; PMID:27787547; https://doi.org/10.1001/jamaoncol.2016.4419
- Paolicchi E, Gemignani F, Krstic-Demonacos M, Dedhar S, Mutti L, Landi S. Targeting hypoxic response for cancer therapy. Oncotarget 2016; 7:13464-78; PMID:26859576; https://doi.org/10.18632/oncotarget.7229
- Lock FE, McDonald PC, Lou Y, Serrano I, Chafe SC, Ostlund C, Aparicio S, Winum JY, Supuran CT, Dedhar S. Targeting carbonic anhydrase IX depletes breast cancer stem cells within the hypoxic niche. Oncogene 2013; 32:5210-9; PMID:23208505; https://doi.org/10.1038/onc.2012.550
- Shin HJ, Rho SB, Jung DC, Han IO, Oh ES, Kim JY. Carbonic anhydrase IX (CA9) modulates tumor-associated cell migration and invasion. J Cell Sci 2011; 124:1077-87; PMID:21363891; https://doi.org/10.1242/jcs.072207
- Radvak P, Repic M, Svastova E, Takacova M, Csaderova L, Strnad H, Pastorek J, Pastorekova S, Kopacek J. Suppression of carbonic anhydrase IX leads to aberrant focal adhesion and decreased invasion of tumor cells. Oncol Rep 2013; 29:1147-53; PMID:23291973; https://doi.org/10.3892/or.2013.2226
- Pan JS, Huang L, Belousova T, Lu L, Yang Y, Reddel R, Chang A, Ju H, DiMattia G, Tong Q, et al. Stanniocalcin-1 inhibits renal ischemia/reperfusion injury via an AMP-activated protein kinase-dependent pathway. J Am Soc Nephrol 2015; 26:364-78; PMID:25012175; https://doi.org/10.1681/ASN.2013070703
- Ohkouchi S, Ono M, Kobayashi M, Hirano T, Tojo Y, Hisata S, Ichinose M, Irokawa T, Ogawa H, Kurosawa H. Myriad functions of stanniocalcin-1 (STC1) cover multiple therapeutic targets in the complicated pathogenesis of Idiopathic Pulmonary Fibrosis (IPF). Clin Med Insights Circ Respir Pulm Med 2015; 9:91-6; PMID:26740747; https://doi.org/10.4137/CCRPM.S23285
- Koizumi K, Hoshiai M, Ishida H, Ohyama K, Sugiyama H, Naito A, Toda T, Nakazawa H, Nakazawa S. Stanniocalcin 1 prevents cytosolic Ca2+ overload and cell hypercontracture in cardiomyocytes. Circ J 2007; 71:796-801; PMID:17457011; https://doi.org/10.1253/circj.71.796
- Rezapour S, Bahrami T, Hashemzadeh S, Estiar MA, Nemati M, Ravanbakhsh R, Feizi MA, Kafil HS, Pouladi N, Ghojazadeh M, et al. STC1 and NF-kappaB p65 (Rel A) is constitutively activated in colorectal cancer. Clin Lab 2016; 62:463-9; PMID:27156337; https://doi.org/10.7754/Clin.Lab.2015.150827
- Du YZ, Gu XH, Cheng SF, Li L, Liu H, Hu LP, Gao F. The oncogenetic role of stanniocalcin 1 in lung adenocarcinoma: A promising serum candidate biomarker for tracking lung adenocarcinoma progression. Tumour Biol 2016; 37:5633-44; PMID:26577859; https://doi.org/10.1007/s13277-015-4431-x
- Su J, Guo B, Zhang T, Wang K, Li X, Liang G. Stanniocalcin-1, a new biomarker of glioma progression, is associated with prognosis of patients. Tumour Biol 2015; 36:6333-9; PMID:25783529; https://doi.org/10.1007/s13277-015-3319-0
- Ito Y, Zemans R, Correll K, Yang IV, Ahmad A, Gao B, Mason RJ. Stanniocalcin-1 is induced by hypoxia inducible factor in rat alveolar epithelial cells. Biochem Biophys Res Commun 2014; 452:1091-7; PMID:25251473; https://doi.org/10.1016/j.bbrc.2014.09.060
- Durukan Tolvanen A, Westberg JA, Serlachius M, Chang AC, Reddel RR, Andersson LC, Tatlisumak T. Stanniocalcin 1 is important for poststroke functionality, but dispensable for ischemic tolerance. Neuroscience 2013; 229:49-54; PMID:23159313; https://doi.org/10.1016/j.neuroscience.2012.10.062
- Yeung BH, Law AY, Wong CK. Evolution and roles of stanniocalcin. Mol Cell Endocrinol 2012; 349:272-80; PMID:22115958; https://doi.org/10.1016/j.mce.2011.11.007
- Shi X, Wang J, Qin Y. Recombinant adeno-associated virus-delivered hypoxia-inducible stanniocalcin-1 expression effectively inhibits hypoxia-induced cell apoptosis in cardiomyocytes. J Cardiovasc Pharmacol 2014; 64:522-9; PMID:25490418; https://doi.org/10.1097/FJC.0000000000000146
- Abaza HM, Elmougy MI, El Maraghy HM, Mahmoud HM. Stanniocalcin1 gene expression in patients with acute leukemia: Impact on response to therapy and disease outcome. Int J Lab Hematol 2016; 38:81-9; PMID:26547904; https://doi.org/10.1111/ijlh.12445
- Dai D, Wang Q, Li X, Liu J, Ma X, Xu W. Klotho inhibits human follicular thyroid cancer cell growth and promotes apoptosis through regulation of the expression of stanniocalcin-1. Oncol Rep 2016; 35:552-8; PMID:26531219; https://doi.org/10.3892/or.2015.4358
- Han J, Jeon M, Shin I, Kim S. Elevated STC1 augments the invasiveness of triplenegative breast cancer cells through activation of the JNK/cJun signaling pathway. Oncol Rep 2016; 36:1764-71; PMID:27459971; https://doi.org/10.3892/or.2016.4977
- Tafreshi NK, Bui MM, Bishop K, Lloyd MC, Enkemann SA, Lopez AS, Abrahams D, Carter BW, Vagner J, Grobmyer SR, et al. Noninvasive detection of breast cancer lymph node metastasis using carbonic anhydrases IX and XII targeted imaging probes. Clin Cancer Res 2012; 18:207-19; PMID:22016510; https://doi.org/10.1158/1078-0432.CCR-11-0238
- Law AY, Ching LY, Lai KP, Wong CK. Identification and characterization of the hypoxia-responsive element in human stanniocalcin-1 gene. Mol Cell Endocrinol 2010; 314:118-27; PMID:19628018; https://doi.org/10.1016/j.mce.2009.07.007
- Chafe SC, Lou Y, Sceneay J, Vallejo M, Hamilton MJ, McDonald PC, Bennewith KL, Moller A, Dedhar S. Carbonic anhydrase IX promotes myeloid-derived suppressor cell mobilization and establishment of a metastatic niche by stimulating G-CSF production. Cancer Res 2015; 75:996-1008; PMID:25623234; https://doi.org/10.1158/0008-5472.CAN-14-3000
- Kim BR, Shin HJ, Kim JY, Byun HJ, Lee JH, Sung YK, Rho SB. Dickkopf-1 (DKK-1) interrupts FAK/PI3K/mTOR pathway by interaction of carbonic anhydrase IX (CA9) in tumorigenesis. Cell Signal 2012; 24:1406-13; PMID:22430125; https://doi.org/10.1016/j.cellsig.2012.03.002
- Richmond A. Nf-kappa B, chemokine gene transcription and tumour growth. Nat Rev Immunol 2002; 2:664-74; PMID:12209135; https://doi.org/10.1038/nri887