ABSTRACT
The tumor suppressor gene TP53 is the most frequently mutated gene in human papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSCC). It represents a known transcription factor that controls different microRNAs (miRNA) and target genes involved in the regulation of cellular stress, apoptosis and response to DNA damage. We used The Cancer Genome Atlas database to investigate the difference in transcriptome and proteome levels between mutated and wild-type TP53 HPV-negative HNSCC. Using different databases and an extensive literature review, we built the transcriptional and post-transcriptional network regulated by TP53. TP53 mutation was associated with poor overall survival in 203 HPV-negative patients compared to 40 patients with TP53 wild-type tumors. Using the enrichment analysis, we found that UHRF1BP1 and SESN1 mRNA were linked to prognosis in the TP53 mutated group. This is also the case for miR-377-3p, an important miRNA regulator of SESN1. Our study shows that SESN1 mRNA, UHRF1BP11 mRNA and miRNA-377-3p levels are prognostically relevant in HPV-negative HNSCC patients. This finding may help with patient stratification and the development of potential new therapeutic targets to treat patients with HNSCC.
Introduction
In many human cancers, chromosome instability, poor prognosis and poor response to cancer therapy are associated with alterations to the TP53 gene.Citation1-5 Numerous studies have confirmed that TP53 is the most frequently mutated gene in head and neck squamous cell carcinoma (HNSCC).Citation6-9
TP53 regulates the transcription of numerous target genes involved in cell cycle control, DNA repair, senescence and apoptosis.Citation10,11 It is able to prevent cancer formation by stopping damaged cells from propagating through proliferation. The TP53 wild-type is therefore commonly referred to as the “guardian of the genome” due to its ability to ensure genome stability.
Several types of TP53 mutations have been described:
Most TP53 mutations found in human tumors are missense mutations or point mutations whereby a single nucleotide change causes substitution of a different amino acid. Point mutations at a DNA-binding domain (DBD) block the normal regulation of target genes and thus allow TP53 mutants to exert oncogenic activities.Citation12-15 Fifty of these have been classified as hotspot mutationsCitation16 due to their high prevalence in cancer. Beyond these known mutations, other potential mutations should be researched and identified by programs which estimate the probability of impact of physico-chemical modifications on amino acids and proteins on gene functionnality.Citation17
Truncating or nonsense mutations are point mutations in a sequence of DNA that result in a premature stop codon. When the mutated sequence is generated into a protein, the protein is incomplete and consequently usually nonfunctional.
Frameshift mutations cause insertion or deletion of a number of nucleotides in a DNA sequence which can result in the modification of the reading frame and modification of translation. This is contrary to inframe mutations which do not introduce a shift in the triplet reading frame.
TP53 mutations can also be classified as disruptive and non-disruptive, based on the degree of disturbance of protein structure predicted from the crystal structure of the p53-DNA complexes.Citation18 Especially disruptive mutations seem to be related to decreased overall survival when compared to wild-type TP53.Citation18,19 Gross et al showed that the frequent association between TP53 mutation and loss of chromosome 3p is directly related to decreased survival.Citation20 Furthermore, the classical TP53 target genes, which are normally activated by TP53 wild type (WT), are repressed by TP53 mutation or vice-versa.Citation21,22
To further investigate the difference in mRNA, miRNA, and protein expression levels of TP53 target genes in HPV-negative patients with distinct TP53 status, we constructed the transcriptional and post-transcriptional network regulated by TP53 using freely available databases.
We identified two TP53 targeted genes, namely SESN1 and UHRF1BP1, as significantly enriched in patients who were TP53 mutated. Together with miR-377-3p (a down-regulator of SESN1), there seems to be an impact on prognosis. Whereas SESN1 and miR-377-3p, are implicated in DNA repair, the role of UHRF1BP1 is less investigated. Functionally, both genes are known to be dependent on TP53 wild-type activity.
Results
Prognostic relevance of TP53 status in HPV negative HNSCC
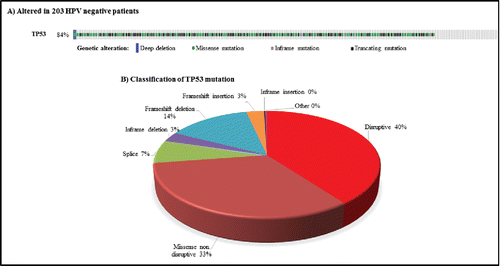
We downloaded the TP53 mutation landscape of HPV-negative HNSCC patients using the cBio Cancer Genomics Portal tool (). Eighty-four percent of patients (203/243 patients) had at least one TP53 alteration. Among these, 86 patients had (at least) a truncating mutation, 96 patients a hotspot mutation, and 119 a medium mutation. Forty percent of the TP53 mutations have been identified as disruptive according to Poeta et al.Citation18
Figure 1. TP53 status in 243 patients with human papillomavirus-negative squamous cell carcinoma of the head and neck from The Cancer Genome Atlas Network7: A) Oncoprint screen shot from cBioportal. B) Pie chart showing different TP53 mutations (%).
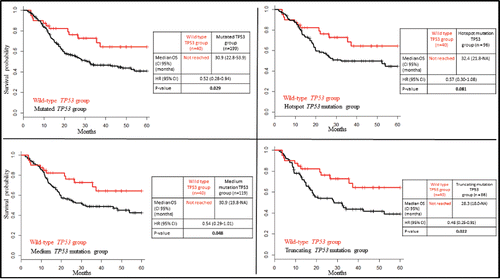
Overall survival analyses are reported in . Survival plots showed comparatively poor overall survival for patients in the mutated TP53 group compared to patients bearing wild-type TP53, and this was statistically significant. Median survival was 30.9 months (CI 95%; 22.8–53.9) for the mutated TP53 group, but was not reached for the TP53 wild-type group (HR 0.52; 95% CI 0.28–0.94; p = 0.02). The truncating mutation and the medium mutation TP53 subgroups also had a significantly poorer prognosis compared to the wild-type TP53 group (p = 0.02 and 0.05, respectively), whereas the hotspot mutation subgroup did not differ significantly from the wild-type TP53 group (p = 0.08) ().
TP53 regulatory network
We used TRRUST to identify the gene regulated by TP53.Citation23
A total of 159 genes, regulated by TP53 through 166 interactions (53 activation, 61 repression, 52 unknown) were identified. Furthermore, we found 113 additional target genes regulated by TP53 after an extensive literature review.Citation20-25 The TP53 target genes shared between the different sources are shown in Supplementary Fig. 1.
Sixteen miRNAs regulated by TP53 were extracted from the TransmiR database.Citation26 An extensive literature search of TP53 dependent miRNA revealed 13 additional candidates.Citation31-33 All miRNAs regulated by TP53 are listed in Supplementary Table S1.
Enrichment and prognostic relevance of TP53 target genes and miRNAs
We used enrichment analysis, available on cBioportal, to identify genes and proteins related to TP53 function, and to evaluate whether the expression of these genes has prognostic relevance either in the TP53 mutated or wild-type group.
The protein expression analysis in The Cancer Genome Atlas was restricted to 160 proteins of which 28 were TP53 target genes (Supplementary Table S3). Only the enhancer zeste homolog 2 (EZH2) protein was expressed at a higher level in the TP53 mutated tumors compared to the wild-type TP53 group (p = 0.04) (Fig. S3). However, protein expression of EZH2 was not linked to overall survival (Fig. S3).
At mRNA expression level, a total of 43 genes were found to be significantly enriched between the TP53 wild-type and the mutated groups: 21 of them had a higher expression level in TP53 mutated tumors, and 22 had a higher expression level in TP53 wild-type patients. Only two genes, sestrin 1 (SESN1) and ubiquitin-like containing PHD and RING finger domains 1-binding protein 1 (UHRF1BP1) were prognostic for overall survival in HPV-negative patients (Supplementary Table S2 and Fig. S2).
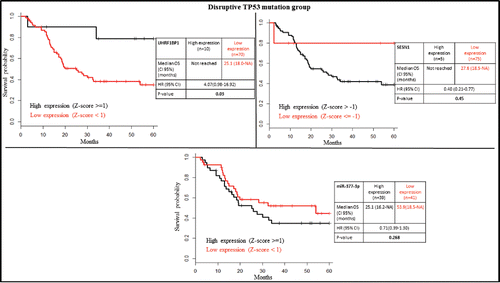
SESN1 is known to be activated by wild-type TP53,Citation26,34,35 and UHRF1BP1 is downregulated by wild-type TP53.Citation36 Accordingly, we found a lower expression of SESN1 and a higher expression of UHRF1BP1 in the TP53 mutated group compared to TP53 wild-type (Supplementary Table S2). Low mRNA expression of SESN1 (Z-score ≤ (−1)) was associated with poor overall survival, whereas high mRNA expression of UHRF1BP1 (Z-score ≥ 1) was associated with good overall survival in patients bearing TP53 mutated tumors (). This difference was still preserved inside the TP53 disrupted group (). However, the expression of these genes could not predict overall survival in patients with TP53 wild-type tumors (). Of note is that only a few patients (n = 40) were available for analysis in the wild-type group. If we focus on patients with disruptive TP53 mutation, only UHRF1BP1 was prognostically significant (). Additionally, using the entire cohort of HPV-negative patients, SESN1 and UHRF1BP1 were still prognostically significant (Fig. S2).
Figure 3. Kaplan-Meier survival plots according to gene expression level of UHRF1BP1, SESN1 and miR-377-3p in the TP53 mutated group.
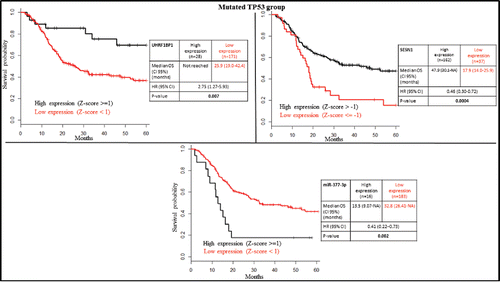
Figure 4. Kaplan-Meier survival plots according to gene expression level of UHRF1BP1, SESN1 and miR-377-3p in the TP53 wild-type group.
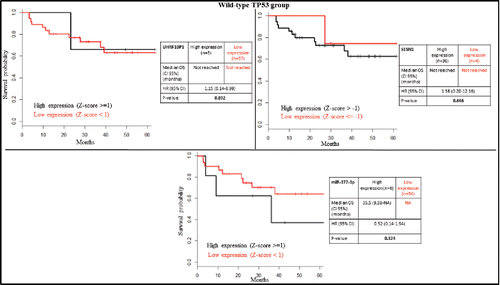
Figure 5. Kaplan-Meier survival plots according to gene expression level of UHRF1BP1, SESN1 and miR-377-3p in the TP53 disruptive group.
We looked further for miRNA regulators of UHRF1BP and SESN1 to investigate the prognostic relevance of these miRNAs. Several miRNAs regulators were reported in miRTarBase for SESN1 but not for UHRF1BP1.
SESN1 is experimentally repressed by seven different miRNAs (let-7a-5p, miR-21-5p, miR-24-3p, miR-154-5p, miR-26b-5p, miR-375, and miR-377-3p). We investigated the prognostic relevance of their high expression (Z-score ≥ 1) in HPV-negative patients. High miR-377-3p expression was associated with poor prognosis in TP53 mutated but not in TP53 wild-type patients, or patients with TP53 disruptive mutations (, and ).
Univariate analysis was used to assess the prognostic significance of clinical factors (age > 70 years, stage I, II, III and IV, gender, tumor localization, smoking and alcohol) on overall survival of the TP53 mutated population. This analysis showed poor prognosis for patients >70 years or females ().
Table 1. Univariate overall survival analysis in patients with TP53 mutated status.
A multivariate Cox regression analysis based on significant clinical (patient age and gender) and biological factors (SESN1, UHRF1BP1 and miR-377-3p) identified SESN1, UHRF1BP1 and gender as independent prognostic factors in TP53 mutated patients ().
Table 2. Multivariate Cox regression analysis based on the significant variable determined in .
Discussion
In this study, we investigated the prognostic value of TP53, as well as that of genes relating to the TP53 regulatory network, in HNSCC. Accumulating evidence suggests that TP53 alterations are significantly associated with poor prognosis and treatment resistance in this disease.Citation16,33,34 Our study confirms previous findings suggesting that patients with TP53 wild-type tumors have increased overall survival rates when compared to those with TP53 mutations.Citation18,37-39
Using enrichment analysis, we investigated genes and proteins implicated in the regulatory network of TP53, and evaluated the potential implications of these genes on oncologic outcome. At the protein expression level, this analysis was limited by the number of available proteins in TCGA. However, we identified increased proteomic expression of EZH2 in TP53 mutated tumors compared to wild-type tumors (p = 0.04). Although poor survival outcome and decreased sensitivity to cisplatin-based chemotherapy is related to EZH2 expression in patients with HNSCC,Citation40 we were unable to confirm a significant prognostic value of EZH2 expression in this dataset (Fig. S3). This may be related to the limited number of patients available for this analysis (Fig. S3).
At the transcriptional level, we identified two genes, SESN1 and UHRF1BP1, as being implicated in significant changes to overall survival in the TP53 mutated HNSCC population (). The same conclusions can be drawn when analyzing the whole HPV-negative patient cohort, including TP53 wild-type and mutated patients (Fig. S2), even when the data is statistically less significant.
SESN1, which encodes a member of the Sestrin family, is associated with autophagy related genes and activated by TP53. This gene plays a role in the cellular response to DNA damage and oxidative stress. The potential functional role of SESN1 is to repair damaged cells in the G1 cell cycle checkpoint.Citation38 In addition, SESN1 is an important regulator of homeostasis through the suppression of the mechanistic target of rapamycin complex 1 (mTORC1) kinase.Citation41-43 In a recent paper, Cordani et al demonstrated that the depletion of mutant TP53 determined an increase of SESN1 in a breast cancer cell line. Furthermore, these investigators demonstrated that low SESN1 expression and low expression of other autophagy related genes is significantly associated with poor prognosis in TP53 mutant breast cancer patients.Citation44 This paves the way for the hypothesis that mTOR inhibitors may be of interest in patients with TP53 mutations and low SESN1 expression.
UHRF1BP1, an ubiquitin-like containing PHD and RING finger domains 1-binding protein 1, encodes a highly conserved protein with unknown function. A coding variant in this gene was found to be associated with systemic lupus erythematosus.Citation45 Ugoni and colleagues found that UHRF1BP1 is one of the members of the ICBP90 complex, and that overexpression of UHRF1BP1 might induce inhibition of cell growth like a tumor suppressor.Citation46 The role of this protein and its prognostic relevance in other tumor types requires further clarification.
MiR-377-3p is an important down-regulator of SESN1 that directly targets the 3′-untranslated region of this gene. Higher miR-377-3p expression showed significantly poorer overall survival in TP53 mutated patients but not in TP53 disruptive mutation and wild-type patients (, and ).
Corroborating our study, Wen et al. found that gastric tumors expressing a high expression of this miRNA were also associated with worse prognosis.Citation47 Another study showed that downregulation of miR-377 was associated with best prognosis in an intestinal type of periampullary adenocarcinoma.Citation48 Interestingly, miR-377 was detectable by RT-PCR in peripheral plasma and urine samples.
The results from this purely bioinformatic exploratory study suggest potential new biomarker candidates which can predict overall survival in TP53 mutated HPV-negative HNSCC patients. The stratification of patients may have potential clinical implications that require further investigation and confirmation in experimental settings.
Materials and methods
The prognostic relevance of TP53 mutation in HPV negative HNSCC
We used cBio Cancer Genomics Portal (http://www.cbioportal.org)Citation49,50 to download the mutational profiles of 243 HPV-negative HNSCC produced by The Cancer Genomic Atlas network (TCGA). This study met the publication guidelines requested by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). TCGA published dataCitation7 were used in the analyses because of the availability of clinical and biologic characteristics other than TP53 status.
Different types of TP53 mutations were individualized. The mutated TP53 group included different patient subgroups that had at least one of the following TP53 mutations:
| (i) | The truncating mutation TP53 subgroup – defined as patients bearing a tumor harboring a nonstop, nonsense, frameshift and splice site mutations in TP53.Citation49,50 | ||||
| (ii) | The hotspot mutation TP53 subgroup – defined according to the criteria published by Chang and colleagues.Citation16 | ||||
| (iii) | The medium mutation TP53 subgroup – defined as patients bearing a tumor harboring a TP53 missense mutation with at least a medium predicted functional impact. For this subgroup, the functional impact was calculated by the mutation assessor tool which captures the evolutionary conservation of a residue in a protein family and its subfamilies using combinatorial entropy measurement,Citation17 as used by cBio Cancer Genomics Portal. | ||||
| (iv) | Disruptive mutations were defined as DNA sequence alterations that introduce a STOP sequence resulting in disruption of p53 protein production or any DNA sequence alteration which: a) occurs within the L2 or L3 binding domains (codons 163–195 or 236–251), and b) replaces an amino acid from one polarity/charge category with an amino acid from another category, as described in Poeta et al.Citation18 | ||||
With regards to the proteome, 185 HPV-negative HNSCC samples investigated in TCGA using reverse-phase protein arrays (RPPA) were included in this work.Citation7 Altogether, 160 proteins were investigated in these samples.
TP53 regulatory network and enrichment analyses
To construct the regulatory network of TP53, we used the “transcriptional regulatory relationships unraveled by sentence-based text-mining” (TRRUST) database (http://www.grnpedia.org/trrust), which is a manually curated database of human transcriptional regulatory network.Citation23 To expand the transcriptional regulatory network, we looked for other transcriptional interactions based on an extensive literature review.Citation24-29 Then, we retrieved the TP53-miRNA regulatory interactions from the TransmiR database (http://cmbi.bjmu.edu.cn/transmir),Citation30 and compiled this data with that from the literature.Citation31-33
For all 243 HPV-negative HNSCC patients, we evaluated changes in the expression of TP53 target genes at mRNA and protein levels using “enrichment analysis”, as available in cBio Cancer Genomics Portal (version June 2016).Citation49,50 We considered targets to be significantly enriched if the p-value calculated with the unpaired t-test was ≤ 0.05.
We investigated the post-transcriptional network of significantly enriched TP53 target genes through the miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw/), a reference database for experimentally validated miRNA-target interactions.Citation51
Survival analyses
Overall survival (OS) was defined as the time from study entry to death or to last follow-up (1–60 months). We used the Kaplan-Meier methodCitation52 to estimate OS in patients with TP53 mutated status, and derived hazard ratios (HRs) using a stratified Cox proportional hazards model.
For the OS analysis, we used the z-score to indicate the number of standard deviations the gene expression is either above or below the mean away from the mean expression of a gene in patients with TP53 mutated tumors. The z-scores related to the enriched mRNA and protein expression levels, measured respectively by RNA seq V2 RSEM (a normalized value outputted by the RSEM software,Citation53), or RPPA, were downloaded from cBio Cancer Genomics Portal (version June 2016). We downloaded the expression of miRNA from TCGA (https://tcga-data.nci.nih.gov/docs/publications/hnsc_2014/) level 3 (post-normalized data), and we calculated their corresponding z-scores.Citation54
A log-rank test was used to compare groups. We considered a p-value ≤ 0.05 to be statistically significant.
The Cox proportional hazard ratio model was completed using the “coxph()” function in R programming to investigate the relationship between different molecular factors (SESN1, UHRF1BP1 and miR-377-3p) and clinical prognostic factors (patient age and gender). These factors were retained based on their statistically significant expression (p-value ≤ 0.05) on univariate analysis. In more detail, we used the Cox proportional hazard method to test the following covariates: SESN expression (high expression (Z-score > −1) versus low expression (Z-score < = −1)), UHRF1BP1 expression (high expression (Z-score > = 1) versus low expression (Z-score < 1)), miR-377-3p expression (high expression (Z-score > = 1) versus low expression (Z-score < 1)), patients age (< = 70 versus > 70), gender (female versus male).
Disclosure
The authors have no conflicts of interest to disclose.
2017CBT10129R1-file002.docx
Download MS Word (24.8 KB)Acknowledgement
The authors wish to thank Aileen Eiszele for writing assistance.
References
- Ganci F, Sacconi A, Bossel BM, Manciocco V, Sperduti I, Strigari L, Covello R, Benevolo M, Pescarmona E, Domany E, et al. Expression of TP53 mutation-associated microRNAs predicts clinical outcome in head and neck squamous cell carcinoma patients. Ann Oncol. 2013;24:3082-8. https://doi.org/10.1093/annonc/mdt380. PMID:24107801
- Hanel W, Moll UM. Links between mutant p53 and genomic instability. J Cell Biochem. 2012;113:433-9. https://doi.org/10.1002/jcb.23400. PMID:22006292
- Liu J, Ma Q, Zhang M, Wang X, Zhang D, Li W, Wang F, Wu E. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: Evidence from a systematic review and meta-analysis. Eur J Cancer. 2012;48:2328-38. https://doi.org/10.1016/j.ejca.2012.03.001. PMID:22459764
- Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM, Fitzgerald AL, Giri U, Ang KK, Myers JN. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res Off J Am Assoc Cancer Res. 2012;18:290-300. https://doi.org/10.1158/1078-0432.CCR-11-2260
- Walerych D, Napoli M, Collavin L, Sal GD. The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012;33:2007-17. https://doi.org/10.1093/carcin/bgs232. PMID:22822097
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie T-X, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154-7. https://doi.org/10.1126/science.1206923. PMID:21798897
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576-82. https://doi.org/10.1038/nature14129. PMID:25631445
- Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond J, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3:770-81. https://doi.org/10.1158/2159-8290.CD-12-0537. PMID:23619168
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157-60. https://doi.org/10.1126/science.1208130. PMID:21798893
- Giono LE, Manfredi JJ. The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol. 2006;209:13-20. https://doi.org/10.1002/jcp.20689. PMID:16741928
- Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis-the p53 network. J Cell Sci. 2003;116:4077-85. https://doi.org/10.1242/jcs.00739. PMID:12972501
- Brosh R, Rotter V. When mutants gain new powers: News from the mutant p53 field. Nat Rev Cancer. 2009;9:701-13. PMID:19693097.
- Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26:1268-286. https://doi.org/10.1101/gad.190678.112 PMID:22713868.
- Muller PA, Vousden KH. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell. 2014;25:304-17. https://doi.org/10.1016/j.ccr.2014.01.021 PMID:24651012.
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622-29. https://doi.org/10.1002/humu.20495 PMID:17311302.
- Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, Gao J, Socci ND, Solit DB, Olshen AB, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34(2):155–63. advance online publication. Available from: http://www.nature.com/nbt/journal/vaop/ncurrent/full/nbt.3391.htmlhttps://doi.org/10.1038/nbt.3391
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118.https://doi.org/10.1093/nar/gkr407. PMID:21727090
- Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552-61. https://doi.org/10.1056/NEJMoa073770. PMID:18094376
- Masica DL, Li S, Douville C, Manola J, Ferris RL, Burtness B, Forastiere AA, Koch WM, Chung CH, Karchin R. Predicting survival in head and neck squamous cell carcinoma from TP53 mutation. Hum Genet. 2015;134:497-507. https://doi.org/10.1007/s00439-014-1470-0. PMID:25108461
- Gross AM, Orosco RK, Shen JP, Egloff AM, Carter H, Hofree M, Choueiri M, Coffey CS, Lippman SM, Hayes DN, et al. Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nat Genet. 2014;46:939-43. https://doi.org/10.1038/ng.3051. PMID:25086664
- Vikhanskaya F, Lee MK, Mazzoletti M, Broggini M, Sabapathy K. Cancer-derived p53 mutants suppress p53-target gene expression–potential mechanism for gain of function of mutant p53. Nucleic Acids Res. 2007;35:2093-104. https://doi.org/10.1093/nar/gkm099. PMID:17344317
- Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene. 2004;23:5759-69. https://doi.org/10.1038/sj.onc.1207706. PMID:15208672
- Han H, Shim H, Shin D, Shim JE, Ko Y, Shin J, Kim H, Cho A, Kim E, Lee T, et al. TRRUST: A reference database of human transcriptional regulatory interactions. Sci Rep. 2015;12(5):11432. Available from: http://www.nature.com/srep/2015/150612/srep11432/full/srep11432.html?WT.ec_id = SREP-20150616
- Garritano S, Inga A, Gemignani F, Landi S. More targets, more pathways and more clues for mutant p53. Oncogenesis. 2013;2:e54.https://doi.org/10.1038/oncsis.2013.15. PMID:23817466
- Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, Resnick MA. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res. 2013;41(15):7286-301. https://doi.org/10.1093/nar/gkt504. PMID:23775793
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402-12. https://doi.org/10.1038/nrm2395. PMID:18431400
- Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, Lathan C, Marcoux JP, Du J, Okuda K, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051-60. https://doi.org/10.1158/0008-5472.CAN-11-1340 PMID:21791641
- Tamura K. Development of cell-cycle checkpoint therapy for solid tumors. Jpn J Clin Oncol. 2015;45(12):1097-102. PMID:26486823
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207-19. https://doi.org/10.1016/j.cell.2005.10.043. PMID:16413492
- Wang J, Lu M, Qiu C, Cui Q. TransmiR: A transcription factor–microRNA regulation database. Nucleic Acids Res. 2010;38:D119-22. https://doi.org/10.1093/nar/gkp803. PMID:19786497
- Garibaldi F, Falcone E, Trisciuoglio D, Colombo T, Lisek K, Walerych D, Del Sal G, Paci P, Bossi G, Piaggio G, et al. Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex. Oncogene. 2016;35(29):3760-70. https://doi.org/10.1038/onc.2016.51. PMID:26996669
- Krell J, Stebbing J, Carissimi C, Dabrowska AF, de Giorgio A, Frampton AE, Harding V, Fulci V, Macino G, Colombo T, et al. TP53 regulates miRNA association with AGO2 to remodel the miRNA–mRNA interaction network. Genome Res. 2016;26:331-41. https://doi.org/10.1101/gr.191759.115. PMID:26701625
- Zhang Y, Dai J, Deng H, Wan H, Liu M, Wang J, Li S, Li X, Tang H. miR-1228 promotes the proliferation and metastasis of hepatoma cells through a p53 forward feedback loop. Br J Cancer. 2015;112:365-74. https://doi.org/10.1038/bjc.2014.593. PMID:25422913
- Budanov AV, Shoshani T, Faerman A, Zelin E, Kamer I, Kalinski H, Gorodin S, Fishman A, Chajut A, Einat P, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21:6017-31. https://doi.org/10.1038/sj.onc.1205877. PMID:12203114
- Peeters H, Debeer P, Bairoch A, Wilquet V, Huysmans C, Parthoens E, Fryns JP, Gewillig M, Nakamura Y, Niikawa N, et al. PA26 is a candidate gene for heterotaxia in humans: Identification of a novel PA26-related gene family in human and mouse. Hum Genet. 2003;112:573-80. PMID:12607115
- Arima Y, Hirota T, Bronner C, Mousli M, Fujiwara T, Niwa S, Ishikawa H, Saya H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells Devoted Mol Cell Mech. 2004;9:131-42. https://doi.org/10.1111/j.1356-9597.2004.00710.x
- Bertheau P, Turpin E, Rickman DS, Espié M, de Reyniès A, Feugeas JP, Plassa LF, Soliman H, Varna M, de Roquancourt A, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLOS Med. 2007;4:e90.https://doi.org/10.1371/journal.pmed.0040090. PMID:17388661
- Lindenbergh-van der Plas M, Brakenhoff RH, Kuik DJ, Buijze M, Bloemena E, Snijders PJF, Leemans CR, Braakhuis BJM. Prognostic significance of truncating TP53 mutations in head and neck squamous cell carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2011;17:3733-41. https://doi.org/10.1158/1078-0432.CCR-11-0183
- Molleví DG, Serrano T, Ginestà MM, Valls J, Torras J, Navarro M, Ramos E, Germà JR, Jaurrieta E, Moreno V, et al. Mutations in TP53 are a prognostic factor in colorectal hepatic metastases undergoing surgical resection. Carcinogenesis. 2007;28:1241-6. https://doi.org/10.1093/carcin/bgm012. PMID:17259658
- Chang JW, Gwak SY, Shim GA, Liu L, Lim YC, Kim JM, Jung MG, Koo BS. EZH2 is associated with poor prognosis in head-and-neck squamous cell carcinoma via regulating the epithelial-to-mesenchymal transition and chemosensitivity. Oral Oncol. 2016;52:66-74. https://doi.org/10.1016/j.oraloncology.2015.11.002. PMID:26604082
- Evangelista M, Baroudi ME, Rizzo M, Tuccoli A, Poliseno L, Pellegrini M, Rainaldi G. Alkaline phosphatase-positive immortal mouse embryo fibroblasts are cells in a transitional reprogramming state induced to face environmental stresses. Genet Epigenetics. 2015;7:33-41.
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451-60. https://doi.org/10.1016/j.cell.2008.06.028. PMID:18692468
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9:1-8. https://doi.org/10.1016/j.celrep.2014.09.014. PMID:25263562
- Cordani M, Oppici E, Dando I, Butturini E, Dalla Pozza E, Nadal-Serrano M, Oliver J, Roca P, Mariotto S, Cellini B, et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol Oncol. 2016;10:1008-29. https://doi.org/10.1016/j.molonc.2016.04.001. PMID:27118659
- Zhang Y, Yang W, Mok CC, Chan TM, Wong RWS, Mok MY, Lee KW, Wong SN, Leung AMH, Lee TL, et al. Two missense variants in UHRF1BP1 are independently associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2011;12:231-4. https://doi.org/10.1038/gene.2010.66. PMID:21326321
- Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601-10. https://doi.org/10.1038/sj.onc.1208053. PMID:15361834
- Wen X, Wu JQ, Peng W, Feng JF, Tang JH. MicroRNA-377 predicts poor clinical outcome of gastric cancer and induces tumorigenesis by targeting multiple tumor-suppressor genes. Oncol Rep. 2015;34:203-10. https://doi.org/10.3892/or.2015.3981. PMID:25998046
- Sandhu V, Bowitz Lothe IM, Labori KJ, Lingjærde OC, Buanes T, Dalsgaard AM, Skrede ML, Hamfjord J, Haaland T, Eide TJ, et al. Molecular signatures of mRNAs and miRNAs as prognostic biomarkers in pancreatobiliary and intestinal types of periampullary adenocarcinomas. Mol Oncol. 2015;9:758-71. https://doi.org/10.1016/j.molonc.2014.12.002. PMID:25579086
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-04. https://doi.org/10.1158/2159-8290.CD-12-0095. PMID:22588877
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.https://doi.org/10.1126/scisignal.2004088. PMID:23550210
- Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239-247. https://doi.org/10.1093/nar/gkv1258. PMID:26590260
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457-81. https://doi.org/10.1080/01621459.1958.10501452
- Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.https://doi.org/10.1186/1471-2105-12-323 PMID:21816040
- Sauro J. What's a Z-score and why use it in usability testing? Meas Usability Quant Usability Stat Six Sigma. 2004.