ABSTRACT
Exosomes released from cancer cells support metastasis and growth of recipient cells and increase their resistance to chemotherapy. Therapeutic targeting of exosomes is a promising area in cancer research. Our aim is to test the effect of the mast cell stabilizer ketotifen on exosomes release from cancer cells and how this can modify their response to doxorubicin. Exosomes release from three cancer cell lines (MCF7, HeLa and BT549) was assessed by scan electron microscope and exosomes quantification kit. Doxorubicin export within exosomes was monitored flurometrically and cellular sensitivity to doxorubicin ± ketotifen was measured by sulphorhodamine-B and colony formation assays. The three cell lines release different amounts of exosomes with the highest quantity released from BT549 followed by MCF7 and then HeLa. Ketotifen (10 µmol L−1) reduced exosomes release in all three cell lines with different efficiency (HeLa>MCF7>BT549). Doxorubicin export via exosomes was highest in BT549, lower in HeLa and lowest in MCF7 cells. Pretreatment with ketotifen sensitized the cells to doxorubicin (HeLa>MCF7>BT549) with a sensitization factor of 27, 8 and 1.25 respectively. Increased sensitivity of cells to doxorubicin by ketotifen was proportional to its effect on exosomes release. Our data is the first report of ketotifen modulating exosomes release from cancer cells and opens the avenue for exosomes-targeting cancer therapy. The differential effects of ketotifen on doxorubicin exosomal export in the cell lines studied, suggests an opportunity of pharmacological enhancement of doxorubicin anti-tumor activity in some but not all cancer types.
| Abbreviations | ||
| ER | = | estrogen receptor |
| PR | = | progesterone receptor |
| FBS | = | fetal bovine serum |
| SEM | = | scan electron microscope |
| PBS | = | Phosphate buffered saline |
| CD81 | = | cluster of differentiation 81 |
| SDS-PAGE | = | sodium dodecyl sulphate polyacrylamide gel electrophoresis |
| PBS-T | = | phosphate buffered saline – tween |
| HRP | = | horseradish peroxidase |
| KETO | = | ketotifen |
| DOX | = | doxorubicin |
| SRB | = | SulphoRhodamine-B |
Introduction
Exosomes are nano-sized (40–100 nm diameter) lipid bilayer membrane-containing vesicles produced by different if not all types of cells. Exosomes derive from the endolysosomal pathway and contain many molecular constituents from their parent cells including proteins, DNA, and RNA. They are released from normal and malignant cells and are present abundantly in different body fluids including blood, urine, saliva, ascites and amniotic fluid.Citation8,4 Regardless of their origin, exosomes may share analogous protein constituents, those can be categorized into three groups: genuine protein raft, proteins like cytoskeleton and heat shock proteins (HSP).Citation24 Exosomes released from different types of cells can be taken up by neighboring cells or by distant organs and are thought to induce various functions in target cells, including immune modulation, induction of proliferation and tolerance.Citation24,13 Exosomes released from malignant cells appear to have specific biological activity leading to enhanced proliferation of recipient cells, tumor development, and metastasis as well as reduced immune surveillance of cancer cells and resistance to cancer therapy.Citation28 Thus, targeting of tumor cell exosomes is an attractive strategy for advanced tumor therapies.
The release of exosomes from disease or normal cells is controlled by various factors which may include ischemia, cellular stress, calcium, calcium ionophores, heat, phosphatidylinositol 3-kinsase, pH, phorbol esters, and loss of cellular attachments.Citation17,15 Hypoxia, for example, enhances the release of exosomes which in turn stimulate angiogenesis.Citation17 Inhibitors of phosphatidylinositol 3-kinases have been shown to reduce production of exosomes confirming a role of these kinases in exosome production.Citation28 Similarly as in traditional exocytosis mechanisms, exosome release seems to be controlled by intracellular calcium (Ca2+) levels.Citation20 Furthermore, the inter- and intra-cellular pH may affect exosome release. Reduction of the micro-environmental pH increases the release of exosomes and their cellular uptake.Citation18 It is also evident that oncogenes and tumor suppressors can control the release of cancer exosomes.Citation26 It has been described that the P53-regulated protein tumor suppressor-activated pathway 6 (TSAP6) can promote exosome release under cellular stress.Citation27
Different exosome-mediated mechanisms are being recognized as contributors to anticancer drug resistance. These include the transfer of proteins such as P-glycoprotein and other multi-drug resistance (MDR)-associated proteins or genetic materials such as DNA, RNA and microRNA.Citation1 Moreover, exosomes may also serve as vehicles for the export of drugs from intracellular compartments such as the nucleus.Citation7
Doxorubicin, a potent anthracycline antibiotic targeting DNA, is extensively used for the treatment of solid and hematological malignancies.Citation3 Doxorubicin resistance is one of the challenges arising during the typical course of cancer chemotherapy. Different mechanisms are proposed for resistance to doxorubicin such as decreased uptake in or increased export from cancer cells, enhanced DNA repair, qualitative/quantitative changes in topoisomerase II enzyme.Citation9
The first generation antihistamine ketotifen is used as mast cell stabilizer and acts as a store-operated calcium channel blocking agent.Citation12 It is previously demonstrated to have cytotoxic effects on breast cancer and leukemia cells.Citation30 Moreover, ketotifen was also shown to reduce the expression of some proteins such as CDC42, Rac and Rho, which are associated with increased invasiveness of cancer cells. This effect is believed to be mediated by a block of calcium entry into the cells.Citation2,5,14 In the present study, we investigated whether ketotifen would alter exosome release from cancer cells and whether this activity may enhance the anti-tumor effects of doxorubicin in three different cancer model cell lines, namely HeLa (in vitro model for cervix carcinoma), MCF7 (in vitro model for breast cancer) and BT549 (in vitro model for breast cancer). The ultimate goal is to open the door for developing pharmacological agents targeting exosome release and their role in supporting the growth of cancer cells and their resistance to cancer therapy.
Results
Exosome isolation and characterization
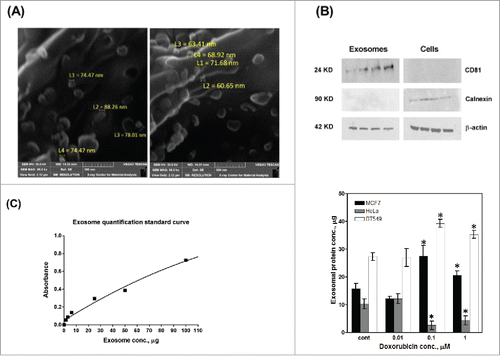
Exosomes were isolated from three cancer cell lines; MCF7, HeLa and BT549 (with or without doxorubicin treatment). Exosome isolation was confirmed by inspection of exosomes' morphology and measuring their sizes using SEM micrographs (), and by analyzing exosomal (CD81 and β-actin) and cellular (calnexin and β-actin) proteins by Western blot analysis. As anticipated, CD81 protein was detected only in the total protein isolated from the exosome pellets, whereas calnexin was found in proteins isolated from cells, but not from exosomes (). β-actin was used as a housekeeping protein present in both cells and exosomes. To further confirm the isolation of exosomes, an exosome quantification kit (ExoTEST Kit, Hansa BioMed, Estonia) was used to quantify exosomes isolated from MCF7, HeLa and BT549 cells (). 10 – 28 µgtotal exosomal protein were isolated from control cells. Treatment with doxorubicin increased the exosomes released from MCF7 cells whereas treatment with doxorubicin reduced the amount of exosomes released from HeLa cells.
Figure 1. Isolation, characterization and quantification of exosomes. Exosomes were isolated from exponentially growing MCF7 and HeLa cells. Exosome isolation was confirmed by electron microscopy (A) and Western blot analyses of CD81 (exosome specific) and calnexin (cellular protein not contained in exosomes); β–actin (cellular and exosomal protein) (B). (C) Standard curve based quantification of exosome release from MCF7 and HeLa cells under increasing doxorubicin concentrations; n = 3 separate experiments per treatment.
Effect of ketotifen on exosome release from cancer cells
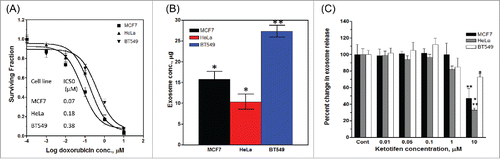
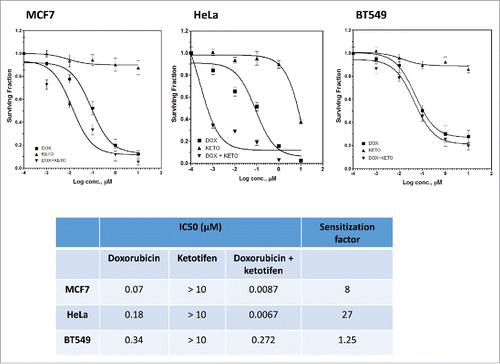
The three cancer cell lines used in the present study showed different sensitivity to doxorubicin () and exhibited differences in the amount of exosome released (). The BT549 cells that is most resistant to doxorubicin showed the highest amount of exosome released. HeLa cells showed the lowest amount of exosmes released whereas MCF7 cells (most sensitive to doxorubicin) showed an intermediate amount. Exponentially growing cells treated with different concentrations of ketotifen for 24 h showed no change in the quantity of exosome release at low concentrations of ketotifen (<1 µmol L−1). At 1 µmol L−1 ketotifen, the exosome quantity was reduced by 20% in HeLa and BT549 cells with no effect on exosomes released from MCF7 cells whereas at 10 µmol L−1 ketotifen, the reduction in exosome release was 70, 45 and 30% in HeLa, MCF7 and BT549 cells, respectively (). Based on this result, 10 µmol L−1ketotifen was selected for downstream experiments.
Figure 2. Effect of ketotifen on exosome release from cancer cell lines. The three cell lines show different sensitivity to doxorubicin (A), and quantitative differences in exosome release (B). Exosome release (in percent of control cells) from MCF7, BT549 and HeLa in the presence of increasing ketotifen concentrations (C); n = 3 separate experiments per treatment *P< 0.05; **P< 0.01; ***P< 0.001 vs. Cont.
Doxorubicin efflux from cancer cells through exosomes
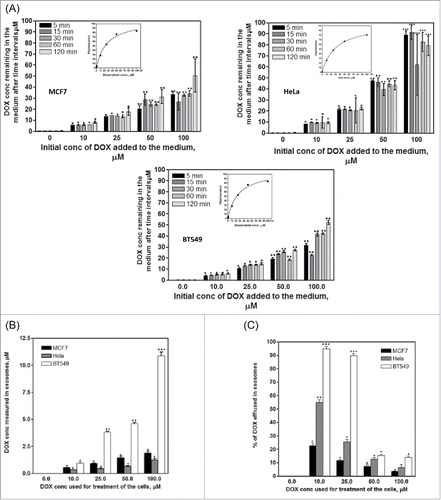
Induction of chemo-resistance by exosomes may involve extracellular export of anticancer drugs by exosomes. We investigated the role of exosomes as mediators of doxorubicin export from cancer cells and how this may affect the response of cancer cells to doxorubicin. The concentration of doxorubicin remaining in the medium of HeLa cells is higher than that measured for the other two cell lines at all time intervals and at all doxorubicin concentrations, which indicates less doxorubicin uptake by HeLa cells than MCF7 and BT549 cells (). Exosomes were then isolated from the three cell lines after treatment with different concentrations of doxorubicin and doxorubicin concentration in the isolated exosomes was measured fluorometrically. Doxorubicin concentration in exosomes isolated from BT549 cells was much higher than in exosomes from MCF7 and HeLa cells (). BT549 cells showed the highest percent of exosomal doxorubicin export, with lower export in HeLa and lowest export in MCF7 cells ().
Figure 3. Exosomal doxorubicin export from cancer cell lines. (A) Doxorubicin uptake by the three cell lines. Cells were incubated with different concentrations of doxorubicin (0–100µM) and at different time intervals, 200µl medium were taken and the fluorescence was measured and converted into doxorubicin concentration using the standard curve (inset). Doxorubicin uptake was obtained by subtracting the amount of doxorubicin remaining in the medium from the original doxorubicin concentration added to the medium. Significance of difference from cells treated with exosomes from control cells is described as *P< 0.05; **P< 0.01: ***P< 0.001. (B) Concentration of doxorubicin efflux from cancer cells within exosomes. Cells were treated with different concentrations of doxorubicin for 2h and exosomes released from the cells overnight were collected and analyzed for doxorubicin content. Significance of difference from cells treated with exosomes from control cells is described as *P< 0.05; **P< 0.01: ***P< 0.001. (C) Percent of doxorubicin efflux in exosomes in the three cell lines was calculated using the amount of exosomes taken by each cell line from (A). Each point is the mean ± SEM of 3 separate experiments. Significance of difference from cells treated with exosomes from control cells is described as * P < 0.05; **P < 0.01: *** P < 0.001
Effect of ketotifen on doxorubicin efflux from cancer cells through exosomes
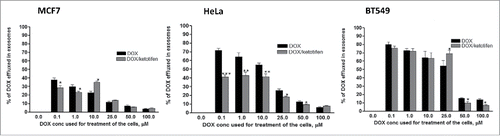
Ketotifen at 10 µmol L−1 reduced exosome release in the three cell lines by 30 to 70% (). We next measured whether ketotifen would reduce exosomal doxorubicin export. Cells were treated with different concentrations of doxorubicin (0.1–100 µmol L−1) for 2 h and after washing doxorubicin, half of the flasks were incubated for 48 h with medium containing ketotifen (10 µmol L−1) and the other half was incubated with ketotifen-free medium. At the end of the incubation period, exosomes were collected from the medium and the concentration of doxorubicin in exosomes was measured. Ketotifen reduced the percent of exosomal doxorubicin release mainly in HeLa cells and showed variable results in MCF7 and BT549 cells (). In HeLa cells, the inhibitory effect of ketotifen was highest at 0.1 and 10 µmol L−1 doxorubicin and absent at 100 µmol L−1 doxorubicin. In the other two cell lines, ketotifen appears affective at low doxorubicin concentrations (0.1 and 1 µmol L−1) in MCF7 cells, while in BT549 cells ketotifen is affective only at highest doxorubicin concentrations (50 and 100 µmol L−1). These results correlate with the effect of ketotifen on the sensitivity of the three cell lines to doxorubicin. These findings suggest that inhibition of doxorubicin release via exosomes and its accumulation within the cells contribute to the mechanisms underlying the increased doxorubicin cytotoxicity under ketotifen co-administration.
Figure 4. Effect of ketotifen on doxorubicin release from cancer cells through exosomes. Cells were treated with doxorubicin (0–100 µM) for 2h followed by ketotifen (or medium only) for 16 h. Exosomes were collected and concentration of doxorubicin within exosomes was quantified. Each point is the mean ± SEM of 3 separate experiments. * P < 0.05; ** P < 0.01: *** P < 0.001
Effect of ketotifen on the sensitivity of HeLa, MCF7 and BT549 cells to doxorubicin
We investigated the effect of modulating exosome release by ketotifen on the response of the three cancer cell lines to the anti-proliferative effects of doxorubicin using SRB assay. Ketotifen alone up to 10 µM did not affect the survival of the three cell lines. Exponentially growing cells were treated with 10 µmol L−1 ketotifen followed by different concentrations of doxorubicin (0.001 – 10 µmol L−1) for 48 h. Pretreatment with ketotifen reduced the doxorubicin's IC50 from 0.18 to 0.0067 µmol L−1 for Hela, from 0.07 to 0.0087 µmol L−1 for MCF7 cells, and from 0.34 to 0.272 µmol L−1 for BT549 cells with a sensitizing factor of 27, 8 and 1.25 respectively (). This result was consistent with the effect of ketotifen on exosome release () where inhibition of exosome release by ketotifen was highest in HeLa cells followed by MCF7 and was lowest in BT549 cells.
Figure 5. Effect of ketotifen on sensitivity of MCF7, BT549 and HeLa cells to doxorubicin. Cells were exposed to either vehicle (DMSO; baseline), ketotifen (KETO), doxorubicin (DOX) alone (0.01 – 10 µmol L−1), or combination of ketotifen and doxorubicin (DOX + KETO) for 48 h as indicated. Cells were fixed and subsequently stained with sulphorhodamine-B (SRB) and the optical density was measured spectrophotometrically. The table summarizes IC50 and sensitization factor for the indicated interventions in indicated cancer cell lines.
Effect of exosomes isolated from doxorubicin-treated cells on the colony-forming ability of MCF7 cells
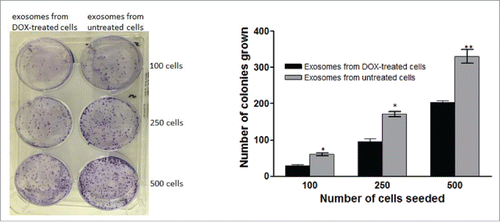
To confirm the activity of doxorubicin in exosomes released from doxorubicin-treated cells, different volumes of exosomes isolated from doxorubicin-treated MCF7 cells and exosomes from control MCF7 cells were added to MCF7 cells. After 14 day in culture a reduction of colony formation was observed in the doxorubicin containing exosome treated group ().
Figure 6. Effect of exosomes isolated from doxorubicin-treated cells on the colony-forming ability of breast cancer (MCF7) cells. Exosomes were isolated from doxorubicin treated or control cells and were added to cells plated in 6-well plate and the cells were kept for 14 days till colony formation. Each point is the mean ± SEM of 3 separate experiments. Significance of difference from cells treated with exosomes from control cells is described as * P < 0.05; ** P < 0.01: *** P < 0.001
Discussion
Exosomes were long considered as byproduct of membrane biosynthesis and shedding. Only recently, their role in cell-cell communication has been discovered.Citation28 Recent studies have explored exosome biology in more detail and discovered multiple important biological functions. The demonstration that exosomes released from normal cells are qualitatively and quantitatively different from those released from cancer cells,Citation25, 16 carries the hope for developing agents that may selectively target exosomes from cancer cells. In the present study, we show that exosomes are involved in the export of doxorubicin from cancer cells and that the mast cell stabilizer ketotifen reduces exosome release and thereby increases cellular doxorubicin levels, leading to enhanced anti-tumor activity especially in HeLa cells.
Development of agents that may reduce release of exosomes from cancer cells or reduce their uptake by the recipient cells may greatly affect the management and progression of cancer. Actually there are now some ways to reduce the effect of exosomes in patients by removing the tumor exosomes from patient's blood by a hemopurifier using antibodies that capture exosomes by binding to their surface molecules. However, this technique is difficult to apply because it is associated with many complications such as the risk of blood clotting or infection and the patient should be hospitalized. Therefore, development of pharmacological agents that reduce exosomal release, uptake or effects is an attractive alternative.
Different cancer cells have shown to release large amounts of exosomes in-vitro and in-vivo. Some previous studies have shown that vesicles, including exosomes, released from cancer cells are a potential mechanism for expulsion of anticancer drugs and induction of chemoresistance in malignant cells.Citation21 Chen et al reported that the vacuolar protein sorting 4a may be involved in the expulsion of anticancer drugs from malignant cells through exosomes and that disruption of this protein reduces doxorubicin efflux in exosomes.Citation6
Quantitative analysis of exosome release from cancer cells used in the present study shows that cells releasing higher amounts of exosomes are less sensitive to doxorubicin than cells releasing less exosomes. This may indicate a role of exosome release in response of cancer cells to doxorubicin most probably through modulation of doxorubicin intracellular accumulation.
Our finding is similar to the previous demonstration by Shedden et al.,Citation21 who showed the existence of doxorubicin containing exosomes near the cell membrane of doxorubicin-treated MCF7 cells. This concept is confirmed in the present study by showing that reduction of exosomal doxorubicin release from cancer cells by ketotifen enhanced the response of these cells to the cytotoxic effects of doxorubicin and was further strengthened by the demonstration that ketotifen reduces doxorubicin efflux from cancer cells through exosomes. This result is in line with the previous demonstration of a correlation between the rate of vesicle shedding and resistance to doxorubicin in different cancer cell lines.Citation21
Exosome release was shown to be regulated by calcium-dependent mechanisms and inhibitors of calcium entry into the cells reduce exosome release.Citation20 Ketotifen was reported previously to block calcium influx into cellsCitation22 and this may explain the reduction in exosomal release in presence of ketotifen. In the present study, 10 µmol L−1 ketotifen reduced the exosomal release of doxorubicin in HeLa cells much more than MCF7 and BT549 cells. This effect correlates with the effect of ketotifen on the response of the three cell lines to doxorubicin, which confirms the relationship between exosome release, doxorubicin export and response of cancer cells to doxorubicin.
Another possible mechanism by which Ketotifen may modulate the response of cancer cells to doxorubicin is through modulation of multiple-drug resistance (MDR) gene expression. Zhang and Berger have shown that ketotifen reverses MDR1-mediated resistance in cancer cells through down-regulation of p-glycoprotein expression.Citation29 We speculate the existence of a link between p-glycoprotein expression and extracellular exosome release from cancer cells. Alternatively, p-glycoprotein may play a role in exosome release from cancer cells and its down-regulation by ketotifen is responsible for ketotifen's effect on exosome release. This speculation may be supported by the work of Shedden et al who showed a positive association between exosomes shedding and resistance of cancer cells to different anticancer drugs belonging to different classes and acting by different mechanisms.
Ketotifen has been shown to suppress the migration and invasion of cancer cells.Citation14 This effect may also be attributed to its ability to inhibit exosome release because exosomes are known to enhance the metastatic potential of cancer cells.Citation28 This effect plus our demonstration in the present study that ketotifen enhances the cytotoxic effects of anticancer drugs supports the suggestion of using ketotifen as a repurposed drug for management of cancer in combination with other anticancer agents.
Results of the present study is the first report of that ketotifen modulates exosome release and exosomal doxorubicin export from cells. We consider this finding as important evidence in support of the design of in vivo and eventually patient studies on the use of ketotifen as a supportive drug in the treatment of malignant tumors.
Material and methods
Cell lines, culture conditions and drugs
Three human cancer cell lines from different histological origins were used in this study; Cervix carcinoma (HeLa), estrogen receptor (ER) and progesterone receptor (PR) double positive invasive ductal breast carcinoma (MCF7), and double negative invasive ductal breast carcinoma (BT549). All cell lines were purchased from the American Type Culture Collection (ATCC) and were maintained in Roswell Park Memorial Institute Medium (RPMI-1640 Gibco, Massachusetts, USA) and Dulbecco's Modified Eagle's Medium (DMEM: Gibco, Massachusetts, USA) supplemented with 10% fetal bovine serum (Gibco, Massachusetts, USA) and penicillin/streptomycin (Gibco, Massachusetts, USA). All incubations were done at 37 °C in a humidified atmosphere of 5% CO2. Mycoplasma was tested at 3 months intervals. For experiments involving exosomes isolation, cells were incubated in medium supplemented with exosome-free serum 24 h before the experiment and during the whole procedure.
Doxorubicin (cat #0025316409) and ketotifen (cat # 34580148) were obtained from Sigma Aldrich (MO, USA).
Preparation of exosome-free medium
The exosome-free fetal bovine serum (FBS) was prepared by ultracentrifugation of fetal bovine serum (FBS) (Gibco, Massachusetts, USA) at 120,000 g, 4 °C for 18 hours followed by filteration through 0.22 micron filter (Whatman Part No. 6900–2502, UK). The exosome-free medium was prepared by adding 10% exosome-free Fetal Bovine Serum (Gibco, Massachusetts, USA) and 1% penicillin/streptomycin (Gibco, Massachusetts, USA) to the RMPI-1640 and DMEM media.
Isolation of exosomes
Exosomes were isolated from the culture medium using the Total exosome isolation kit from Life Technologies (Invitrogen, Massachusetts, USA). Briefly, the cell-free culture medium was transferred to a falcon tube and total exosome isolation reagent was added in a ratio of 2:1. After through mixing, the samples were incubated at 4°C overnight. The mixture was centrifuged at 10,000 g for 1 hour at 4°C. The supernatant was discarded and the exosome pellet was re-suspended in 1X PBS. This exosome solution was kept at −20 °C for further analysis and at −70 °C for long-term storage.
Exosome characterization
(A) By scanning electron microscope (SEM)
Exosomes characterization was done initially by SEM using the method previously described.Citation23 The exosomes suspended in PBS were spread on a carbon adhesive tape sticking to an aluminum pin stub. Evaporation was done using a vacuum concentrator at 30°C, and coating with gold/palladium was done by sputtering (Quorum Technology Mini Sputter Coater, SC7620, UK). The SEM scans were done on a VEGA3 XM microscope (Tescan, Bruno, Czech Republic) at 30kV.
(B) By western blot
Three different antibodies that are differentially expressed in cells and/or exosomes were used to confirm the availability of exosomes: including beta-actin (β-Actin (13E5) rabbit mAb #4970, cell signaling, Boston, USA), CD 81 Anti-TAPA1 antibody [TS81] (Abcam, Massachusetts, USA, ab59477) and Calnexin (C5C9) rabbit (mAb #2679, Cell signaling Boston, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Thermo Scientific.
Western blots were performed as previously described.Citation11 Briefly, exosomes were lysed in reducing sample buffer [0.25 M Tris–HCl (pH 6.8), 40% glycerol, 8% SDS, 5% 2-mercaptoethanol and 0.04% bromophenol blue] and boiled for 10 minutes at 95°C. Proteins were resolved by SDS-PAGE (SDS-polyacrylamide gel electrophoresis), transferred to polyvinylidene fluoride membranes, blocked in 5% non-fat powdered milk in PBS-T (0.5% Tween-20) and probed with antibodies. All proteins were resolved under fully denaturing and reducing conditions, apart from CD81, which was resolved under non-reducing conditions. Protein bands were detected using X-ray film and enhanced chemiluminescence reagent (Amersham ECL Select).
Exosomes quantification
Exosomes released from the cells into the culture medium were quantified using the ExoTEST Kit (Hansa BioMed, Estonia) according to the manufacturer instructions (Exotest ready to Use Kit designed to capture exosomes onto ELISA plate and quantify them by colorimetric, luminometric or fluorimetric detection from any biological sample). Exosome standard was reconstituted and used for the construction of a calibration curve. The ELISA plate with 100 µl test samples loaded per well was incubated at room temperature with shaking for 30 minutes. After washing 3 times with washing buffer, 100 μl of primary antibody (1/500 ratio) was added to each well and incubated at room temperature while shaking for 15 minutes (2–3 rotations per second) followed by 2 hours incubation at 37°C in humid chamber. The plate was washed again with the washing buffer and 100 μl of diluted HRP-conjugated was added to each well. The plate was incubated at room temperature while shaking for 15 minutes (2–3 rotations per second) for 2 hours at 37°C in humid chamber. 100 μl of Substrate Chromogenic Solution was added to each well and incubated uncovered at room temperature in the dark for 5–10 minutes. The plate was monitored until a blue color was visible. At this point, the reaction was stopped by adding 100 μl of stop solution to each well. The absorbance was recorded at 450 nm within 10 minutes with a Fluorimeter (Thermo Fischer Scientific, USA) and the actual amount of exosomes was obtained from the standard curve.
Measurement of doxorubicin uptake by the cells
Doxorubicin is a fluorescent drug therefore all quantitative doxorubicin measurements were done by fluorometry using Varioscan fluoremetr (Thermo scientific, USA) at 488 nm excitation and 606 nm emission wavelengths. The cell culture was initiated with 1 million cells in each tissue culture flask. 27 tissue culture flasks for each cell line were used and treatment was started when the flasks were 50–70% confluent (48 hours after cell seeding). Doxorubicin in different concentrations was added in exosome-free medium in 24 flasks whereas in the remaining 3 flasks, only exosomes free medium (without drug) was added and kept as control. After addition of doxorubicin, 200 μl of the medium was sampled at different time intervals (5, 15, 30, 60, & 120 minutes) and the amount of doxorubicin remaining in the medium was calculated by measuring the fluorescence and converting it into its corresponding concentration using a doxorubicin fluorescence standard curve. Cellular uptake of doxorubicin was determined by subtraction of the actual doxorubicin concentration from the initial doxorubicin concentration shortly after its first addition.
Doxorubicin fluorescence standard curve
The absorbance of increasing concentrations (0, 10, 25, 50 and 100 µmol L−1) of doxorubicin at 488 nm excitation and 606 nm emission wavelengths was used to construct the doxorubicin standard curve.
Determination of doxorubicin export through exosomes and its modulation by ketotifen
After treatment of the cells with different concentrations of doxorubicin for 2 h, the medium was removed and the cells were washed once with PBS, exosome- and doxorubicin-free medium was added to all flasks and ketotifen (10 µmol L−1) was added to half of the flasks. The medium was collected from all flasks after 3 days and kept for exosomes isolation. Quantification of doxorubicin efflux through exosomes was determined fluorometrically by dissolving the isolated exosome pellet (after different treatments) in PBS and measurement of doxorubicin fluorescence in a Varioskan fluorometer (Thermo Fisher). The absorbance is converted to its corresponding doxorubicin concentration using the doxorubicin standard curve. Dividing the amount of doxorubicin in exosomes by the amount of intracellular doxorubicin yields the percent (fraction) of doxorubicin exported through exosomes.
Chemo sensitivity assay
(A) SulphoRhodamine-B (SRB) assay
Cytotoxicity of doxorubicin (Sigma, USA), Ketotifen (Sigma, USA) and their combination was determined using SulphoRhodamine-B (SRB) method as previously described.Citation19 Cells were seeded at a cell density of 5 × 103 cells per well in 96-well plates. After overnight incubation, the cells were treated with ketotifen alone (0.001 – 10 µmol L−1), doxorubicin alone (0.001–10 µmol L−1) or their combination. In combination experiments, cells were treated with doxorubicin for 24 h and after washing doxorubicin, ketotifen (10 µmol L−1) was added to the cells. DMSO was added to the control cells and each treatment was done in triplicates. Fixation of the cells was done after 48 hours. The wells were washed, staining was done with 0.4% SRB and then again washed with 1% acetic acid. The solubilization of the dye was done with 10 mM Tris base (pH 10.5). The measurement of the optical density was achieved spectrophotometrically at 564 nm with the help of an ELISA microplate reader (Thermo scientific, USA). The sigmoidal concentration–response curve fitting model (Graph Pad, Prism software) was used to calculate the IC50 values.
(B) Colony formation assay
The effect of exosomes isolated from doxorubicin-treated cells and from untreated cells on the survival of MCF7 cells was examined using colony formation assay.Citation10 Different number of cells (100, 250 and 500) were cultured in 6-well plates. After attachment, cells were treated with exosomes isolated from doxorubicin-treated cells or from untreated cells. Plates were incubated for 14 days to allow cells to form colonies. At the end of the incubation period, formed colonies were fixed with 70% ethanol, stained with crystal violet. Colonies containing 50 cells or more were counted.
Statistical analysis
All results are presented as mean ± SEM of at least three independent experiments. Curves, data fitting and statistical analysis were done using Graph Pad Prism (San Diego, USA). Significance of difference from the control cells is described as *P < 0.05; **P < 0.01: ***P < 0.001.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions
FMK performed cell culture, exosome isolation, Western blot and analysis of doxorubicin content within exosomes EMS Performed doxorubicin uptake experiment, analysis of results and revision of the manuscript HA performed and analyzed exosome visualization by electron microscopy and revised the manuscript RH preparation of the manuscript WHZ set the idea of the work and preparation of the manuscript RAE designed the study, followed the experiments, analyzed the results and wrote the paper. All authors approved the final version of the manuscript
Acknowledgments
Al Jalila Foundation Research Center (AJFRC) supported this work, project number AJF201612
Additional information
Funding
References
- Akao Y, Iio A, Itoh T, Noguchi S, Itoh Y, Ohtsuki Y, Naoe T. Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages. Mol Ther. 2011;19:395–9. doi:10.1038/mt.2010.254.
- Aspenström P. Integration of signalling pathways regulated by small GTPases and calcium. Biochim Biophys Acta – Mol Cell Res. 2004;1742:51–58. doi:10.1016/j.bbamcr.2004.09.029.
- Bartlett NL, Lum BL, Fisher GA, Brophy NA, Ehsan MN, Halsey J, Sikic BI. Phase I trial of doxorubicin with cyclosporine as a modulator of multidrug resistance. J Clin Oncol. 1994;12:835–842. doi:10.1200/JCO.1994.12.4.835.
- van den Boorn JG, Dassler J, Coch C, Schlee M, Hartmann G. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev. 2013;65:331–335. doi:10.1016/j.addr.2012.06.011.
- Bray K, Gillette M, Young J, Loughran E, Hwang M, Sears JC, Vargo-Gogola T. Cdc42 overexpression induces hyperbranching in the developing mammary gland by enhancing cell migration. Breast Cancer Res. 2013;15:R91. doi:10.1186/bcr3487.
- Chen VY, Posada MM, Blazer LL, Zhao T, Rosania GR. The Role of the VPS4A-Exosome Pathway in the Intrinsic Egress Route of a DNA-Binding Anticancer Drug Pharm Res. 2006;23:1687–1695.
- Corcoran C, Rani S, O'Brien K, O'Neill A, Prencipe M, Sheikh R, Webb G, McDermott R, Watson W, Crown J, et al. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7:e50999. doi:10.1371/journal.pone.0050999.
- Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ, Advani RJ, et al. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113 Pt 19:3365–74.
- El-Awady RA, Hersi F, Al-Tunaiji H, Saleh EM, Abdel-Wahab AH, Al Homssi A, Suhail M, El-Serafi A, Al-Tel T. Epigenetics and miRNA as predictive markers and targets for lung cancer chemotherapy. Cancer Biol Ther. 2015;16:1056–1070. doi:10.1080/15384047.2015.1046023.
- El-Awady RA, Saleh EM, Dahm-Daphi J. Targeting DNA double-strand break repair: is it the right way for sensitizing cells to 5-fluorouracil? Anticancer Drugs. 2010;21:277–287. doi:10.1097/CAD.0b013e328334b0ae.
- El-Awady RA, Saleh EM, Ezz M, Elsayed AM. Interaction of celecoxib with different anti-cancer drugs is antagonistic in breast but not in other cancer cells. Toxicol Appl Pharmacol. 2011;255:271–286. doi:10.1016/j.taap.2011.06.019.
- Franzius D, Hoth M, Penner R. Non-specific effects of calcium entry antagonists in mast cells. Pfl{ü}gers Arch. 1994;428:433–438. doi:10.1007/BF00374562.
- Katakowski M, Buller B, Zheng X, Lu Y, Rogers T, Osobamiro O, Shu W, Jiang F, Chopp M. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2016;335:201–204. doi:10.1016/j.canlet.2013.02.019.
- Kim HJ, Park MK, Kim SY, Lee CH. Novel suppressive effects of ketotifen on migration and invasion of MDA-MB-231 and HT-1080 cancer cells. Biomol Ther. 2014;22:540–546. doi:10.4062/biomolther.2014.081.
- Koumangoye RB, Sakwe AM, Goodwin JS, Patel T, Ochieng J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS One. 2011;6:e24234. doi:10.1371/journal.pone.0024234.
- Palma J, Yaddanapudi SC, Pigati L, Havens MA, Jeong S, Weiner GA, Weimer KM, Stern B, Hastings ML, Duelli DM. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125–9138. doi:10.1093/nar/gks656.
- Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic Tumor Cell Modulates Its Microenvironment to Enhance Angiogenic and Metastatic Potential by Secretion of Proteins and Exosomes. Mol Cell Proteomics. 2010;9:1085–1099. doi:10.1074/mcp.M900381-MCP200.
- Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi:10.1074/jbc.M109.041152.
- Saleh EM, El-Awady RA, Anis N. Predictive markers for the response to 5-fluorouracil therapy in cancer cells: Constant-field gel electrophoresis as a tool for prediction of response to 5-fluorouracil-based chemotherapy. Oncol Lett. 2012;5:321–327.
- Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi:10.1074/jbc.M301642200.
- Shedden K, Xie XT, Chandaroy P, Chang YT, Rosania GR. Expulsion of Small Molecules in Vesicles Shed by Cancer Cells. Cancer Res. 2003;63:4331LP–4337.
- Soboloff J, Zhang Y, Minden M, Berger SA. Sensitivity of myeloid leukemia cells to calcium influx blockade: Application to bone marrow purging. Exp Hematol. 2002;30:1219–1226. doi:10.1016/S0301-472X(02)00893-7.
- Szajnik M, Derbis M, Lach M. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet 2013;4(3):1–11.
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi:10.1038/ncb1596.
- Di Vizio D, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, Mulholland D, Rotinen M, Hager MH, Insabato L, et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am J Pathol. 2012;181:1573–1584. doi:10.1016/j.ajpath.2012.07.030.
- Yu JL, May L, Lhotak V, Shahrzad S, Shirasawa S, Weitz JI, Coomber BL, Mackman N, Rak JW. Oncogenic events regulate tissue factor expression in colorectal cancer cells : implications for tumor progression and angiogenesis. Blood. 2015;105:1734–1742. doi:10.1182/blood-2004-05-2042.
- Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi:10.1158/0008-5472.CAN-05-4579.
- Zhang HG, Grizzle WE. Exosomes: A novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28–41. doi:10.1016/j.ajpath.2013.09.027.
- Zhang Y, Berger SA. Ketotifen reverses MDR1-mediated multidrug resistance in human breast cancer cells in vitro and alleviates cardiotoxicity induced by doxorubicin in vivo. Cancer Chemother Pharmacol. 2003;51:407–414.
- Zhang Y, Crump M, Berge SA. Purging of contaminating breast cancer cells from hematopoietic progenitor cell preparations using activation enhanced cell death. Breast Cancer Res Treat. 2002;72:265–278. doi:10.1023/A:1014965726663.
