ABSTRACT
Advances in multiplex immunohistochemistry (IHC) techniques and digital pathology platforms allow quantification of multiple proteins at same tissue section and produce continuous data. TGF-β signaling plays crucial and complex roles in colorectal cancer (CRC). We here aimed to investigate clinical pathological relevant of proteins involved in TGF-β signaling at CRC tissues. Multiplex fluorescent IHC was used to quantitative analysis. The levels of eight proteins (TGF-β1, TGFBRI, TGFBRII, SMAD4, SMAD2/3, p-SMAD2/3, SMAD1/5/9, and p-SMAD1/5/9) were determined in TMA sections. Quantitative analysis was carried out by a scoring system by InForm software. It revealed that TGF-β signaling was hyper active. The levels of TGF-β1, TGFBRI, TGFBRII, SMAD4, SMAD1/5/9 and p-SMAD2/3 were significantly increased in cancer tissues when compared their levels in normal tissues. Furthermore, the levels of eight proteins in stroma were significantly lower than the levels that in cancer tissues. Clinical pathological relevant analysis exhibited that TGF-β signaling inclined to suppress the progression of tumor. SMAD1/5/9, TGFBRII, SMAD2/3 were confirmed as significant predictors for overall survival. In conclusion, we established a method based on multispectral imaging to extensively explore the clinical relevant of TGF-β signaling proteins. These results provided an opportunity to consider the novel application for proteins involving TGF-β signaling that used as diagnostic or prognostic biomarkers to conduct tumor therapy.
Introduction
Transforming growth factor (TGF-β) signaling pathway is aberrant in colorectal cancer (CRC), and exhibits a paradoxical roles.Citation1 A large amount of data supported an early tumor-suppressive role for TGF-β. It was also shown that TGF-β signaling within the carcinoma cell could promote tumor progression and metastasis at late stage.Citation2 However, the mechanisms are still not known how TGF-β signaling switched from tumor suppressor to tumor promoter.
TGF-β signaling pathway has been well studied. TGF-β, acts as one of the most important components in TGF-β signaling pathway, consists by three isoforms (TGF-β1, TGF-β2, and TGF-β3). Among of them, TGF-β1 is the most abundant and well-studied isoform.Citation3 There are two typical TGF-β receptor, TGF-β type I and type II receptors (TGFBRI and TGFBRII), with active serine/threonine kinase.Citation4 The activated TGF-β can bind to TGFBRII, then phosphorylates TGFBRI. Activated TGFBRI propagates the signal by phosphorylating intracellular downstream effectors, SMAD2 and SMAD3 (SMAD2/3).Citation5 Once binding, SMAD4 will be recruitment by phosphorylated-SMAD2/3 and translocate to the nucleus to activate TGF-β-responsive target genes.Citation6 SMAD1/5/9 are targets of bone morphogenetic protein (BMP) receptor, regarded as non-canonical SMAD-independent pathways.Citation7 SMAD4 coupling with phosphorylated SMAD1/5/9 can mediate an antitumor function.Citation8
Recently, the treatment with drugs targeting TGF-β signaling pathway had been developed.Citation9 The challenge is to identify the group of patient where TGF-β signaling needed to be inhibited or enhanced for delineating the tumor suppressive versus the tumor promoting roles of TGF-β signaling pathway.Citation9,Citation10
So far, there are relatively few studies investigating the relationships between clinical characteristics and changes of the proteins involved in TGF-β signaling. In light of this, we attempted to analyze the proteins involved in TGF-β signaling by using multiplex immunofluorescent staining. Tumor sections from 90 CRC patients were co-stained with antibodies against to TGF-β1, TGFBRI, TGFBRII, SMAD4, SMAD2/3, p-SMAD2/3, SMAD1/5/9, and p-SMAD1/5/9. For high throughput analysis, the multispectral imaging and analysis system (Vectra-Nuance-InForm®) from Perkin Elmer were employed to analyze the tumor sections labeled with multiple fluorophores. To figure the clinical relevant, the levels of those eight proteins were further analyzed to develop relationships between the characteristics in patients with CRC.
Results
Multiplex fluorescent IHC determines eight proteins involved TGF-β signaling
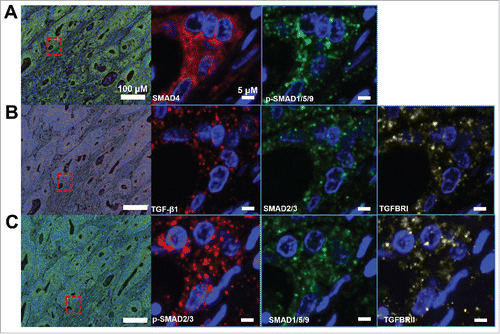
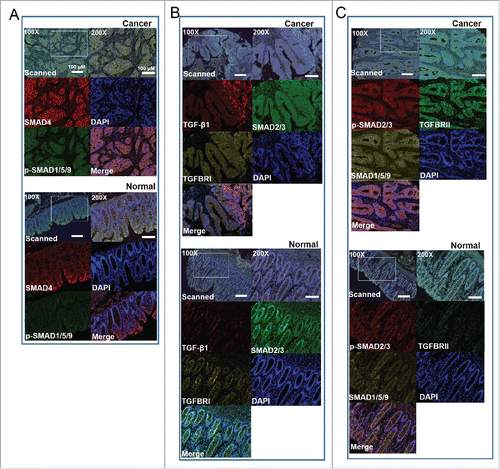
Eight proteins (TGF-β, TGFBRI, TGFBRII, SMAD4, SMAD2/3, p-SMAD2/3, SMAD1/5/9, and p-SMAD1/5/9), represented the most important factors in TGF-β signaling, were chosen for multiplex fluorescent IHC on TMA slides containing 180 tissues (cancer vs. adjacent normal tissues) sections from 90 patients with CRC. A four-color fluorescent IHC kit was applied and it enabled to simultaneous stain three proteins and nuclei on one TMA slide. So, three slides were used to stain eight proteins. SMAD4 and p-SMAD1/5/9 were successfully stained in cancer and adjacent normal tissues and unmixed into three spectral channels (Opal570, Opal670, and DAPI) in one slides (). Antibodies against to TGF-β1, SMAD2/3, and TGFBRI were applied to determine their expression in another TMA slide. The fluorescence of TGF-β1, SMAD2/3, TGFBRI and nuclei unmixed to four spectral channels (Opal570, Opal 670, Opal620, and DAPI) (). In following, unmixable signals of p-SMAD2/3, SMAD1/5/9 and TGFBRII were also obtained and included four spectral channels (Opal570, Opal 670, Opal620, and DAPI) ().
Figure 1. Multispectral IHC stained eight proteins involved in TGF-β signaling on TMA slides of colorectal cancer (CRC). A. Representative images for SMAD4 and p-SMAD1/5/9 staining in cancer tissues (upper) and paired normal tissues (bottom). The upper images showing the raw scanned images. B. Triple staining shows TGF-β1, SMAD2/3, and TGFBRI in cancer tissues (upper) and paired normal tissues (bottom). C. Images show the triples staining for SMAD2/3, SMAD1/5/9 and TGFBRII in cancer tissues (upper) and paired normal tissues (bottom). Nuclei were counterstained with DAPI.
Subcellular localization of stained TGF-β signaling proteins
We reviewed a common region in same section of three slides to show the subcellular localization of stained proteins. As expected, the subcellular localizations were coincided with the functions of those protein in activing TGF-β signaling. SMAD4, SMAD1/5/9, and SMAD2/3 mainly located in cytoplasm (). TGFBRI and TGFBRII were found to be expressed around the cellular membrane (). Meanwhile, p-SMAD2/3 and p-SMAD1/5/9 distributed in cytoplasm and near or in the nuclei. Besides, TGF-β1 mostly located in the extracellular component. In particular, except SMAD4, most stained proteins were accumulated in the specific cellular component. Interestingly, TGF-β1, TGFBRI and TGFBRII were co-localized in the same cellular component ().
Scoring and comparing the levels of eight TGF-β signaling proteins
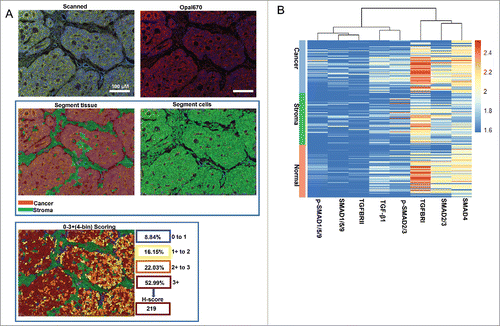
To determine the levels of eight TGF-β signaling proteins, we explored a scoring system based on InForm software and the pipeline is showing in . The mean H-score by calculating three to six regions for each patient represented the expression levels of proteins. Unsupervised hierarchical clustering demonstrated that eight TGF-β signaling proteins can be classified into two subgroups (SMAD4, SMAD2/3 and TGFBRI; p-SMAD1/5/9, SMAD1/5/9, p-SMAD2/3, TGFBRII and TGF-β1) ().
Figure 3. Scoring the stained proteins. A. Pipeline for tissues and cells segment by InForm software. The image of raw multiple fluorescent channels merged was separated into two fluorescent channels (Opal670 and DAPI) (upper). Segment tissue was divided into cancer and stroma based on the fluorescent signal intensity of DAPI (middle panel). Representative image of 0–3+(4-bin) scoring system (bottom). This score system can be used to calculate H-score with cell stains. Difference colors displays four levels (0∼1, 1∼2, 2∼3, 3∼) and percentage positivity of cells with each bin of stained protein. H-score was automatic calculated by InForm software. B. Unsupervised hierarchical clustering of eight TGF-β signaling proteins classified the 90 patients with CRC.
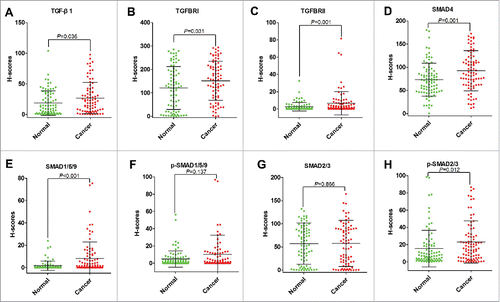
Next, we compared their levels among cancer tissue, stroma tissues and normal tissues. All of them were shown higher levels in cancer tissues than normal tissues. The levels of TGF-β1, TGFBRI, TGFBRII, SMAD4, SMAD1/5/9, and p-SMAD2/3 were significantly increased in cancer tissues (P<0.05) (). The levels of p-SMAD1/5/9 and SMAD2/3 were increased but did not reach the statistical significance (). Moreover, we analyzed the difference of eight proteins between cancer and stroma. All the molecular involving TGF-β signaling pathway, were significantly higher in cancer when compared their expression in stroma tissues (Figure S1 A, B, C, D, E, F, G, H). In combine with above findings, it indicated that TGF-β signaling was highly active in cancer tissues.
The correlation between eight proteins and clinicopathological characteristics
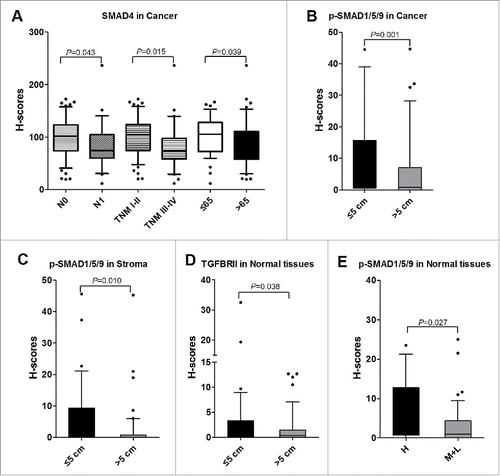
The correlation between eight proteins and nine clinical factors (as listed in Tables S1) were analyzed. In cancer tissues, the levels of SMAD4 were significantly higher in CRC patients at N0 stage than those patients whom were judged as N1 stage (). The levels of SMAD4 were decreased for those advanced (TNM III-IV) patients in compared with whom at early stage (TNM I-II) (). In addition, SMAD4 was found to be significantly downregulated for older subjects (>65 y old) (). Furthermore, the expressions of p-SMAD1/5/9 were significantly higher in cancer tissues obtained from CRC patients with tumor diameter ≤ 5cm (). In stroma, p-SMAD1/5/9 was also proved to be significantly increased for CRC patients whom had tumor diameter ≤ 5cm (). In normal tissues, levels of TGFBRII for CRC patients with tumor diameter ≤ 5cm were significantly higher than whom had tumor diameter > 5 cm (). Moreover, levels of p-SMAD1/5/9 were increased in normal tissues from high differentiation CRC patients than those moderate or low differentiation CRC patients (). Above all, the inactivation of TGF-β signaling pathway inclined to make a clue of more aggressive cancer status.
Figure 5. Correlations between TGF-β signaling and clinicopathological characteristics. A. The levels of SMAD4 associated with N stage, TNM stage, and age (years) in cancer tissues. B. The levels of p-SMAD1/5/9 were increased in CRC patients with tumor volume ≤ 5 cm than those whom tumor volume > 5cm in cancer tissues. C. In stroma, the levels of p-SMAD1/5/9 were increased in CRC patients with tumor volume ≤ 5 cm than those whom tumor volume > 5 cm. D. In normal tissues, TGFBRII was correlated with tumor volume. E. p-SMAD1/5/9 was associated with differentiation in normal tissues. H, High; M, Moderate; L, Low.
Prognostic significance of TGF-β signaling proteins
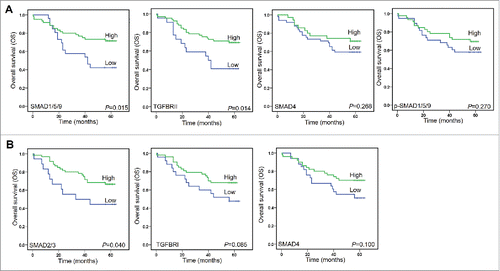
There were 34 (37.78%) patients died during following-up period (Table S1). On univariate analysis, two TGF-β signaling proteins (SMAD1/5/9 and TGFBRII) in cancer tissue, one of them in normal tissue (SMAD2/3), and four clinical factors (TNM, T stage, N stage, and Tumor size) were all confirmed as prognostic factors for OS (Table S3). In cancer tissues, Kaplan-Meier analysis also demonstrated that CRC patients whose tumors featured high SMAD1/5/9 and TGFBRII had a longer OS time than those whose tumors featured low SMAD1/5/9 and TGFBRII (). Meanwhile, high levels of SMAD4 and p-SMAD1/5/9 predicted a longer OS time than those had low SMAD4 and p-SMAD1/5/9. However, those differences failed to reach statistical differences on OS rates (). Kaplan-Meier analysis proved that there were significantly difference in OS for CRC patients who shown high or low expression of SMAD2/3 in normal tissues (). Although there were no statistical differences for TGFBRI and SMAD4 when used as factors to predict the OS, the high levels for both proteins indicated a longer OS times (). In combining with those evidences, we might conclude that the activation of TGF-β signaling would prolong OS time.
Figure 6. Kaplan-Meier analyzed overall survival (OS) rates. A. SMAD1/5/9, TGFBRII, SMAD4, and p-SMAD1/5/9 as prognostic factors in cancer tissues. B. SMAD2/3 and TGFBRI, and SMAD4 as prognostic factors in normal tissues.
We further evaluated the genetic alterations of SMAD1, ∼2, ∼3, ∼4, ∼5, ∼9, TGF-β1, TGFBRI, and TGFBRII based on the dataset from The Cancer Genome Atlas (TCGA). TP53, KRAS, APC, and EGFR (ERBB1) had been shown more genetic alterations and also been analyzed in our study (Figure S2). APC was shown to the highest genetic alteration rates (73%). The genetic alteration rates for KRAS, TP53, EGFR were 44%, 53%, 5%, respectively. The genetic alteration rate for SMAD4 was 15% and was recognized as the highest alteration rates molecular of TGF-β signaling. Secondly, we detected the prognostic relevant of mRNA levels for those genes. The enrolled TCGA dataset contained normalized values of colon cancer tissues by RNA-sequencing from 454 patients (Figure S3). SMAD2, ∼6, ∼7, ∼9, TGFBRI and TGFBRII were proved to be negatively associated with prognosis. SMAD3, ∼4, and TGF-β1 were positively associated with prognosis. Those results partially supported our conclusion that the activated TGF-β signaling benefited cancer patients and prevent tumor progression at early stage.
Discussion
Technologies such as multiplex IHC have been in the forefront of development because they significantly enrich the data extracted from tumor tissue and allow for relationship analysis between proteins levels and tumor progression.Citation11,Citation12 At present study, we established a method based on multispectral imaging to extensively explore the clinical relevant of TGF-β signaling in CRC patients. These results provide an opportunity to consider the novel application for proteins involving TGF-β signaling that used as diagnostic or prognostic biomarkers to conduct tumor therapy.
As noted, TGF-β signaling plays complex and often contradictory roles in cancer.Citation5 Its tumor suppressive roles is most commonly attributed to inhibition of cell proliferation and negatively associated with prognosis.Citation13 Our findings just revealed a consistent conclusion that high levels of TGF-β signaling proteins (eg. SMAD1/5/9 and TGFBRII) predicted a more benefited prognosis for CRC patients. Meanwhile, TGFBRII also confirmed by other study that was a prognostic biomarkers for CRC patients.Citation14 Here, we took a glance on the relevant between prognosis and levels of TGF-β signaling proteins in normal tissue and shown the expression of SMAD2/3 was associated with prognosis. It was usually ignored by other study.
Hyper active of TGF-β signaling had been determined by Lampropulos P. et al with IHC staining in CRC.Citation14 Interestingly, we found the hyper active TGF-β signaling exhibited a role on preventing tumor progression. The dysregulation of oncogene and gene mutations acts as two major factors to prevent the inhibition tumor progression by TGF-β signaling. For instance, inhibition tumor growth by activating TGF-β signaling could be greatly diminished by MYC.Citation5 Mutations were common in the genes that coding TGF-β signaling proteins.Citation15 Specially, those mutations major effected TGFBRII and SMAD4.Citation16,Citation17 TGFBRII mutations were detected nearly 74% of CRC patients with microsatellite instability (MSI) and did not associated with the patients outcome.Citation18 Approximately 20–30% of patients with CRC had SMAD4 mutations.Citation19 Moreover, alterations in SMAD4 preferentially occurred as late events while TP53 mutations preferentially occurred early stage of carcinogenesis.Citation20
In particular, we want to emphasize SMAD4 of which was traditionally recognized as tumor suppressor.Citation21 Our findings supported this consensus that SMAD4 prevented tumor progression because it was negative associated with TNM and N stage. A meta-analysis revealed that SMAD4 loss was associated with a poor overall survival.Citation22 However, SMAD4 was recently proved to exert a tumor-promoting role in hepatocellular carcinoma (HCC).Citation8 This contradictory effects could be the results of gene mutations. SMAD4 mutation did not change levels of proteins mutation but exhibited reversibly functions.Citation23 Still, we lack effective techniques to distinguish mutated and wild type SMAD4 in proteins levels. Furthermore, one of most important function for SMAD4 is nuclear translocation to act as transcription factors. But SMAD4 binding to DNA sequences have a lower affinity.Citation24 Therefore, high affinity transcription factor is a needed for SMAD4 activation gene transcription. For instance, we previously identified PSG9 that directly binding to SMAD4 and promoted its nuclear translocation, subsequently activation genes expression associated with angiogenesis.Citation25
Usually, SMAD1/5/9 and SMAD2/3 existed as complexes and conducted canonical and mixed TGF-β/BMP signaling respectively in cytoplasm.Citation26 In addition, phosphorylated SMAD1/5/9 and SMAD2/3 complexes were distributed in nucleus. Both phosphorylated complexes had no prognosis relevant and this finding consisted with a previous report.Citation14 One limitation of present study was that InForm software was unfit to segment nuclei and cytoplasm, we thus failed to explore pathological features significance and prognostic relevant for nuclear p-SMAD2/3 and p-SMAD1/5/9.
In conclusion, our study for the first time report the ability to perform multiplex IHC with TGF-β signaling proteins to identify their clinical relevant. Importantly, this method provides the ability to simultaneously quantity analysis levels for multiple factors on a single tissue section. Additionally, high levels of TGF-β signaling proteins were proved to prevent tumor progress. The loss of SMAD1/5/9 and TGFBRII predicted a poor prognosis for CRC patients. In view of this, multiplex IHC determined TGF-β signaling proteins could be used as one of tools to conduct therapy by targeting TGF-β signaling.
Material and methods
Patients
The HCol-Ade180Sur-07 tumor tissue microarray (TMA) (Shanghai OUTDO Biotech Co., Shanghai, China) consisted of 90 paired colorectal adenocarcinoma tissues and matched normal mucosa. Patients underwent surgery from January 2009 to October 2009, and the follow-up information was available from February 2009 to May 2014. All sample donors provided informed consent, and the study was conducted under the approval of the Institutional Ethics Committee. All tissue samples were collected from patients of colon cancer for whom was not received any chemotherapy or radiotherapy before surgery. All procedures were performed in accordance with the relevant guidelines and regulations. The pathological characteristics of 90 subjects were summarized in Supplementary Table S1.
Immunostaining of TMA
For staining eight proteins in TGF-β signaling, three same TMA slides were used to immunostaining analysis. The TMA slides were stained using PerkinElmer Opal™ 4-color fIHC Kit (PerkinElmer, Hopkinton, MA, USA).
For staining eight proteins in TGF-β signaling, three same TMA slides were used to immunostaining analysis. The TMA slides were stained using PerkinElmer Opal™ 4-color fIHC Kit (PerkinElmer, Hopkinton, MA, USA). Briefly, the deparaffinization protocol was run (xylene 15 min two times, 100% ethanol, 95% ethanol, 85% ethanol, and 75% ethanol for 10 min) until to rinse in water for 10 min. After that, slide was pretreated with antigen retrieval solution (provided in Opal kit) by microwaving method (2 min on 100% power, followed by 20 min on 20% power), followed by 10 min blocking in 10% goat serum in TBS and rinsed in water. Then, three slides were stained respectively with primary antibody against TGF-β1, p-SMAD2/3, and SMAD4 for 1 hour. After washing the slides with 1XTBST for three times, the slides were incubated with horseradish perodidase labeled mouse/rabbit secondary antibody (Zhongshan Golden Bridge, Beijing, China) for 10 min. Tyramide (TSA)-conjugated fluorophore (Opal570) was added to slides at 1:100 dilution in Amplification plus buffer and incubated for 10 min at RT. TSA was vacuumed off, and slides were washed three times. For staining SMAD2/3, SMAD1/5/9, and p-SMAD1/5/9, three slides were then treated with antigen retrieval solution and TSA-conjugated fluorophore (Opal670) as mentioned above. Next, TSA-conjugated fluorophore (Opal520) was applied to stain TGFBRI and TGFBRII with same procedures. The detail information for primary antibodies were summarized in Supplementary Table S2. At last, all slides were counterstained with Spectral DAPI to show nuclei and cover slips followed by sealing with mounting medium supplied with antifade solution (Applygen Technologies Inc. Beijing, China).
Multispectral imaging
Multispectral images were taken with Vectra Imaging System (Perkin Elmer) by 20X objective lens. Nuance system (Perkin Elmer) was used to build libraries of each spectrum (570, 670,620 and DAPI) and unmix multispectral images with high contrast and accuracy.
Scoring multispectral images
InForm 2.1.1 software (Perkin Elmer) was used to batch analysis of multispectral images from the experiment. The images were loaded into InForm to build single color compensation library and substrate background using the image deriving from a slide without staining in the same exposure settings. For scoring, three to six representative regions of interest for high powered (200X) imaging from every cases were selected. To build algorithm for segment tissues and cells, a few representative multispectral images from the experiment were loaded into InForm software. Then, the segmented tissues of parenchymal neoplasms and mesenchyme and cells were trained according to DAPI signals intensity. After, the detected tissues compartments were selected and quantified for each stained proteins in slides. 0–3+(4-bin) scoring system was used to quantify proteins levels. This score system can be used to calculate H-score with cell stains. It included four levels (0∼1, 1∼2, 2∼3, 3∼), score results were shown by the percentage positivity of cells with each bin. H-score was calculated using the percentages in each bin and ranges from 0 to 300. Batch process all remaining multispectral images from the experiment.
TCGA dataset analysis and Statistics
Molecular data from human colorectal cancers were generated by TCGA Research Network (http://www.cbioportal.org/). The significance of the data from patient specimens was determined by the Mann-Whitney U test. Overall survival (OS) rates were assessed by the Cox proportional hazards regression model and Kaplan-Meier test, and the log-rank test was used to plot survival curves. P<0.05 was considered to be significant.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Authors’ contributions
LY obtained funding and participated in the study design and coordination.LY, ZL, JJT, and HWD performed IHC assays. LY, ZL, and XJZ analyzed data.
2017CBT10296R1-file002.docx
Download MS Word (1.2 MB)Acknowledgments
We would like to acknowledge Ms Na Gao, from Respiratory Medicine Department, Beijing Chaoyang Hospital, Capital Medical University, who selfless provided Opal™ 4-color fIHC kit for this study.
Additional information
Funding
References
- Katz LH, Likhter M, Jogunoori W, Belkin M, Ohshiro K, Mishra L. TGF-beta signaling in liver and gastrointestinal cancers. Cancer Lett. 2016;379:166–72.
- Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160:963–76.
- Chen J, Katz LH, Munoz NM, Gu S, Shin JH, Jogunoori WS, Lee MH, Belkin MD, Kim SB, White JC, et al. Vitamin D Deficiency Promotes Liver Tumor Growth in Transforming Growth Factor-beta/Smad3-Deficient Mice Through Wnt and Toll-like Receptor 7 Pathway Modulation. Sci Rep. 2016;6:30217.
- Yin S, Fan Y, Zhang H, Zhao Z, Hao Y, Li J, Sun C, Yang J, Yang Z, Yang X, et al. Differential TGFbeta pathway targeting by miR-122 in humans and mice affects liver cancer metastasis. Nat Commun. 2016;7:11012.
- Dews M, Tan GS, Hultine S, Raman P, Choi J, Duperret EK, Lawler J, Bass A, Thomas-Tikhonenko A. Masking epistasis between MYC and TGF-beta pathways in antiangiogenesis-mediated colon cancer suppression. J Natl Cancer Inst. 2014;106:dju043.
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–76.
- Zhang YH, Cheng F, Du XT, Gao JL, Xiao XL, Li N, Li SL, Dong DL. GDF11/BMP11 activates both smad1/5/8 and smad2/3 signals but shows no significant effect on proliferation and migration of human umbilical vein endothelial cells. Oncotarget. 2016;7:12063–74.
- Hernanda PY, Chen K, Das AM, Sideras K, Wang W, Li J, Cao W, Bots SJ, Kodach LL, de Man RA, et al. SMAD4 exerts a tumor-promoting role in hepatocellular carcinoma. Oncogene. 2015;34:5055–68.
- Katz LH, Li Y, Chen JS, Munoz NM, Majumdar A, Chen J, Mishra L. Targeting TGF-beta signaling in cancer. Expert Opin Ther Targets. 2013;17:743–60.
- Iyer S, Wang ZG, Akhtari M, Zhao W, Seth P. Targeting TGFbeta signaling for cancer therapy. Cancer Biol Ther. 2005;4:261–6.
- Feng Z, Puri S, Moudgil T, Wood W, Hoyt CC, Wang C, Urba WJ, Curti BD, Bifulco CB, Fox BA. Multispectral imaging of formalin-fixed tissue predicts ability to generate tumor-infiltrating lymphocytes from melanoma. J Immunother Cancer. 2015;3:47.
- Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–7.
- Chun HK, Jung KU, Choi YL, Hong HK, Kim SH, Yun SH, Kim HC, Lee WY, Cho YB. Low expression of transforming growth factor beta-1 in cancer tissue predicts a poor prognosis for patients with stage III rectal cancers. Oncology. 2014;86:159–69.
- Lampropoulos P, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. Prognostic significance of transforming growth factor beta (TGF-beta) signaling axis molecules and E-cadherin in colorectal cancer. Tumour Biol. 2012;33:1005–14.
- Pinheiro M, Pinto C, Peixoto A, Veiga I, Lopes P, Henrique R, Baldaia H, Carneiro F, Seruca R, Tomlinson I, et al. Target gene mutational pattern in Lynch syndrome colorectal carcinomas according to tumour location and germline mutation. Br J Cancer. 2015;113:686–92.
- Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6.
- de Miranda NF, van Dinther M, van den Akker BE, van Wezel T, ten Dijke P, Morreau H. Transforming Growth Factor beta Signaling in Colorectal Cancer Cells With Microsatellite Instability Despite Biallelic Mutations in TGFBR2. Gastroenterology. 2015;148:1427–37 e8.
- Shima K, Morikawa T, Yamauchi M, Kuchiba A, Imamura Y, Liao X, Meyerhardt JA, Fuchs CS, Ogino S. TGFBR2 and BAX mononucleotide tract mutations, microsatellite instability, and prognosis in 1072 colorectal cancers. PLoS One. 2011;6:e25062.
- Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–103.
- Braxton DR, Zhang R, Morrissette JD, Loaiza-Bonilla A, Furth EE. Clinicopathogenomic analysis of mismatch repair proficient colorectal adenocarcinoma uncovers novel prognostic subgroups with differing patterns of genetic evolution. Int J Cancer. 2016;139:1546–56.
- Yamamoto T, Kawada K, Itatani Y, Inamoto S, Okamura R, Iwamoto M, Miyamoto E, Chen-Yoshikawa TF, Hirai H, Hasegawa S, et al. Loss of SMAD4 Promotes Lung Metastasis of Colorectal Cancer by Accumulation of CCR1+ Tumor-associated Neutrophils through CCL15-CCR1 Axis. Clin Cancer Res. 2016.
- Voorneveld PW, Jacobs RJ, Kodach LL, Hardwick JC. A Meta-Analysis of SMAD4 Immunohistochemistry as a Prognostic Marker in Colorectal Cancer. Transl Oncol. 2015;8:18–24.
- Demagny H, De Robertis EM. Point mutations in the tumor suppressor Smad4/DPC4 enhance its phosphorylation by GSK3 and reversibly inactivate TGF-beta signaling. Mol Cell Oncol. 2016;3:e1025181.
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84.
- Yang L, Hu S, Tan J, Zhang X, Yuan W, Wang Q, Xu L, Liu J, Liu Z, Jia Y, et al. Pregnancy-specific glycoprotein 9 (PSG9), a driver for colorectal cancer, enhances angiogenesis via activation of SMAD4. Oncotarget. 2016.
- Flanders KC, Heger CD, Conway C, Tang B, Sato M, Dengler SL, Goldsmith PK, Hewitt SM, Wakefield LM. Brightfield proximity ligation assay reveals both canonical and mixed transforming growth factor-beta/bone morphogenetic protein Smad signaling complexes in tissue sections. J Histochem Cytochem. 2014;62:846–63.