ABSTRACT
Multidrug resistance (MDR) represents a major hindrance to the efficacy of cancer chemotherapeutics. While surgical resection, radiation, and chemotherapy can be used to reduce tumor size, the subsequent appearance of drug resistant cells is a frequent problem. One of the main contributors to the development of MDR is increased expression of multi-drug resistant protein 1 (MDR1), also known as P-glycoprotein (P-gp). P-gp is a membrane-associated efflux pump that can efficiently remove internalized taxane-base chemotherapeutics thus preventing drug accumulation and maintaining cellular viability. Consequently, investigation into the molecular mechanisms responsible for regulation of P-gp expression is necessary to facilitate treatment of MDR tumors. Using molecular and biochemical approaches, we identified that the micro-RNA, miRNA149, contributes to the development of MDR within malignant mesothelioma cells by regulating the expression of MDR1.
Introduction
Mesothelioma is a cancer arising from the mesothelium, which is a thin lining covering many types of internal organs. However, the overwhelming majority of mesothelioma originates within lung tissues.Citation1 This cancer type is often aggressive with a comparatively low response to chemotherapeutic interventions.Citation2 The most potent of which is the combination of cisplatin with pemetrexed though patient response has been shown to be less than fifty percent.Citation3 Consequently, other chemotherapeutic strategies have been investigated including the use of paclitaxel (Taxol).Citation4
Recent studies utilizing paclitaxel found that drug distribution is incredibly heterogeneous within the cancer tissue resulting in uneven efficacy.Citation5 While this in and of itself could be detrimental to chemotherapeutic interventions, it also provides cancer cells an opportunity to acquire chemotherapeutic resistances via exposure induced increases in drug effluxing membrane proteins.Citation6,Citation7 The first described and fully characterized drug efflux protein was P-glycoprotein (P-gp).Citation8,Citation9 Subsequent to this discovery, another distantly related ATP-binding cassette (ABC) transporter protein was identified, multidrug resistant protein 1 (MRP1).Citation10 P-gp and MRP1 have been observed in many human cancers, including mesothelioma, and have been associated with reductions in patient outcome and survival.Citation11 As such, understanding the molecular mechanisms that regulate efflux-related drug resistance is critical especially in an environment where drug delivery is not uniform.
MicroRNAs (miRNAs) are an abundant class of small non-coding RNAs approximately 23 nucleotides in length that are endogenously expressed in mammalian cells.Citation12 MiRNAs regulate gene expression post-transcriptionally by binding via partial complementation to the 3′ untranslated regions (3′UTR) of messenger RNAs (mRNAs), resulting in translational suppression and mRNA degradation.Citation10,Citation12-15 Since their discovery in C. elegans in the early 1990s, several miRNAs have been shown to affect cancer progression by regulating tumorigenicity, cell cycle, apoptosis, and metastasis.Citation16-18 Moreover, it is now suggested that alterations in miRNA expression profiles can prognosticate cancer development and indicate disease stage.Citation19-25 One of the first suggestions that miRNA could modulate drug resistance came from the observation that breast cancer patients with increased sensitivity to anthracycline treatment had a deletion in chromosome 11q that encoded miRNA miR125b.Citation26 The involvement of miRNAs in chemo-sensitivity and chemo-resistance was further corroborated by a systematic study showing a significant correlation between miRNA expression profiles and drug potency.Citation27 Since this report, drug resistance related miRNAs have gained attention due to the therapeutic potential. However, the precise role of miRNAs in the development of chemotherapeutic resistance in mesothelioma remains largely unexplored.
In this report, we investigated whether miRNAs can mediate mesothelioma resistance to the Paclitaxel analog Simotaxel. We first identified a group of differentially expressed miRNAs that mediate taxane resistance in a cellular model with acquired resistance. Then we demonstrated that miRNA149 plays an important role in regulating P-gp expression levels. Taken together our results suggest that miRNAs can modulate malignant mesothelioma taxane resistance.
Results
P-glycoprotein expression confers taxane resistance to malignant mesothelioma cells
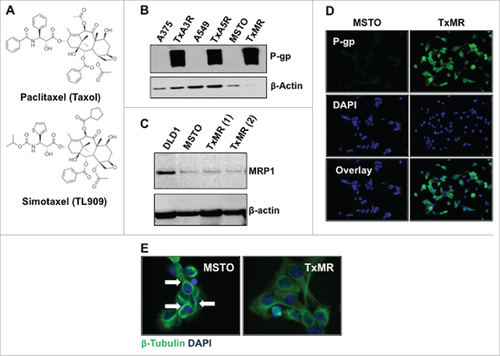
To begin our investigation we determined if exposing malignant mesothelioma cells (MSTO-211H) to increasing concentrations of the paclitaxel analog Simotaxel over time would confer drug resistance (). Briefly, cells were exposed to their IC50 (half maximal inhibitory concentration) until cell death stopped and surviving cells began to recover and divide. The drug concentration was then doubled and the process repeated to increase resistance. This was also conducted upon the A375 malignant mesothelioma cell line and the A549 lung carcinoma cell line to demonstrate reproducibility. Under constant exposure conditions, cells did indeed develop resistance. The resistant cell line, which we termed TxMR, was maintained in 25nM Simotaxel, and expressed high levels of the transmembrane transporter P-glycoprotein (P-gp), but not the related transporter, multidrug resistant protein 1 (MRP1) (). Moreover, TxMR cells did not display the typical tubulin bundling indicative of Simotaxel exposure (). To further demonstrate that P-gp overexpression was responsible for the development of taxane resistance, we sequenced the genes encoding β-tubulin (target molecule for taxanes) and ABCB1 (encodes P-gp). We found that no functional mutations were present within the TxMR cells. Taken together these results indicate that TxMR cell drug resistance is related to the up-regulation of P-gp expression.
Figure 1. P-gp expression confers resistance to Simotaxel in drug-selected cells. (A) Paclitaxel (Taxol) and Simotaxel structures. (B) Western blot analysis of 3 different sensitive and Simotaxel resistant cancer cell lines. Protein lysates were probed against P-gp (upper panel) and β-Actin (loading control, bottom panel) (C) Western blot indicating the levels of MRP1 in a control cells, DLD1 (Dukes' type C, colorectal adenocarcinoma, ATCC® CCL-221™), Parental sensitive MSTO cells and two independent clones of the resistant MSTO cells, TxMR. (D) Immunofluorecent staining of P-go in MSTO (upper panel) and TxMR cells (bottom panel). DAPI was used for nuclei staining. (E) Immunofluorescent staining of β-tubulin illustrates typical effects of taxane treatment upon the cellular microtubule (MT) network. MT bundling is observed the Simotaxel sensitive parental MSTO cells. However, this effect is not induced within resistant TmXR cells.
TxMR cells display differential microRNA expression profiles when compared to parental cells
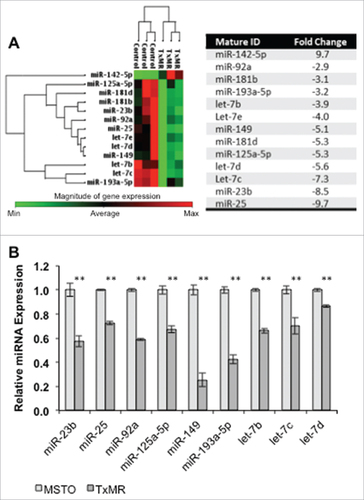
While it has been shown within other cancer models that P-gp related drug resistance could be induced in response to increasing chemotherapeutic treatment doses, the regulatory factors that modulate this overexpression remain elusive. As such, we wanted to investigate if microRNAs (miRNAs), which post-transcriptionally regulate protein expression, were also affected by Simotaxel treatment. We analyzed miRNA expression in our TxMR cells and parental MSTO cells using the RT2 miRNA PCR Arrays (SABioscences™, Qiagen®) consisting of a panel of over eighty (80) cancer related microRNAs. We found that 12 miRNAs displayed over 2-fold reduction in expression within the TxMR cells when compared to parental MSTO cells (). Changes in miRNA expression were independently validated via qPCR ().
Figure 2. Differential expression of miRNAs in TxMR. (A) Heat map with cluster analysis of miRNAs expression in MSTO vs TxMR was generated with results produced by RT2 miRNA PCR Arrays (SABioscences, Qiagen). We identified a group of thirteenCitation13 miRNAs with differential expression higher than 2-fold (with p-values < 0.05 by Student's t-test) in TxMR cells when compared to MSTO (left panel). A compiled list of their differential fold expression is shown (righ panel). (B) Independent validation in triplicate of the RT2 miRNA PCR array data using qPCR. The relative amount of each miRNA was normalized to U6 snRNA and the fold change for each miRNA from TxMR relative to MSTO was calculated using the 2−ΔΔCT method. ##p < 0.05.
Overexpression of miRNA149 down regulates P-gp protein levels
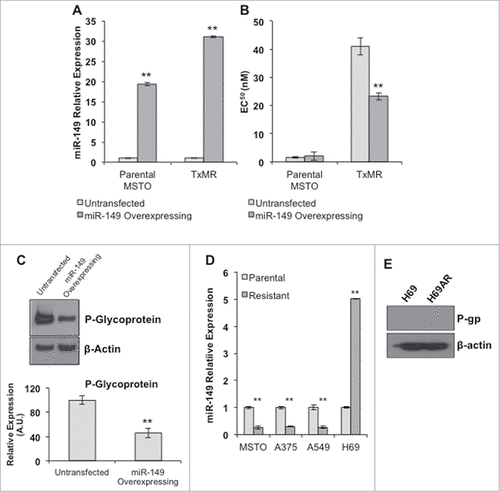
We next determined if overexpression of the downregulated miRNAs, particularly miRNA149, could rescue chemotherapeutic sensitivity within the TxMR cells. miRNA149 was of particular interest due to being highly down regulated, over 5- fold, and also because it has been implicated in chemo sensitivity. In addition, this miRNA was selected because of an in silico analysis using the TargetScanHuman algorithm [28] found miRNA149 as a potential regulator of P-gp. To address this question, TxMR and parental MSTO cells were stably transfected with pcDNA3.1+ containing the miRNA149 and subsequently selected for miRNA149 overexpressing cells using G418 (). We next utilized the MTT assay to determine if miR149 overexpression reduced resistance. As shows, increasing miR149 expression reduced overall resistance to Simotaxel in the TxMR. Unsurprisingly, no difference was observed within the parental MSTO cells since P-gp expression was unaltered. We then determined if reductions in TxMR cell resistance was indeed related to decreases in P-gp. Western blot analysis showed that miRNA149 transfection did reduce P-gp protein expression by over 50 percent (). Finally, we determined if this relationship extended to other resistant cell types. As such, we examined three additional cell lines, A375, A549, and H69. Quantitative PCR revealed that miR149 was down regulated in all P-gp overexpression cell lines but not in resistant H69 (). While the A375 and A549 cell lines developed resistance by increasing P-gp expression in a similar fashion to our TxMR cells (), H69 cells were determined to attain drug-resistance through increased expression of multi-drug resistance protein 1 (MRP1) and not P-gp ().Citation28 Taken together, these results indicate that miRNA149 downregulation plays a critical role in P-gp induced drug resistance.
Figure 3. MiR149 regulates P-glycoprotein expression, which impacts drug resistance. (A) MiRNA149 expression within stably transfected parental MSTO and TxMR cells was determined via qPCR. Results were normalized to U6 and compared to un-transfected cells. (B) MTT analysis was performed on transfected and un-transfected cells. Overexpression of miRNA149 greatly increased TxMR sensitivity to Simotaxel. No change in sensitivity was observed within parental MSTO cells. (C) Western blot analyses revealed that overexpression of miRNA149 significantly reduced P-gp expression within TxMR cells. (D) Quantitative PCR was performed to determine miRNA149 expression within additional Simotaxel resistant cells. Cells expressing high levels of P-gp (resistant MSTO, A375, A549) displayed significantly reduced levels of miRNA149 when compared to parental controls. Resistant H69 cells (H69AR), which express MRP1 and not P-gp, displayed increased levels of miRNA149. (E) Western blot indicating the levels of P-gp levels in the H69 and H69AR cancer cell lines. β-actin was used as loading control. ##P < 0.05.
Discussion
A major hurdle to chemotherapeutic treatment is the development of drug resistance. While this may be overcome by changes in patient treatment strategies, for instance inclusion of alternative chemotherapeutics or a wholesale change to radiotherapy, the risk for cancer cells to develop additional resistances remains. Consequently, understanding the means by which drug resistance develops is critical to the promotions of successful patient outcomes. In this study we focused on the role miRNAs may play in the development of drug resistance via overexpression of the transmembrane pump P-glycoprotein and whether they can confer resistance to a variety of chemotherapeutics. Using malignant mesothelioma cells that have been developed experimentally to acquire Simotaxel resistance we found that miRNA149 is a potential regulator of P-gp overexpression.
miRNA149 has been identified as a potent tumor suppressor within a variety of cancer types including breast, laryngeal squamous cell carcinoma, and glioblastoma.Citation29-31 In most of these cancer types the suppressive ability seems to be linked to inhibition of proliferation where reduced levels of miRNA149 leads to unchecked proliferation. Moreover, increasing miRNA149 expression can induce apoptosis in both neuroblastoma and HeLa cells by inhibiting RAC-alpha serine/threonine-protein kinase 1 (Akt1) and Transcription factor E2F1 expression.Citation32 In regards to lung cancer progression, miRNA149 can block epithelial-to-mesenchymal transition (EMT) of non-small-cell lung cancer cells (NSCLC cells) via suppression of Forkhead box M1 (FOXM1).Citation33 Consequently, miRNA149 has a diverse role in the preventing cancer progression though it is this miRNA's ability to modulate chemo resistance that is of interest.
In relation to drug resistance, a similar study utilizing the A549 cell line with Cisplatin, a chemotherapeutic that induces apoptosis in proliferating cells via induction of DNA crosslinking, demonstrated that resistance cells increased expression of miRNA149.Citation34 Moreover, investigations into oral squamous cell carcinoma demonstrated that miR149 is also increased within Cisplatin resistant cells.Citation35 However, other studies utilizing Paclitaxel, the parental compound of Simotaxel, and Adriamycin (Doxorubicin) show that resistance correlated with decreased miRNA149 expression.Citation36,Citation37 It should be noted that these studies indicated that miRNA149 modulated resistance through MyD88 and GlcNAc N-deactylase/N-sulfotransferase-1 (NDST1) respectively. Consequently, it would seem that miR149 expression is chemotherapeutic dependent. One explanation for the observed difference in miRNA149 expression between Cisplatin resistance cells and those resistant to other chemotherapeutics is that Cisplatin resistance is not dependent upon P-glycoprotein expression.Citation38 Conversely, resistance to Paclitaxel, Simotaxel and Adriamycin is P-gp dependent.Citation39,Citation40 Moreover, it was observed that Cisplatin resistance induced down-regulation of P-gp within lung cancer cell lines.Citation41 It would seem then that the mechanism by which resistance is acquired, influences whether miRNA149 is up- or down-regulated. This supports the observation that Cisplatin resistant tumors remain sensitive to Paclitaxel since Cisplatin-induced up-regulation of miRNA149 would down-regulate P-gp maintaining Paclitaxel sensitivity.Citation42
In conclusion, our results demonstrate miRNA149 directly influences P-gp overexpression supporting the notion that this miRNA is not only a tumor suppressor but also a chemo-modulator. Moreover, the suggested dependence upon how resistance develops makes miRNA149 an ideal candidate for identifying alternative therapeutic interventions and prognostication.
Materials and methods
Cell lines and culture
The human cancer cell lines used in this report were obtained from the American Type Culture Collection (ATCC). The human mesothelioma cell line MSTO-211H (CRL-2081), the human malignant melanoma cell line A-375 (CRL-1619) and the human carcinoma lung cell lines: A549 (CCL-185), H69 (ATCC HTB-119) and H69AR (CRL-11351) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (Invitrogen). Cells were propagated according to ATCC guidelines and maintained in a 37°C incubator with 5% CO2 atmosphere. Resistant mesothelioma cells (TxMR), malignant melanoma cells (TxA3R) and carcinoma lung cells (TxA5R) were established by exposing cells to a concentration of the Paclitaxel analog, Simotaxel, equal to their respective IC50 until cell death stopped and surviving cells began to recover and divide. The concentration was then doubled and the process repeated to increase resistance. After 18 months the TxMR, dubbed TXSimR cells were resistant and kept in 25 nM of Simotaxel. After 8 months A-375 cells were established to be resistant to 32 nM Simotaxel and after 6 months A549 cells were established to be resistant to 4 nM Simotaxel.
Stably expressing miRNA cell lines were established by transfecting MSTO and TXMR cells with pcDNA 3.1+ plasmids expressing the miRNA 149. Cells were then selected with G418 (Sigma Aldrich, St. Louis, MO). Sensitive cells were maintained in the absence of Simotaxel while resistant cells were propagated in the presence of the drug. Cells were maintained in RPMI supplemented with 10% FBS and penicillin/streptomycin in at 37°C in 5% CO2 incubator.
Western blotting
At appropriate time points cells were harvested, by centrifugation, washed in cold Phosphate Saline Buffer (PBS), and protein lysate was generated using EDTA buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 5% β-ME, 10 mM EDTA). Samples were run on NuPAGE 4–12% Bis-Tris Gels (Life Technologies, Carlsbad, CA) and transferred onto PDVF membranes (GE Healthcare, Chicago, IL). After blocking in 5% milk in PBS with 0.05% Tween (PBS-T) for 30 minutes at room temperature, membranes were probed against P-glycoprotein (SC-13131, Santa Cruz Biotechonology, Inc., Dallas, TX) and β-actin (A5316, Sigma-Aldrich, Saint Louis, MO), which was used as a loading control overnight at 4°C in 1% milk in PBS-T. Specific secondary HRP-conjugated antibodies were used at 1:2500 dilution (Santa Cruz Biotechonology, Inc., Dallas, TX). Blots were then developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Inc., Waltham, MA) and exposed to x-ray film.
Immunocytochemistry
Microtubule bundling via fluorescent microscopy was performed using immunocytochemistry analysis: Cells in complete tissue culture medium were added to wells of slide chambers and allowed to grow and attach overnight in 4-chamber slides. Drug was added at specified concentration and cells incubated for one doubling time (usually 18 to 22 hours). Cells were fixed in a phosphate buffered saline (PBS) solution containing 10% Formaldehyde/ 3% sucrose for 10 minutes at room temperature. Cells were washed in PBS and permeabilized in 2% triton X-100/PBS solution for 10 min at room temperature, incubated with a 1:1000 dilution of mouse anti-β tubulin (Sigma cat# T5168) for 60 min at 37°C, followed by incubation with 1:1000 dilution of FITC conjugated goat anti-mouse antibody (Sigma cat# F4018) for 60 min at 37°C. DAPI (4′-6-Diamidino-2-phenylindole) solution was added and incubated at room temp for 2 min. Slides were then washed with PBS and allowed to air dry. Cover slips were mounted to slides and the slides were examined using fluorescence microscopy.
MTT cell proliferation assay
Half-maximal inhibitory concentration (IC50) was determined using the metabolic dimethyl thiazolyl diphenyl tetrazolium salt (MTT) assay, which measures conversion of MTT into formazan by mitochondrial succinate dehydrogenase and other oxidoreductases [26]. 2.5 × 103 cells/well were seeded into 96-well plates and allowed to reach 70–80% confluence. Serial dilutions of Simotaxel, synthesized by Dr. Robert A. Holton's laboratory (Florida State University, Tallahassee, FL), were then used to treat cells. After 72 hrs, MTT (M5655, Sigma) was added for a final concentration of 1 mg/mL. Plates were incubated for 60 mins and then centrifuged. Supernatant was removed and 200 μL DMSO added to each well. After 6 mins of shaking, absorbance was measured in a Synergy HT plate reader (Biotek, Winooski, VT) at 600 nm.
Quantitative real time PCR
To gain insights into possible miRNAs related mechanisms for this acquired MDR resistance phenotype we profiled the expression of over eighty (80) cancer related miRNAs between the MSTO and TxMR cells. RNAs from these cells, in three independent replicates, were extracted using RNA-Bee reagent (Tel-Test, Inc) and enhanced for small RNA molecules using miRNeasy kit from Qiagen. cDNAs were synthesized using RT2 miRNA First Strand Kit from Qiagen and used as templates for qPCR on RT2 miRNA PCR Arrays (SABioscences™, Qiagen). We performed independent validation in triplicate of the RT2 miRNA PCR array data using qPCR with TaqMan probes (Applied Biosystems, Life Technologies). Synthesis of cDNA in this case was done using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Individual miRNA levels were confirmed using TaqMan probes (Applied Biosystems). The relative amount of each miRNA was normalized to U6snRNA and the fold change for each miRNA from TxMR relative to MSTO was calculated using the 2-ΔΔCT method.Citation43
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors acknowledge and thank Dr. Robert A. Holton (Florida State University Research Foundation, Tallahassee, FL) for supporting this study and providing Paclitaxel and its analog Simotaxel. DARZ was partially supported by Genentech Grant G-47608 while submitting this manuscript.
Additional information
Funding
References
- Robinson BM. Malignant pleural mesothelioma: an epidemiological perspective. Ann Cardiothorac Surg. 2012 Nov;1(4):491–6.
- Bech C, Sørensen JB. Chemotherapy induced pathologic complete response in malignant pleural mesothelioma: a review and case report. J Thorac Oncol. 2010 May;5(5):735–40. doi.org/10.1097/JTO.0b013e3181d86ea9.
- Vogelzang NJ, Rusthoven JJ, Symanowski J, Denham C, Kaukel E, Ruffie P, Gatzemeier U, Boyer M, Emri S, Manegold C, et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J Clin Oncol. 2003 Jul 15;21(14):2636–44. doi.org/10.1200/JCO.2003.11.136.
- Kanai O, Fujita K, Nakatani K, Mio T. Repetitive responses to nanoparticle albumin‐bound paclitaxel and carboplatin in malignant pleural mesothelioma. Respirol Case Rep. 2016 Mar;4(1):28–31.
- Giordano S, Zucchetti M, Decio A, Cesca M, Fuso Nerini I, Maiezza M, Ferrari M, Licandro SA, Frapolli R, Giavazzi R, et al. Heterogeneity of paclitaxel distribution in different tumor models assessed by MALDI mass spectrometry imaging. Sci Rep. 2016 Dec 21;6:39284. doi.org/10.1038/srep39284.
- Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–85. doi.org/10.1038/sj.onc.1206948.
- Leslie EM, Deeley RG, Cole SP. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol Appl Pharmacol. 2005;204:216–37. doi.org/10.1016/j.taap.2004.10.012.
- Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–9. doi.org/10.1016/0092-8674(86)90595-7.
- Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–398. doi.org/10.1146/annurev.pharmtox.39.1.361.
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNA. Genome Res. 2009;19:92–105. doi.org/10.1101/gr.082701.108.
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi.org/10.1038/nrc706.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi.org/10.1016/S0092-8674(04)00045-5.
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi.org/10.1101/gad.1399806.
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104:9667–72. doi.org/10.1073/pnas.0703820104.
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi.org/10.1126/science.1149460.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi.org/10.1016/0092-8674(93)90529-Y.
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi.org/10.1038/nrg2634.
- Hammond SM. RNAi, microRNAs and human diseases. Cancer Chemother Pharmacol. 2006;58:s63–8. doi.org/10.1007/s00280-006-0318-2.
- Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–56. doi.org/10.1200/JCO.2009.24.0317.
- Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–41. doi.org/10.1158/1078-0432.CCR-09-1736.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi.org/10.1038/nature03702.
- Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293–300. doi.org/10.1089/dna.2006.0554.
- Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, Mitchell JB. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4:e6377. doi.org/10.1371/journal.pone.0006377.
- Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi.org/10.1038/nrc1840.
- Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–92. doi.org/10.4161/cc.7.16.6453.
- Climent J, Dimitrow P, Fridlyand J, Palacios J, Siebert R, Albertson DG, Gray J W, Pinkel D, Lluch A, Martinez-Climent JA. Deletion of chromosome 11q predicts response to anthracycline-based chemotherapy in early breast cancer. Cancer Res. 2007;67:818–26. doi.org/10.1158/0008-5472.CAN-06-3307.
- Blower PE, Verducci JS, Lin S, Zhou J, Chung JH, Dai Z, Liu CG, Reinhold W, Lorenzi PL, Kaldjian EP, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6:1483–91. doi.org/10.1158/1535-7163.MCT-07-0009.
- Mirski SE, Cole SP. Antigens associated with multidrug resistance in H69AR, a small cell lung cancer cell line. Cancer Res. 1989;49:5719–24.
- Bischoff A, Huck B, Keller B, Strotbek M, Schmid S, Boerries M, Busch H, Müller D, Olayioye MA. miR149 functions as a tumor suppressor by controlling breast epithelial cell migration and invasion. Cancer Res. 2014 Sep 15;74(18):5256–65. doi.org/10.1158/0008-5472.CAN-13-3319.
- Xu Y, Lin YP, Yang D, Zhang G, Zhou HF. Clinical Significance of miRNA149 in the Survival of Patients with Laryngeal Squamous Cell Carcinoma. Biomed Res Int. 2016;2016:8561251. doi.org/10.1155/2016/8561251.
- Ghasemi A, Fallah S, Ansari M. MicroRNA-149 is epigenetically silenced tumor-suppressive microRNA, involved in cell proliferation and downregulation of AKT1 and cyclin D1 in human glioblastoma multiforme. Biochem Cell Biol. 2016 Dec;94(6):569–76. doi.org/10.1139/bcb-2015-0064.
- Lin RJ, Lin YC, Yu AL. miRNA149# induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol Carcinog. 2010 Aug;49(8):719–27.
- Ke Y, Zhao W, Xiong J, Cao R. miRNA149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochem Res Int. 2013;2013:506731. doi.org/10.1155/2013/506731.
- Wang Q, Zhong M, Liu W, Li J, Huang J, Zheng L. Alterations of microRNAs in cisplatin-resistant human non-small cell lung cancer cells (A549/DDP). Exp Lung Res. 2011 Sep;37(7):427–34. doi.org/10.3109/01902148.2011.584263.
- Ghosh RD, Ghuwalewala S, Das P, Mandloi S, Alam SK, Chakraborty J, Sarkar S, Chakrabarti S, Panda CK, Roychoudhury S. MicroRNA profiling of cisplatin-resistant oral squamous cell carcinoma cell lines enriched with cancer-stem-cell-like and epithelial-mesenchymal transition-type features. Sci Rep. 2016 Apr 5;6:23932. doi.org/10.1038/srep23932.
- Zhan Y, Xiang F, Wu R, Xu J, Ni Z, Jiang J, Kang X. MiRNA-149 modulates chemosensitivity of ovarian cancer A2780 cells to paclitaxel by targeting MyD88. J Ovarian Res. 2015 Jul 30;8:48. doi.org/10.1186/s13048-015-0178-7.
- He DX, Gu XT, Li YR, Jiang L, Jin J, Ma X. Methylation-regulated miRNA149 modulates chemoresistance by targeting GlcNAc N-deacetylase/N-sulfotransferase-1 in human breast cancer. FEBS J. 2014 Oct;281(20):4718–30. doi.org/10.1111/febs.13012.
- Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi.org/10.1146/annurev.pharmtox.48.080907.180426.
- Pires MM, Emmert D, Hrycyna CA, Chmielewski J. Inhibition of P-glycoprotein-mediated paclitaxel resistance by reversibly linked quinine homodimers. Mol Pharmacol. 2009 Jan;75(1):92–100. doi.org/10.1124/mol.108.050492.
- Bao L, Haque A, Jackson K, Hazari S, Moroz K, Jetly R, Dash S. Increased expression of P-glycoprotein is associated with doxorubicin chemoresistance in the metastatic 4T1 breast cancer model. Am J Pathol. 2011 Feb;178(2):838–52. doi.org/10.1016/j.ajpath.2010.10.029.
- Wang J, Wang H, Zhao L, Fan S, Yang Z, Gao F, Chen L, Xiao GG, Molnár J, Wang Q. Down-regulation of P-glycoprotein is associated with resistance to cisplatin and VP-16 in human lung cancer cell lines. Anticancer Res. 2010 Sep;30(9):3593–8.
- Stordal B, Pavlakis N, Davey R. Treating cisplatin-resistant cancer: A systematic analysis of oxaliplatin or paclitaxel salvage chemotherapy. In: Bonetti A, Leone R, Muggia FM, Howell SB, editors. Platinum and other heavy metal compounds in cancer chemotherapy. Cancer drug discovery and development. Totowa (NJ): Humana Press; 2009.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi.org/10.1006/meth.2001.1262.
