ABSTRACT
Osteosarcoma (OS) is one of the most common primary bone tumors and has a high disablity rate and case-fatality rate. The protracted stagnancy of the chemotherapy program and surgical technology for OS treatment prompted us to focus on the mechanisms of cancer carcinogenesis progression in OS.
Nucleoside diphosphate kinase B (NME2) is a type of nucleoside diphosphate kinase that plays an important role in cellular processes. In this study, we report overexpression of NME2 in OS cell lines and correlate this overexpression with the clinicopathologic features of osteosarcoma. We used si-NME2 to downregulate expression of NME2 in OS cell lines. The results of the CCK8 and clone forming assays show that NME2 promotes OS cell line proliferation. Western blot assays show that deregulation of NME2 results in enhanced the expression of c-Myc, which promotes OS proliferation.
KEYWORDS:
Introduction
Osteosarcoma (OS) is one of the most common primary bone tumors. Its incidence in the population is only 3 per one million.Citation1 Although the incidence of OS is very low compared to lung cancer and breast cancer, tumor-related death and the rate of amputation is very high in the pediatric age group.Citation2 With early metastasis and a powerful ability to proliferate, OS has become a troublesome disease for orthopedicians. The stagnancy of the chemotherapy program and surgical technology in OS treatment spurred us to focus on the mechanisms of cancer progression in OS.
NME2 was initially considered a metastasis suppressor akin to NME1.Citation3 In a previous study, NME2 was located in the cytoplasm and nucleus;Citation4 it could bind to DNA and act as a transcription factor.Citation5 It was also reported to have a major role in the synthesis of nucleoside triphosphates.Citation6 With more research on NME2, it was found that NME2 also played a very important role in tumorigenesis. In studies on hepatocellular carcinoma (HCC), the overexpression of NME2 was found to promote the proliferation of HCC cell linesCitation7. In lung and cervical cancer, NME2 also plays an positive role in cell proliferation and promotes tumor growth.Citation8 However, the role of NME2 in osteosarcoma is still unknown.
The proto-oncogene c-MYC is a transforming member of the Myc family. It has 3 exons and 2 introns.Citation9 In many cancers, the deregulation of c-Myc is the key to tumorigenesis. The overexpression of c-Myc plays an important role in many cellular processes, such as proliferation,Citation10 the cell cycle,Citation11 differentiation and apoptosis.Citation12 c-Myc is also a downstream protein in many tumor related pathways. In Wnt/beta-catenin signaling, c-Myc is targeted by beta-catenin, which leads to abnormal proliferation and tumorigenesis.Citation13 In the Notch pathway, c-MYC targeting by NOTCH1 induces a series of transcriptional pathways promoting leukemic cell growth.Citation14 As a key of oncogene, c-MYC has been reported in many types of cancers, such as lung cancer,Citation15 breast cancerCitation16 and liver cancer.Citation17 In OS, c-MYC has been reported to be an important oncogene in OS cell proliferation.Citation18
In this study, we report that NME2 is an oncogene in OS that is associated with its clinicopathologic features. Downregulation of NME2 could reduce the proliferation of OS cells and arrest the OS cell cycle in the G2/M phase. The deregulation of NME2 enhanced the expression of c-Myc and affected the proliferation of osteosarcoma.
Results
1. NME2 is overexpressed in OS cell lines.
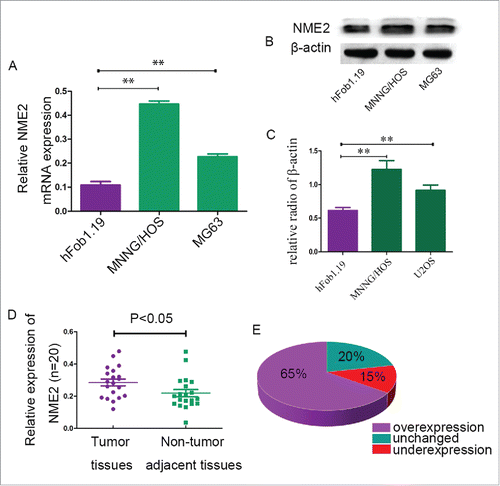
RT-PCR technology was used to detect the mRNA expression of NME2 in human immortalized osteoblast cells (hFob1.19) and osteosarcoma cell lines (MNNG/HOS and MG63). The results show that the mRNA of NME2 was overexpressed in OS cell lines by more than a factor of two than in hFob1.19 cells (). Western blot assays showed that NME2 protein expression was higher in MNNG/HOS and MG63 cells than in hFob1.19 cells (). We collected 20 pairs of OS primary frozen tissues and corresponding non-tumor tissues. Meanwhile we detected mRNA expression in 20 sets of fresh tumor tissues and corresponding non-tumor tissues from patients. The NME2 mRNA expression was higher in tumor tissues than in non-adjacent tissues in 65 percent of the patients. In 20 percent of the patients’ tumor tissues, NME2 mRNA expression was unchanged compared to the corresponding non-adjacent tissues. Meanwhile, in 15 percent of the patients, NME2 mRNA expression was lower than in the corresponding non-adjacent tissues (). Student's t-test showed that the NME2 mRNA expression levels in tumor tissues was significantly higher than in the corresponding adjacent non-tumor tissues (P < 0.05).
Figure 1. Compared to the control group, NME2 expression was high in OS cell lines and clinical OS tumor tissues. (A) RT-PCR confirmed that the mRNA expression of NME2 was significantly higher in MNNG/HOS and MG63 cell lines than in hFob1.19 cell lines. (B) NME2 protein was overexpressed in MNNG/HOS and MG63 cell lines compared to hFob1.19. (C) The mRNA expression levels of NME2 in clinical tumor tissues were significantly higher than in the corresponding adjacent non-tumor tissues. (D) RT-PCR demonstrated that the NME2 expression in tumor tissues was 65% higher than in non-tumor tissues.
2. NME2 promotes the proliferation of OS cell lines.
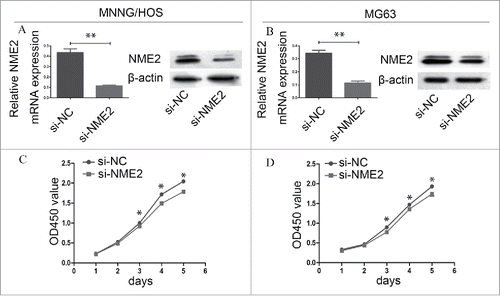
Proliferation is reduced in si-NME2 targeted MNNG/HOS and MG63 cells. We transfected si-NC and si-NME2 into MNNG/HOS and MG63 cells. RT-PCR confirmed that si-NME2 effectively silenced NME2 gene expression in MNNG/HOS and MG63 cells. The NME2 mRNA expression in MNNG/HOS and MG63 was significantly decreased after the transfection of si-NME2 compared with the transfection of si-NC (P < 0.05) (). The expression of NME2 protein was also significantly reduced in si-NME2 targeted MNNG/HOS and MG63 cells (). The CCK8 assay showed that the proliferation ability of si-NME2 targeted OS cells was weakened more than that of si-NC targeted OS cells. In the 3rd day, the mean value of absorbance on si-NC targeted MNNG/HOS and MG63 cells was 0.998 ± 0.022 and 0.896 ± 0.033. On the si-NME2 targeted MNNG/HOS and MG63 cells these value of absorbance was 0.924 ± 0.014 and 0.774 ± 0.024. Error bars represent Standard Deviation. Student's t-test confirmed that significant differences appeared from the 3rd day between si-NC targeted cells and si-NME2 targeted cells in the CCK8 assay (P < 0.05) ().
Figure 2. The downregulation of NME2 expression in osteosarcoma cell lines affects cell proliferation. (A) Compared to si-NC targeted MNNG/HOS cells, NME2 expression was significantly downregulated in si-NME2 targeted MNNG/HOS cells at the mRNA and protein levels. (B) Compared to si-NC targeted MG63 cells, NME2 expression at the mRNA and protein levels was significantly downregulated in the si-NME2 targeted MG63 cells.(C) The proliferation of si-NC targeted MNNG/HOS cells was significantly higher than that of si-NME2 transfected MNNG/HOS cells from the 3rd day. (D) The proliferation of MG63 cells was significantly lower from the 3rd day onwards when transfected with si-NME2 compared to those transfected with si-NC.
3. Downregulation of NME2 could arrest the OS cell cycle in the G2/M phase.
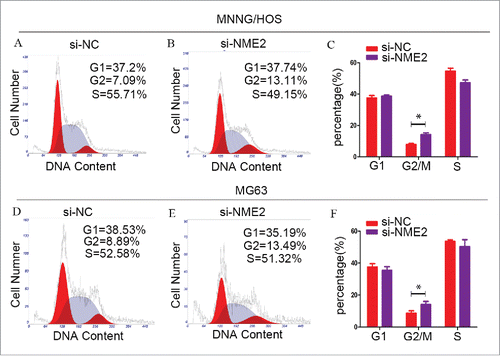
In the MNNG/HOS cell line, when we used si-NME2 to downregulate NME2 expression, the percentage of G2/M phase cells increased from 7.84% ± 0.650% to 14.183% ± 1.060% (). In the MG63 cell line, when NME2 expression is decreased, the percentage of G2 phase cells increases. The percentage of si-NME2 targeted MG63 cells increased from 8.747% ± 1.43% to 14.12% ± 1.962% () (P < 0.05). Error bars represent Standard Deviation. These data demonstrated that the downregulation of NME2 expression could arrest the OS cell cycle in the G2/M phase. NME2 affects the OS cell cycle in the G2/M phase.
Figure 3. Cells were significantly arrested in the G2/M phase, when the expression of NME2 was knocked down in MNNG/HOS and MG63 cells. (A) In the si-NC transfected MNNG/HOS cell line, the cell cycle assay showed that 37.51% ± 1.57% of the cells were in the G1 phase, 7.84% ± 0.65% of the cells were in the G2 phase and 54.65% ± 1.81% of the cells were in the S phase. (B) In the si-NME2 transfected MNNG/HOS cell line, the cell cycle assay showed that 38.61% ± 0.76% of the cells were in the G1 phase, 14.18% ± 1.06% of the cells were in the G2 phase and 47.20% ± 1.78% of the cells were in the S phase. (C) The cell cycle was arrested in the G2/M phase after the transfection of si-NME2 in the MNNG/HOS cell line. (D) In the si-NC transfected MG63 cell line, the cell cycle assay showed that 37.64% ± 2.07% of the cells were in the G1 phase, 8.75% ± 1.43% of the cells were in the G2 phase and 53.61% ± 1.03% of the cells were in the S phase. (E) In the si-NME2 transfected MG63 cell line, the cell cycle assay showed that 35.48% ± 2.36% of the cells were in the G1 phase, 14.12% ± 1.96% of the cells were in the G2 phase and 50.40% ± 4.31% of the cells were in the S phase. (F) The cell cycle was arrested in the G2/M phase after the transfection of si-NME2 in the MG63 cell line. Error bars represent Standard Deviation.
4. NME2 expression was related to MNNG/HOS and MG63 clone formation.
The clone forming assay was used to detect the proliferation ability of MNNG/HOS and MG63 cells. The number of cell colonies was reduced after transfection with si-NME2 (). Compared with the control group, knockdown NME2 in MNNG/HOS and MG63 cells attenuated the proliferation ability of the cells.
Figure 4. The clone forming assay showed that the number of colonies was significantly reduced when the expression of NME2 was knocked down in the MNNG/HOS (A) and MG63 (B) cell lines. (C) NME2 expression in adjacent non-tumor tissues (D) NME2 expression in tumor tissues (E) c-Myc expression in adjacent non-tumor tissues (F) c-Myc expression in tumor tissues. The tumor cells show cytoplasmic staining. All images were captured at 200 × magnification.
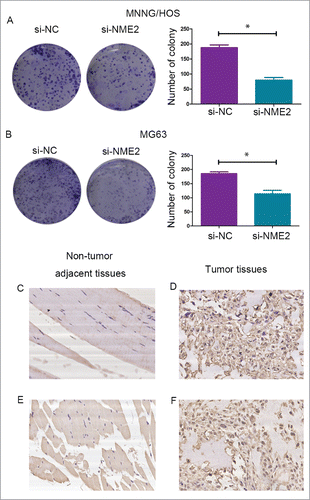
5. Overexpression of NME2 in OS is correlated with the patients’ clinicobiological features.
Immunohistochemical analysis of NME2 expression was performed in 50 sets of tumor and corresponding non-tumor tissues from patients. Tumor tissues had a high positive rate IHC of NME2 compare to correspond adjacent no-tumor tissues. The results are shown in . NME2 expression was correlated to tumor size, Ennecking stage and c-Myc expression (P < 0.05). The age, gender, and pathogenic size had no evident correlation to NME2 expression (). The IHC staining in representative tissues are shown in (). The tumor cells show cytoplasmic staining. All images were captured at 200 × magnification.NME2 maybe a crucial oncogene in osteosarcoma.
Table 1. Relationship between NME2 IHC results and clinical information for 50 OS patients.
6. NME2 could promote c-MYC expression to affect the proliferation of OS.
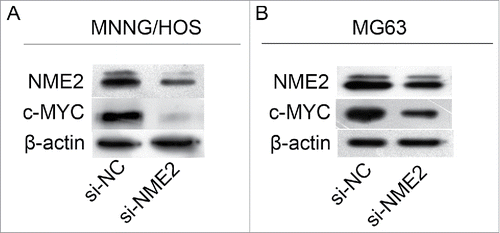
NME2 has been reported as a transcription factor of c-MYC.Citation5 We used si-NME2 to knockdown NME2 expression in the MNNG/HOS and MG63 cell lines. The expression of c-Myc was also reduced with reduced NME2 expression (). Therefore, c-Myc may be downstream of NME2 in MNNG/HOS and MG63 cell lines.
Discussion
Osteosarcoma is a malignant bone tumor that is perplexing to many orthopedicians. Currently, the mainstay of osteosarcoma therapy is still chemotherapy and en bloc resection. Accompanying extensive limb salvage surgery has been carried out in many hospitals. However, the 5 year event-free survival of OS patients was not very promising.Citation19 Targeted therapy has been used successfully in lung cancer and breast cancer. Therefore, understanding the mechanism of OS development may be helpful in developing OS targeted therapies.
NME2 is a member of the NDPK family that is reported to play a role in many cellular processes. Previous studies have shown that NME2 could act as a transcription factor,Citation5 bind DNACitation20 and act as a nucleoside diphosphate kinase.Citation21 NME2 plays a role in diverse biological processes. Wieland et al. have reported that NME2 could regulate the synthesis of cAMP in cardiomyocytes.Citation22 Braun et al. have reported that NME2 could promote proliferation, decrease apoptosis and positively regulate the differentiation of keratinocyte cell lines.Citation23 NME2 was initially deemed a metastasis suppresser akin to NME1. With further studies, it was found that NME2 is closely associated with many cancer-related processes. Unlike its homolog NME1, NME2 was discovered as a transcription factor of the well-known oncogene c-MYC, which is involved in determining many cancer cell fates.Citation24
This article is the first to report that NME2 functions as an oncogene in osteosarcoma. Compared to the control group, NME2 was upregulated in OS cell lines and OS patients’ tumor tissues. The overexpression of NME2 was also reported in many other cancer cell lines and cancer tissues. In the HeLa and tumor cell proliferation.Citation25 In chronic myeloid leukemia, NME2 was overexpressed, and this was linked to Bcr-Abl activity as a feature of the disease.Citation26 In giant cell tumors, NME2 overexpression at the protein level was substantiated by immunohistochemical and Western blot analysis.Citation27 These data suggest that NME2 may also play an important role in OS.
To determine the role of NME2 in OS, we used si-RNA to knockdown NME2 expression in OS cell lines. CCK-8 experiments and the clone forming assay showed that the downregulation of NME2 could reduce the ability of MNNG/HOS and MG63 cells to proliferate. In a previous study, NME2 positively regulated epithelial cell proliferation. Similar results were also demonstrated colorectal cancer,Citation28 cervical cancerCitation8 and liver cancer.Citation25 The cell cycle assay affirmed that knocking down the expression of NME2 could arrest MNNG/HOS and MG63 cells in the G2/M phase. To explore the relationship between the high expression of NME2 and the clinical features of OS, we used 50 sets of tumor and corresponding non-tumor tissues from OS patients to determine the expression of NME2 protein. The IHC assay demonstrated that NME2 was correlated with the OS tumor size and Enneking stage. In summary, NME2 is a crucial oncogene in OS. Some studies have shown that NME2, as a DNA binding protein, could bind to the c-MYC gene promoter to activate c-MYC transcription.Citation29 In subsequent research, NME2 was shown to interact with the G4 motif within the nuclease hypersensitive element in the c-MYC promoter.Citation30 To investigate the mechanism of OS cell proliferation induced by NME2, we knocked down the expression of NME2 and used WB to detect changes in the expression levels of c-Myc. The results show that when NME2 was knocked down in the MNNG/HOS and MG63 cell lines, c-Myc expression was also downregulated. Therefore, in OS cell lines, the expression of NME2 was correlated with that of c-Myc.
In previous studies, c-MYC has been characterized as an oncogene that is correlated with cell proliferation,Citation31 differentiationCitation32 and metastasis.Citation33 C-Myc is a downstream targeted protein in many types of proliferation and metastasis linked signaling pathways, such as the Wnt pathway,Citation13 Akt signalingCitation34 and Notch signaling.Citation14 In human non-small-cell lung carcinoma, the overexpression of c-Myc enhances tumor growth.Citation8 In breast cancers, c-Myc has been shown to promote the proliferation of cancer cellsCitation16. These results were confirmed in several cancers including osteosarcoma. Overall, we conclude that NME2 is an OS associated protein that promotes OS cell proliferation by enhancing the expression of c-Myc. Nowadays many oncogene inhibitors has been used in clinical research. CDK4 inhibitor palbociclib was used in breast cancer and resulted in longer progression-free survival.Citation35 In giant cell tumor, RANKL inhibitor denosumab also achieved good treatment effect.Citation36 Claire Bouvard et al. have reported that stauprimide as a staurosporine analog could suppresses c-Myc transcription by inhibiting NME2 nuclear translocation. In xenograft mouse models stauprimide could also inhibits tumor growth.Citation37 According to our research, NME2 may be a new therapeutic target for osteosarcoma.
Materials and methods
Cell culture
The human fetal osteoblast cell line hFob1.19 and the osteosarcoma cell lines MNNG/HOS and MG63 were provided by the Shanghai Institutes of Biological Sciences (CAS). The hFob1.19 cells were cultured in Dulbecco's modified Eagle's medium/Nutrient Mixture F-12 (DMEM/F12) with 3 mg/L G418, 10% fetal calf serum and 1% penicillin-streptomycin at 34°C with 5% CO2. MNNG/HOS and MG63 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum and 1% penicillin-streptomycin at 37°C with 5% CO2.
Transfection
Cells were cultured in 6-well plates in an incubator. After cell growth, when 50% of the space in a well was covered, the cells were subjected to starvation, transfected with 50 nM siRNA using 5 μl Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA), and incubated for 6 hours. Next, the culture medium was discarded, and DMEM with 10% fetal calf serum and 1% penicillin-streptomycin was added. After 48 hours, we collected the transfected cells for subsequent experiments. The sequence of the si-RNA (si-NME2) is as follows: 5’GCT TCG AGC AGA AGG GAT T 3’.
Real-time PCR
Total RNA was extracted from hFob1.19, MNNG/HOS, and MG63 cell lines using TRIzol (Invitrogen, Carlsbad, CA, USA). One milliliter of Trizol reagent was added per 5 × 106 cells, and total RNA was extracted according to the manufacturer's instructions. The reverse transcription mixture contained cDNA, primers, SYBR Green 1 (TaKaRa Bio, Inc.) and reverse transcriptase with the volumes and concentrations based on the manufacturer's instructions. Quantitative real time-PCR was performed using an Applied Biosystems® 7500 PCR instrument. The PCR primers used were as follows. NME2 forward: CCAAAGGGAGCTTGTTTGCC; NME2 reverse: GCCATGGTCCTGGCACTAAA; beta-actin forward: TTGTTACAGGAAGTCCCTTGCC; beta-actin reverse: ATGCTATCACCTCCCCTGTGTG.
Cell counting kit-8 and clone forming assays
The cells were cultured in 96-well plates for 24 hours, 48 hours, 72 hours, 96 hours and 120 hours. They were then incubated in DMEM containing 10 μl CCK8 solution (Dojindo Molecular Technologies, Kumamoto, Japan) for 2 hours. A microplate reader was used to measure the OD values of the wells. For the clone forming assay, the cells were cultured in 6-well plates. Every well was seeded with 1000 cells and cultured for 10 days. The cells were then washed three times with PBS and fixed with methyl alcohol for 30 min. Crystal violet was used to stain the cells in the wells. The assay was repeated thrice.
Cell cycle assay
The distribution of cells across the different cell cycle phases was determined using a fluorescence-activated cell sorter. All cells were collected after 48 h of treatment with siRNA. The results were analyzed using ModFit LT analysis software (BD Biosciences).
Western blot
The cells were collected and lysed with lysis buffer. The BCA assay was used to quantify the total protein. The cell lysates (20 μg) were loaded in the wells of a 10% gel for SDS-PAGE, which was carried out at 120 V. Next, the protein samples were transferred to a PVDF or a nitrocellulose filter membrane (Millipore, Billerica, MA, USA). The fluorescence intensity was quantified by a chemical luminescence intensity analyzer. β-Actin was used as the internal control. The antibodies used were as follows: NME2 (Abcam, 1:1000) c-Myc (Proteintech, 1:500); beta-actin (Sigma-Aldrich, 1:20,000).
Immunohistochemical analysis
Formalin-fixed paraffin-embedded OS tissue sections were collected at the Shanghai Sixth People's Hospital between 2010 and 2015. After a through drying process, the sections were dewaxed in xylene for 15 min. They were then washed with different gradients of alcohol and 0.3% hydrogen peroxide. For antigen retrieval, EDTA solution was used, and the tissue sections were treated at 100°C for 20 min. The sections were washed with PBS, and the tissues were incubated in anti-NME2 antibody for 1 h at room temperature. Hematoxylin was used for counterstaining for 30 min. Scores were assigned on a 5-point scale: 0 (0%–5%), 1 (6%–25%), 2 (26%–50%), 3 (51%–75%), or 4 (>75%). Scores of 0 and 1 were defined as antigen-negative and 2, 3 and 4 as positive. Each section was randomly sampled over 10 optical fields and evaluated by 3 different pathologists.
Immunofluorescence staining
Cells were seeded on glass coverslips overnight and washed 3 times with TBS. Next, 4% paraformaldehyde was used to fix the cells for 30 min. The cells were made permeable by treatment with 0.1% Triton X-100 for 30 min and blocked with 3% BSA for 1 hour. Fixed cells were incubated with primary antibodies for 2 hours at room temperature. Subsequently, the cells were incubated with Alexa-Fluor-labeled secondary antibodies at room temperature for 1 hour. Next, the solution was cleared with PBS, and DAPI was added to the glass coverslips. A total internal reflection fluorescence microscope (TIRFM) was used to observe the fluorescence intensity.
Statistical analysis
Statistical analysis was performed using SPSS software. Student's t-test was performed to compare two groups. P < 0.05 was considered statistically significant. The mean and SD were determined from at least 3 repeated experiments.
Disclosure of potential conflicts of interest
No conflict of interest exits in the submission of this manuscript.
Ethics committee approval
This research was approved by the Ethics Committee of the Shanghai Jiao Tong University Affiliated Sixth People's Hospital (YS-2016-064, 24 February 2016).
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics 2013. CA Cancer J Clin. 2013;63:11–30. doi:10.3322/caac.21166. PMID:17562483.
- Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992-2004). Cancer 2008;112:416–32. doi:10.1002/cncr.23169. PMID:18074355.
- Backer JM, Mendola CE, Kovesdi I, Fairhurst JL, O'Hara B, Eddy RL Jr, Shows TB, Mathew S, Murty VV, Chaganti RS. Chromosomal localization and nucleoside diphosphate kinase activity of human metastasis-suppressor genes NM23-1 and NM23-2. Oncogene 1993;8:497–502. PMID:8381224.
- Valentijn LJ, Koster J, Versteeg R. Read-through transcript from NM23-H1 into the neighboring NM23-H2 gene encodes a novel protein, NM23-LV. Genomics 2006;87:483–9. doi:10.1016/j.ygeno.2005.11.004. PMID:16442775.
- Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, candidate suppressor of tumor metastasis. Science 1993;261:478–80. doi:10.1126/science.8392752. PMID:8392752.
- Schaertl S, Geeves MA, Konrad M. Human nucleoside diphosphate kinase B (Nm23-H2) from melanoma cells shows alteredphosphoryl transfer activity due to the S122P mutation. J Biol Chem 1999;274:20159–64. doi:10.1074/jbc.274.29.20159. PMID:10400630.
- Lee MJ, Xu DY, Li H, Yu GR, Leem SH, Chu IS, Kim IH, Kim DG. Pro-oncogenic potential of NM23-H2 in hepatocellular carcinoma. Exp Mol Med 2012;44:214–24. doi:10.3858/emm.2012.44.3.016. PMID:22192927.
- Tong Y, Yung LY, Wong YH. Metastasis suppressors Nm23H1 and Nm23H2 differentially regulate neoplastic transformation and tumorigenesis. Cancer Lett 2015;361:207–17. doi:10.1016/j.canlet.2015.02.050. PMID:27987529.
- Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 2014;4:a014357. doi:10.1101/cshperspect.a014357. PMID:24384812.
- Liu Z, Gan L, Luo D, Sun C. Melatonin promotes circadian rhythm-induced proliferation through Clock/histone deacetylase3/c-Myc interaction in mouse adipose tissue. J Pineal Res. 2017;62(4). doi:10.1111/jpi.12383. PMID:27987529
- Gruppetta M, Formosa R, Falzon S, Ariff Scicluna S, Falzon E, Degeatano J, Vassallo J. Expression of cell cycle regulators and biomarkers of proliferation and regrowth in humanpituitary adenomas. Pituitary 2017;20:358–71. doi:10.1007/s11102-017-0803-0. PMID:28342098.
- Dakic A, DiVito K, Fang S, Suprynowicz F, Gaur A, Li X, Palechor-Ceron N, Simic V, Choudhury S, Yu S, et al. ROCK inhibitor reduces Myc-induced apoptosis and mediates immortalization of humankeratinocytes. Oncotarget 2016;7:66740–53. doi:10.18632/oncotarget.11458. PMID:27556514.
- Schotanus BA, Kruitwagen HS, van den Ingh TS, van Wolferen ME, Rothuizen J, Penning LC, Spee B. Enhanced Wnt/β-catenin and Notch signalling in the activated canine hepatic progenitor cellniche. BMC Vet Res 2014;10:309. doi:10.1186/s12917-014-0309-1. PMID:25551829.
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 2006;103:18261–6. doi:10.1073/pnas.0606108103. PMID:17114293.
- Romero OA, Verdura S, Torres-Diz M, Gomez A, Moran S, Condom E, Esteller M, Villanueva A, Sanchez-Cespedes M. Sensitization of retinoids and corticoids to epigenetic drugs in MYC-activated lung cancers by antitumor reprogramming. Oncogene 2017;36:1287–96. doi:10.1038/onc.2016.296. PMID:27593925.
- Zhang C, Xu B, Lu S, Zhao Y, Liu P. HN1 contributes to migration, invasion, and tumorigenesis of breast cancer by enhancing MYC activity. Mol Cancer 2017;16:90. doi:10.1186/s12943-017-0656-1. PMID:28490334.
- Zou K, Lu X, Ye K, Wang C, You T, Chen J. Krüppel-like factor 2 promotes cell proliferation in hepatocellular carcinoma through up-regulation of c-myc. Cancer Biol Ther 2016;17:20–6. doi:10.1080/15384047.2015.1108484. PMID:26853883.
- Baker EK, Taylor S, Gupte A, Sharp PP, Walia M, Walsh NC, Zannettino AC, Chalk AM, Burns CJ, Walkley CR. BET inhibitors induce apoptosis through a MYC independent mechanism and synergise with CDK inhibitors to kill osteosarcoma cells. Sci Rep 2015;5:10120. doi:10.1038/srep10120. PMID:25944566.
- Adamopoulos C, Gargalionis AN, Piperi C, Papavassiliou AG. Recent Advances in Mechanobiology of Osteosarcoma. J Cell Biochem 2017;118:232–6. doi:10.1002/jcb.25660. PMID:27463370.
- Kopylov M, Bass HW, Stroupe ME. The maize (Zea mays L.) nucleoside diphosphate kinase1 (ZmNDPK1) gene encodes a humanNM23-H2 homologue that binds and stabilizes G-quadruplex DNA. Biochemistry 2015;54:1743–57. doi:10.1021/bi501284g. PMID:25679041.
- Gilles AM, Presecan E, Vonica A, Lascu I. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the twopolypeptide chains responsible for heterogeneity of the hexameric enzyme. J Biol Chem 1991;266:8784–9. PMID:1851158.
- Wieland T, Hippe HJ, Ludwig K, Zhou XB, Korth M, Klumpp S. Reversible histidine phosphorylation in mammalian cells: a teeter-totter formed by nucleosidediphosphate kinase and protein histidine phosphatase 1. Methods Enzymol 2010;471:379–402. doi:10.1016/S0076-6879(10)71020-X. PMID:20946858.
- Braun S, Mauch C, Boukamp P, Werner S. Novel roles of NM23 proteins in skin homeostasis, repair and disease. Oncogene 2007;26:532–42. doi:10.1038/sj.onc.1209822. PMID:16862176.
- Bouvard C, Lim SM, Ludka J, Yazdani N, Woods AK, Chatterjee AK, Schultz PG, Zhu S. Small molecule selectively suppresses MYC transcription in cancer cells. Proc Natl Acad Sci U S A 2017;114:3497–502. doi:10.1073/pnas.1702663114. PMID:28292893.
- Yao Y, Li C, Zhou X, Zhang Y, Lu Y, Chen J, Zheng X, Tao D, Liu Y, Ma Y. PIWIL2 induces c-Myc expression by interacting with NME2 and regulates c-Myc-mediated tumorcell proliferation. Oncotarget 2014;5:8466–77. doi:10.18632/oncotarget.2327. PMID:25193865.
- Tschiedel S, Bach E, Jilo A, Wang SY, Lange T, Al-Ali HK, Vucinic V, Niederwieser D, Cross M. Bcr-Abl dependent post-transcriptional activation of NME2 expression is a specific and commonfeature of chronic myeloid leukemia. Leuk Lymphoma 2012;53:1569–76. doi:10.3109/10428194.2012.656631; PMID:22251158.
- Wuelling M, Delling G, Kaiser E. Differential gene expression in stromal cells of human giant cell tumor of bone. Virchows Arch 2004;445:621–30. doi:10.1007/s00428-004-1113-2. PMID:15449052.
- Böckelman C, Koskensalo S, Hagström J, Lundin M, Ristimäki A, Haglund C. CIP2A overexpression is associated with c-Myc expression in colorectal cancer. Cancer Biol Ther 2012;13:289–95. doi:10.4161/cbt.18922. PMID:22310977.
- Postel EH, Berberich SJ, Rooney JW, Kaetzel DM. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J Bioenerg Biomembr 2000;32:277–84. doi:10.1023/A:1005541114029. PMID:11768311.
- Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, Basundra R, Kumar A, Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoternuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res 2009;37:172–83. doi:10.1093/nar/gkn919. PMID:19033359.
- Chang YW, Chiu CF, Lee KY, Hong CC, Wang YY, Cheng CC, Jan YH, Huang MS, Hsiao M, Ma JT, et al. CARMA3 Represses Metastasis Suppressor NME2 to Promote Lung Cancer Stemness and Metastasis. Am J Respir Crit Care Med 2015;192:64–75. doi:10.1164/rccm.201411-1957OC. PMID:25906011.
- Song JH, Park E, Kim MS, Cho KM, Park SH, Lee A, Song J, Kim HJ, Koh JT, Kim TS. l-Asparaginase-mediated downregulation of c-Myc promotes 1,25(OH)2 D3 -induced myeloid differentiation in acute myeloid leukemia cells. Int J Cancer 2017;140:2364–74. doi:10.1002/ijc.30662; PMID:28224619.
- Li X, Wu JB, Li Q, Shigemura K, Chung LW, Huang WC. SREBP-2 promotes stem cell-like properties and metastasis by transcriptional activation of c-Myc in prostate cancer. Oncotarget 2016;7:12869–84. doi:10.18632/oncotarget.7331. PMID:26883200.
- Galmozzi E, Casalini P, Iorio MV, Casati B, Olgiati C, Ménard S. HER2 signaling enhances 5′UTR-mediated translation of c-Myc mRNA. J Cell Physiol 2004;200:82–8. doi:10.1002/jcp.20012 doi:10.1002/jcp.20012. PMID:15137060.
- Finn RS, Martin M, Rugo HS, Jones S, Im SA, Gelmon K, Harbeck N, Lipatov ON, Walshe JM, Moulder S, et al. Palbociclib and Letrozole in Advanced Breast Cancer. N Engl J Med. 2016;375:1925–36. doi:10.1056/NEJMoa1607303. PMID:27959613.
- Borkowska A, Goryń T, Pieńkowski A, Wągrodzki M, Jagiełło-Wieczorek E, Rogala P, Szacht M, Rutkowski P. Denosumab treatment of inoperable or locally advanced giant cell tumor of bone. Oncol Lett. 2016;12:4312–18. doi:10.3892/ol.2016.5246. PMID:28101196.
- Claire Bouvard, Sang Min Lim, John Ludka, Nahid Yazdani, Ashley K. Woods, Arnab K. Chatterjee, eter G. Schultz, Shoutian Zhu. Small molecule selectively suppresses MYC transcription in cancer cells. Proc Natl Acad Sci U S A. 2017;114:3497–3502. doi:10.1073/pnas.1702663114. PMID:28292893