 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Objective: The study aimed to investigate the molecular mechanism of miR-144 and CEP55 as well as the influence of their interaction on the cell proliferation, migration, invasion, cell cycle and cell apoptosis in breast cancer.
Methods: In this study, The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/) database was used for microarray analysis. The expressions of miR-144 and CEP55 in 40 adjacent tissues and 36 tumor tissues were examined by western blot, qRT-PCR and immunohistochemistry. The target relationship between miR-144 and CEP55 was predicted and confirmed by TargetScan and luciferase reporter assay. The cell proliferation, cell cycle and cell apoptosis in different groups were detected by MTT and flow cytometry assays, while wound healing and transwell assays were used for the cell migration and invasion tests. The regulatory effects of miR-144 and CEP55 on breast tumor were verified through nude mouse model in vivo experiment.
Results: MiR-144 was down-regulated in breast cancerous tissues and cells, whereas CEP55 expression was up-regulated in breast cancerous tissues. Moreover, there existed a target relationship between miR-144 and CEP55 and negative correlation on their expressions. MiR-144 could down-regulate CEP55 expression, thereby inhibiting proliferation, invasion, migration, retarding cell cycle and accelerating cell apoptosis. MiR-144 could inhibit cell progression through down-regulating CEP55 in vivo.
Conclusion: MiR-144 suppressed cell proliferation, migration, invasion and induced cell cycle arrest and cell apoptosis by repressing CEP55. This might provide a promising therapy for clinical treatment.
Introduction
Over 1 million people are diagnosed with breast cancer worldwide each year.Citation1 Most breast cancers are sporadic, while 5%–10% of the disease are hereditary.Citation2 Currently, tremendous progress was made in the treatment of breast cancer, and commonly used radiotherapy is often administered to destroy breast cancerous cells that may not have been removed during surgery.Citation3 In spite of increasing survival rate, the mechanism of breast carcinogenesis remains complex.Citation4 Recently, genetic factor has become a research hotspot,Citation5 and further studies on genetic level are worthwhile.
MicroRNAs (miRNAs) are small non-coding RNA molecules which regulate the target mRNAs expression.Citation6 Although miRNAs were first ascertained in the 1990s, its potential functions in cancer treatment have not been widely explored until the recent years. It is reported that miRNAs can act as tumor suppressors or promoters, exerting influence on various malignancies.Citation7 Recently, the functions of some miRNAs in breast cancer have been investigated in various types of cancers.Citation8 For instance, miR-214 is identified to be associated with poor prognosis of the triple-negative breast cancer patients.Citation9 The up-regulated expression of miR-29c is related to increased survival rate in breast cancer patients.Citation8 MiR-34 was found to protect breast cancer cells from non-apoptotic cell death.Citation10 MiR-144 is a kind of crucial microRNA that has been identified to be correlated with multiple human diseases and modulates metastasis.Citation11 For instance, MiR-144 was recognized as a tumor suppressor in hepatocellular carcinoma (HCC) and could inhibit HCC cell proliferation, invasion and migration by targeting ZFX.Citation12 A recent study also demonstrated that overexpression of miR-144 could suppress epithelial mesenchymal transition.Citation13 Sun et al. revealed that the miR-144 functioned as a tumor-suppressor in papillary thyroid cancer (PTC).Citation14 However, the deeper molecular mechanism of miR-144 in breast cancer has not been fully elucidated yet.
Centrosomal protein 55 (CEP55), as a microtubule-bundling protein, can be found in centrosome of interphase cells or midbody and plays an important role in cell cycle regulationCitation15,Citation5 It has been found that CEPP 55, whose localization changed significantly during the cell cycle, might be a biomarker of cell cycle.Citation16 Accumulating evidence shows that CEP55 is overexpressed in several types of cancers, such as human colon cancer, lung cancer and breast cancer.Citation5 Overexpression of CEP55 might facilitate cell migration and invasion in mammalian cells.Citation17 Furthermore, CEP55 could also act as a predictive biomarker of poor prognosis in patients with locally advanced esophageal squamous cell carcinoma.Citation18 However, there are few studies focusing on the regulatory mechanism of miR-144-CEP55 pathway in breast cancer.
Our study was aimed at investigating the relationship between miR-144 and CEP55 as well as the influence of their interaction on the cell proliferation, migration, invasion, cell cycle and cell apoptosis in breast cancer. We first supposed that miR-144 could regulate breast cancer progression by targeting CEP55, and then performed the following experiments to verify the hypothesis.
Materials and methods
Cells and tissue specimens
36 human breast cancer tissues and 40 adjacent non-neoplastic tissues were obtained from the First Affiliated Hospital of China Medical University and frozen in liquid nitrogen. All patients were diagnosed in advance and confirmed to receive no radiotherapy or chemotherapy. Written informed consents were obtained from all patients and this experiment was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of China Medical University. Human embryonic kidney cell line HEK-293T and human breast cancer cell lines MCF-7, MDA-MB-231 and SK-BR-3 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human normal breast cell line Hs578Bst was purchased from American Type Culture Collection (Manassas, VA, USA).
Microarray analysis
The data of breast cancer samples were downloaded from TCGA database (https://tcga-data.nci.nih.gov/). The information from this data set was used to identify differentially expressed miRNAs and mRNAs having a fold change (FC) > 2 and P<0.05, as determined by R studio (TIBCO, USA). Targetscan database was used to predict the binding sites of miR-144 on 3′UTR of CEP55. Prognosis data was analyzed and survival curve analysis correlating with miR-144 and CEP55 in breast cancer was plotted by R studio.
Cell culture and transfection
MCF-7, MDA-MB-231 and Hs 578Bst cell lines were incubated with Dulbecco's modified Eagle's medium (DMEM) (10% FBS, Gibco, MD, USA) at 37 °C with 5% CO2. SK-BR-3 was incubated in RPMI-1640 medium (10% FBS, Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C. Cells were transfected with Lipofectamine2000 (Invitrogen, NY, USA) respectively. The cells transfected with scramble and siRNA control were NC group. The cells transfected with CEP55 siRNAs (Silencer®, #AM16708, Invitrogen, NY, USA) were si-CEP55 group. The cells transfected with miR-144 mimics were used as miR-144 group, while those transfected with mir-144 inhibitor acted as mir-144 inhibitor group (mirVana®, #4464084, Invitrogen, NY, USA). The cells in miR-144 inhibitor + si-CEP55 group were co-transfected with miR-144-5p inhibitor and si-CEP55. Fluorescence microscopy was used for observation after 24-hour transfection.
qRT-PCR
After the extraction of the total RNA using TRIzol reagent, TaqMan microRNA Reverse Transcription kit and TaqMan Universal Master Mix was utilized to perform reverse transcription of PCR and real-time PCR according to the manufacturer's instruction. Primers were listed in . StepOne software (Applied Biosystems, CA, USA) was used for data analysis and fold changes were calculated via relative quantification of
method.
Western blot
PCR primer sequences.
The cells were washed with cool phosphate-buffered saline (PBS) and then added with Radio-Immunoprecipitation Assay (RIPA) (Invitrogen, #V89901, USA) containing protease inhibitor. The cells were put on ice for 5 min and supernatant was extracted through centrifugation at 4oC. Protein concentration was determined by the bicinchonininc acid (BCA) protein assay kit (Beyotime, Suzhou, China). Proteins were subjected to electrophoresis and transmembrane before the immunoreaction. Primary antibodies (Rabbit monoclonal CEP55 antibody, 1:10000, Abcam, #ab170414, Cambridge, MA, USA) were first used for the incubation of proteins overnight at 4oC and then secondary antibodies (Goat-Anti-Rabbit IgG H&L (HRP),1:3000,Abcam, #ab6728, Cambridge, MA, USA) were added to incubate membranes for another hour. Quantity One software (Bio-Rad, Hercules, CA, USA) was used to analyze the images after the exposure to enhanced chemiluminescence (ECL) detection (Abcam, Cambridge, MA, USA).
Immunohistochemistry
The same primary and secondary antibodies used in western blot assay were also applied to immunohistochemistry. Paraffin embedding of tissues was performed under standard conditions. The tissues were sliced into 4 μm thickness slices, followed by dewaxing, antigen repair, enzyme inactivation with 3% hydrogen peroxide for 10 min, and washing in PBS. The slices were stained with primary antibodies (1:400), secondary antibodies (1:1000), streptomyces avidin-peroxidase solution and diaminoaniline (DAB, Abcam, #ab64261, Cambridge, MA, USA) for 5 min. Then the slices were stained with hematoxylin, dehydrated, transparentized, and sealed using neutral gum. The observation was performed under microscope. All specimens contained two slices. Positive slices were used as positive control, while the slices treated with PBS were used as a negative control. Cells that manifested brown granules in the cytoplasm and / or nucleus were considered to be positive.
Dual-luciferase reporter assay in vitro
The CEP55 3′UTR sequence predicted to interact with miR-144 or a mutated sequence within the predicted target sites was synthesized and inserted into the pMIR-reporter which contained restriction enzyme sites of Spe I and Hind III. Primers were listed in . After co-transfected with the above-mentioned constructs, the HEK-293T cells were incubated for 48 h. Then they were washed by cold PBS. We added 350 μl pre-cooled harvest buffer at 4oC for cell splitting. Optical density was detected with Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA) according to the manufacturer's instructions.
MTT assay
Primer sequences.
Cells were first cultured in 10% fetal bovine serum (FBS) for 5 days and then added with MTT solution. Following four hours of incubation, the supernatant was discarded and the cells were exposed to 100 μl dimethyl sulfoxide (DMSO) for 10 min. The absorbance at 490 nm was measured in a microplate reader and cell viability was calculated in accordance with OD value.
Annexin V-FITC/PI staining assay
1 × 105 cells were centrifuged and resuspended in 500 μl of 1 × binding buffer. Then Annexin V-fluorescein isothiocyanate (FITC) and Propidium Iodide (PI) (Abcam, Cambridge, MA, USA) were added and incubated with cells for 5 min in the dark at room temperature. Flow cytometer was utilized to analyze the cell apoptosis and CellQuest (BD Biosciences, San Jose, CA, USA) software was used for data analysis.
Flow cytometry (FCM) assay
The effects of CEP55 and miR-144 on breast cancer cell cycle were tested by Tali Cell Cycle Kit (Invitrogen, #A10798, NY, USA). Briefly, the cells were first transfected for 72 h and treated with trypsin, and then washed with washed using cool PBS, followed by fixation with 75% ethanol overnight at 4°C. Lastly, the cells were incubated with 100 μl RNase A for 30 min and 50 μl PI without light for 1 h, respectively.
Wound healing assay
Cells of each group were seeded in the 6-well plates at a density of 1 × 105 cells per well. After transfection overnight, the monolayer of cells was scratched using a sterile 200 ml micropipette tip. The cells were incubated in serum free medium at 37oC, 5% CO2 after washed with PBS for three times and the scratch healing area of cells at 0 h and 24 h was subsequently observed and photographed.
Transwell assay
The upper chambers were coated with 50 μl Matrigel (BD Biosciences, San Jose, CA, USA) at 4oC. Cells were starved for 12 h without serum, washed twice with PBS and resuspended with serum-free BSA (Invitrogen, NY, USA) till cell density reached 105 / L. 500 μl of DMEM high glucose medium (Invitrogen, NY, USA) containing 20% FBS (Invitrogen, NY, USA) was added as a chemokine in the lower chamber. After 8 h incubation, the non-migratory or non-invasive cells were removed using a cotton swab. The chambers were then fixed with 4% paraformaldehyde, washed twice with PBS and stained with 0.1% crystal violet (Thermo Scientific™, #R40052, Waltham, MA, USA). The migrating or invading cells were counted under microscope.
Animal experiment
The transfected MCF-7 cells of five groups were digested with trypsin and suspended with PBS. Twenty male BALB/c mice (4 to 6-week-old) were obtained from Shanghai Experimental Animal Center and maintained under the pathogen-free condition throughout the experiment. The animal experiment was performed following the authenticated animal protocols of Ethical Committee of Animal Welfare of China Medical University. These mice were randomly divided into five groups and the transfected cells (1.5 × 106) were subcutaneously injected into the back of the nude mice. The condition of each mouse was detected every week for 5 weeks. Tumor volume was calculated by the formula [(length + width) / 2]Citation3 × 0.5236. The tumor tissues of dead mice were collected immediately and divided into two groups, one preserved at -70oC and the other fixed with paraformaldehyde for immunohistochemistry.
Statistical analysis
Data analyses were performed through SPSS 21.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad, Software, San Diego, CA, USA). Differences between groups were determined by Student's t-test or one-way ANOVA. The patient survival rate was detected by Kaplan-Meier and the clinical features were examined by chi-square test. The results were presented as mean ± SD and P < 0.05 was considered statistically significant.
Results
MiR-144 was down-regulated in breast cancer tissues and cells
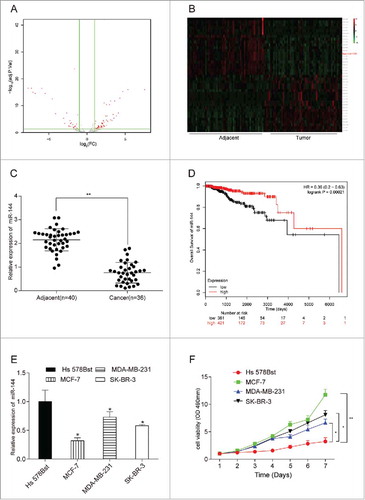
We revealed 67 up-regulated and 31 down-regulated miRNAs based on TCGA database and selected 20 miRNAs with high or low expressions respectively to perform the volcano plot and heat map. The expression of mir-144 was reduced by 4.11 folds in the cancer tissue compared with that in adjacent tissue (). Mir-144 was significantly low-expressed (). MiR-144 mRNA in 40 adjacent tissues and 36 cancer tissues were found low-expressed (, P < 0.01). The survival curve obtained from Kaplan-Meier plotter database (http://kmplot.com/analysis/) also indicated the positive correlation between miR-144 high expression and prolonged life span (, P < 0.01). Breast cancer cell lines MCF-7, MDA-MB-231 and SK-BR-3 displayed a lower expression of miR-144 as well (P < 0.05), in particular MCF-7 cell line presenting the lowest expression of miR-144 (). At the same time, MCF-7 cell line also showed the strongest cell viability among three cell lines (), thus MCF-7 was chosen for the following experiments.
CEP55 was up-regulated in breast cancer tissues and cells
MiR-144 was low-expressed in breast cancer tissues and cells. (A) The volcano plot showed the relationship between fold change and significance of miRNAs expression. (B) The heat map of 20 high-expressed miRNAs and 20 low-expressed ones in breast cancer. (C) The expression level of miR-144 in cancer tissues was significantly low. **P < 0.01, compared with adjacent tissues, number of adjacent tissue = 40, number of cancer tissue = 36. (D) Higher miR-144 expression was related to higher survival rate. (E) MiR-144 expression was down-regulated in breast cancer cells, especially in MCF7. *P < 0.05, compared with normal breast cells. (F) MCF7 cell line had the highest cell proliferation ability. *P < 0.05, **P < 0.01, compared with Hs 578Bst cell line.
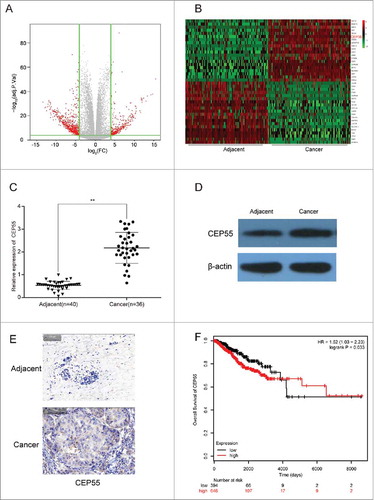
We also identified 473 up-regulated mRNAs and 231 down-regulated mRNAs through TCGA database. A volcano plot of the identified quality-controlled mRNAs (P < 0.05, fold change >2) was presented in . We found that CEP55 exhibited 3.67 times higher expression in breast cancer tissue than in adjacent tissue. Top 20 mRNAs with the highest or lowest expression were respectively selected for heat map analysis (). The results from qTR-PCR and western blot indicated that CEP55 mRNA and protein was overexpressed in breast cancerous tissues compared with adjacent tissues (). Immunohistochemistry assay revealed the same results as the above (). Kaplan-Meier survival analysis verified the positive correlation between decreased CEP55 expression and prolonged life span ().
MiR-144 directly targeted at CEP55
CEP55 was up-regulated in breast cancer tissues and cells. (A) The volcano plot showed the relationship between fold change and significance of mRNAs expression. (B) The heat map showed the top 20 high-expressed and 20 low-expressed mRNAs in breast cancer. (C) QRT-PCR assay was used to detect the mRNA expression level of CEP55. The results showed that the mRNA expression level of CEP55 in cancer tissues was significantly increased.**P < 0.01, compared with adjacent tissues, number of adjacent tissue = 40, number of cancer tissue = 36. (D-E) Western blot and immunohistochemistry assays were used to detect the protein expression level of CEP55. The results revealed that CEP55 protein expression level was up-regulated in breast cancer tissues and cells. (F) Kaplan-Meier method was used to plot the 5-year survival ratio of breast cancer patients. The plot indicated that patients with high expression level of CEP55 had lower survival ratio, compared with those had low-expressed CEP55.
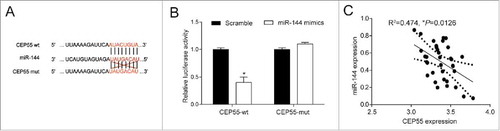
As shown in , CEP55 wild-type (wt) rather than mutated-type might be a target of miR-144. Dual-luciferase reporter assay showed that co-transfection with miR-144 mimics and CEP55 3′UTR-wt significantly reduced the luciferase activity of the cells in comparison with scramble group (P < 0.05), while no significant difference was found between the miR-144 mimics and CEP55 3′UTR-mut co-transfection group and scramble group (P > 0.05) (). Besides, there existed a negative correlation on expression between CEP55 and miR-144 (P < 0.05) ().
MiR-144 could inhibit proliferation, induce cell cycle arrest and promote apoptosis by down-regulating CEP55
MiR-144 directly targeted at CEP55. (A) Bioinformatics method was utilized to predict the potential target of miR-144. The results showed that CEP55 wild-type's binding site matched with miR-144, suggesting that CEP55 wild-type rather than mutated-type was the target of miR-144. (B) Dual-luciferase reporter assay proved that the overexpression of miR-144 significantly down-regulated the expression of CEP55 wild-type. *P < 0.05, compared with scramble group. The expressions of CEP55 mutated-type had no difference in cells of scramble group and miR-144 mimics group. (C) CEP55 was negatively correlated with miR-144.
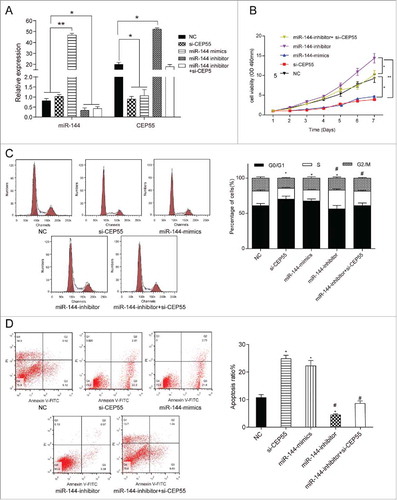
QRT-PCR results displayed that miR-144 mimics contributed to the overexpression of miR-144, whereas miR-144 inhibitor led to down-regulation of miR-144 expression (P < 0.05). The overexpression of miR-144 and knockdown of CEP55 both could down-regulate the CEP55 expression level, while the miR-144 inhibitor could up-regulate the CEP55 expression level, compared with NC group (P < 0.05) (). Furthermore, MTT assay indicated that down-regulation of miR-144 expression significantly increased cell viability, while miR-144 overexpression and knockdown of CEP55 both could decrease cell viability (P < 0.05) (). FCM results suggested that more cells were arrested in G0/G1 phase in miR-144 mimics group and si-CEP55 group compared with NC group, while more cells entered S phase in miR-144-inhibitor group (P < 0.05). Compared with miR-144-mimics group, more cells were arrested at S phase in miR-144-inhibitor group and miR-144-inhibitor+si-CEP55 group (). Annexin V-FITC/PI staining assay showed that apoptosis ratio of cells in miR-144 mimics group and si-CEP55 group was higher than NC group. Compared with miR-144 mimics group and si-CEP55 group, the apoptosis ratio of miR-144 inhibitor group and miR-144 inhibitor + si-CEP55 group was lower. (P < 0.05) (). Taken together, miR-144 inhibited the cell proliferation and induced cell cycle arrest at G0/G1 phase, meanwhile, promoted apoptosis in breast cancer cells through down-regulating CEP55.
MiR-144 overexpression could restrain migration and invasion by down-regulating CEP55
MiR-144 could inhibit proliferation, induce cell cycle arrest and promote apoptosis by down-regulating CEP55. (A) QRT-PCR assay was used to detect the expression level of miR-144 and CEP55 after transfection. The overexpression of miR-144 significantly increased the miR-144 expression level while the inhibition of miR-144 decreased the miR-144 expression level. *P < 0.05, **P < 0.01, compared with NC group and si-CEP55 group. The knockdown of CEP55 and overexpression of miR-144 could down-regulate the CEP55 expression level, while the inhibition of miR-144 could up-regulate CEP55 expression level, *P < 0.05, compared with NC group. (B) The cell proliferation ability was detected by MTT assay. MiR-144 mimics group and si-CEP55 group had the weakest proliferation ability. MiR-144 inhibitor group had the strongest cell proliferation ability, *P < 0.05, compared with NC group and miR-144 inhibitor + si-CEP55 group, **P < 0.01, compared with miR-144 inhibitor group. (C) The cell cycle was detected by FCM assay. More cells of miR-144 mimics group and si-CEP55 group were arrested in G0/G1 phase while more cells were synthesized in S phase in miR-144-inhibitor group. More cells were synthesized in S phase in miR-144-inhibitor group and miR-144-inhibitor+si-CEP55 group compared with miR-144-mimics group and si-CEP55 group. *P < 0.05, compared with NC group. # P < 0.05, compared with si-CEP55 group and miR-144 mimics group. (D) The apoptosis ratio was detected by FCM assay. MiR-144 mimics and si-CEP55 groups had the highest apoptosis ratio, and miR-144 inhibitor group had the lowest ratio. *P < 0.05, compared with NC group. # P < 0.05, compared with si-CEP55 group and miR-144 mimics group.
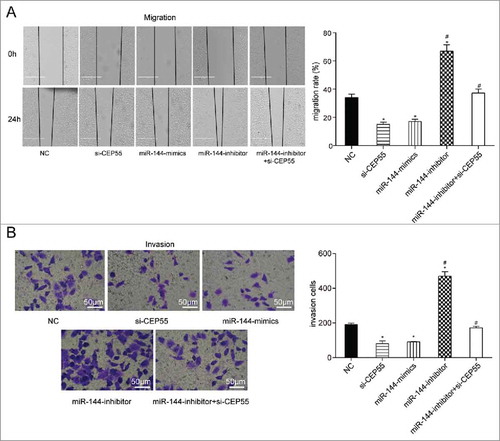
Wound healing assay and transwell invasion assay revealed that the down-regulation of miR-144 considerably accelerated cells mobility and invasion. By contrast, miR-144 overexpression and CEP55 silencing remarkably inhibited cell migration and invasion abilities. Moreover, the cell mobility and invasion capabilities in miR-144 inhibitor + si-CEP55 group were nearly equivalent to those in miR-144 mimics group and si-CEP55 group, while much higher than those in NC group (P < 0.05) (), which indicated that miR-144 could inhibit the migration and invasion of breast cancer cells through targeting CEP55.
MiR-144 could inhibit breast tumor growth through regulating CEP55 in vivo
MiR-144 could restrain migration and invasion by down-regulating CEP55. (A-B) The migration and invasion abilities were detected by wound healing assay (scale bar: 50 μm) and transwell assay (scale bar: 50 μm). MiR-144 mimics and si-CEP55 significantly inhibited the migration and invasion while miR-144 inhibitor significantly promoted the migration and invasion of breast cancer cells. *P < 0.05, compared with NC group and miR-144 inhibitor + si-CEP55 group. # P < 0.05, compared with si-CEP55 group and miR-144 mimics group.
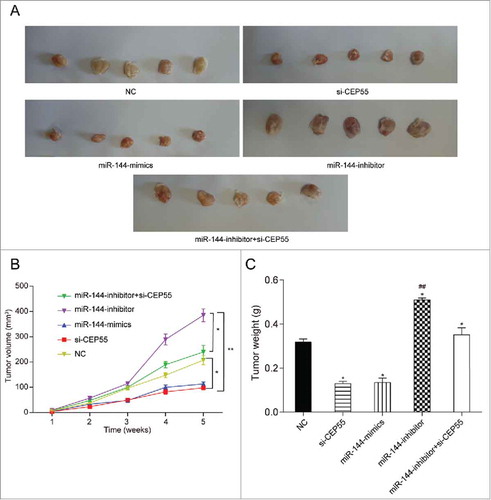
The regulatory effects of miR-144 and CEP55 on breast tumor were further verified through nude mouse model in vivo experiment, which was same as the above results of vitro experiments. Increased miR-144 expression and decreased CEP55 expression contributed to suppressing breast tumor growth, whereas down-regulation of miR-144 expression exerted facilitative influence on tumor growth (). As shown in , the tumor volume of mice treated with si-CEP55 or miR-144 mimics was the smallest, whereas that of the mice treated with miR-144 inhibitor was the biggest among all the groups (P < 0.05). After co-transfected with miR-144 inhibitor and si-CEP55, the tumor weight of the mice notably declined. However, there was no significant difference of tumor volume between miR-144 inhibitor + si-CEP55 group and NC group. Similarly, we also found that the tumor weight of mice in miR-144 inhibitor group was considerably heavier, whereas that in si-CEP55 group and miR-144 mimics group was observably lighter compare with NC group (P < 0.05). Furthermore, no significant distinction was detected between miR-144 inhibitor + si-CEP55 group and NC group in terms of tumor weight (P > 0.05), (). The above results suggested that miR-144 could inhibit breast tumor growth through modulating CEP55 in vivo.
MiR-144 could inhibit cell progression through down-regulating CEP55 in vivo. (A) The tumor samples of miR-144 inhibitor group were the biggest while si-CEP55 and miR-144 mimics groups were the smallest. (B) The measurement of tumor volume showed that miR-144 inhibitor group had the biggest tumor and si-CEP55 and miR-144 mimics groups had the smallest tumor, *P < 0.05, compared with NC group and miR-144 inhibitor + si-CEP55 group, **P < 0.01, compared with miR-144 inhibitor group. (C) The measurement of tumor weight suggested that miR-144 inhibitor induced tumor growth and si-CEP55 and miR-144 mimics showed reverse function. *P < 0.05, compared with NC group, ## P < 0.01, compared with si-CEP55 group and miR-144 mimics group.
Discussion
Our study first identified the low expression of miR-144 and high expression of CEP55 in breast cancer cells, and validated the target relationship between miR-144 and CEP55 as well as their correlation on expression. Meanwhile, we also confirmed the association between miR-144/ CEP55 expression level and patient prognosis. Through vitro and vivo experiments, we finally demonstrated that miR-144 could inhibit cell proliferation, invasion, migration, and induced cell cycle arrest and cell apoptosis by targeting CEP55.
It has been reported that the aberrant expression of miRNAs could result in carcinogenesis or tumorgenesis in various cancers, including breast cancer.Citation19 MiR-214 and MiR-142 are found highly expressed in human breast cancer stem cells.Citation20,Citation21 MiR-411 and miR-520 have been identified to be down-regulated in breast cancer tissues.Citation22,Citation23 Sun et al. disclosed that miR-144 was down-regulated in papillary thyroid cancer tissues.Citation14 In the study, we further verified that miR-144 expression was down-regulated in breast cancer tissues. In addition, Madhavan et al. have revealed that plasma levels of miR-144 was closely associated with progression-free survival, which might be a prognostic markers for metastatic breast cancer.Citation24 In the present study, we found the lower expression of miR-144 was correlated with poorer prognosis of breast cancer patients.
Additionally, aberrantly expressed CEP55 has also been found to play a crucial role in the progression of multiple cancers.Citation5 For instance, CEP55 has been identified to be highly expressed in hepatocellular carcinoma, colon cancer and lung cancer.Citation25-Citation27 Previous studies suggested that CEP55 overexpression accelerated pancreatic cancer cells tumourigenicity.Citation25 Wang et al. revealed that the expression level of CEP55 was significantly up-regulated in breast cancer.Citation5 We also detected that CEP55 was highly expressed in breast cancer tissues and the patients with higher CEP55 expression presented a decreased survival ratio and poor prognosis.
Accumulating evidence has suggested that CEP55 and miR-144 both exert regulatory function on cancer progressions, such as proliferation, cell cycle and apoptosis. CEP55 has been considered as a key regulator of cytokinesis and cell cycle transition.Citation28 Peng et al. demonstrated that CEP55 could promote the proliferation, migration, and invasion in pancreatic cancer cells.Citation25 Chang et al. found that CEP55 played an essential role in regulating the G2/M phase of the cell cycle.Citation15 As for miR-144, it has been reported to be able to inhibit proliferation, enhance apoptosis, and increase autophagy in lung cancer cells.Citation29 For instance, Pan et al. disclosed that miR-144 inhibited cell propagation and promoted cell apoptosis in breast cancer as a tumor suppressor.Citation30 Our study substantiated that miR-144 could suppress proliferation, induce cell cycle arrest and cell apoptosis by targeting CEP55, which was similar to Pan's.
Though we verified our previous hypothesis through vitro and vivo assays, there existed a limitation in the study that the same results may not apply to other cancers due to the complicated regulation network.
Conclusion
To sum up, miR-144 was lowly expressed in breast cancer tissues and cells, while CEP55 was overexpressed in breast cancer. There existed a target relationship between miR-144 and CEP55 and negative correlation on their expressions. Furthermore, decreased expression of miR-144 and increased expression of CEP55 was closely associated with lower survival ratio and poorer prognosis of breast cancer patients. MiR-144 overexpression and knockdown of CEP55 both contributed to inhibiting breast cancer cell propagation, metastasis and tumor growth, and promoting cell apoptosis. MiR-144 functioned as a tumor suppressor that inhibited cell viability, cell migration and invasion, and induced cell cycle arrest and apoptosis in breast cancer via down-regulation of CEP55. The above findings not only revealed the regulatory effects of miR-144-CEP55 interaction on breast cancer cells, but also provided novel therapeutic targets and prognostic biomarkers for the diagnosis and treatment of breast cancer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Ethical approval
This study was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of China Medical University
Author contribution
Jingjing Cai, Fandong Meng and Yuanqin Yin contributed to research conception and design as well as manuscript drafting; Chengguang Sui analysed and interpreted data; Fandong Meng and Jingjing Cai made statistical analysis; Yuanqin Yin and Youhong Jiang revised the manuscript and took the role of funding collectors. In addition, all authors approved final manuscript.
References
- Kroemer G, Senovilla L, Galluzzi L, Andre F, Zitvogel L. Natural and therapy-induced immunosurveillance in breast cancer. Nat Med. 2015;21(10):1128–38. doi:10.1038/nm.3944. PMID:26444637.
- Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DY, Tothill RW, Thorne H, kConFab BDR, Li J, et al. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8(9):e1002894. doi:10.1371/journal.pgen.1002894. PMID:23028338.
- Yu L, Yang Y, Hou J, Zhai C, Song Y, Zhang Z, Qiu L, Jia X. MicroRNA-144 affects radiotherapy sensitivity by promoting proliferation, migration and invasion of breast cancer cells. Oncol Rep. 2015;34(4):52. doi:10.3892/or.2015.4173. PMID:26252024.
- Spanheimer PM, Carr JC, Thomas A, Sugg SL, Scott-Conner CE, Liao J, Weigel RJ. The response to neoadjuvant chemotherapy predicts clinical outcome and increases breast conservation in advanced breast cancer. Am J Surg. 2013;206(1):2–7. doi:10.1016/j.amjsurg.2012.10.025. PMID:23375759.
- Wang Y, Jin T, Dai X, Xu J. Lentivirus-mediated knockdown of CEP55 suppresses cell proliferation of breast cancer cells. Biosci Trends. 2016;10(1):67–73. doi:10.5582/bst.2016.01010. PMID:26902787.
- Chen L, Li Y, Fu Y, Peng J, Mo MH, Stamatakos M, Teal CB, Brem RF, Stojadinovic A, Grinkemeyer M, et al. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PLoS One. 2013;8(1):e54213. doi:10.1371/journal.pone.0054213. PMID:23372687.
- Tang J, Ahmad A, Sarkar FH. The role of microRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci. 2012;13(10):13414–37. doi:10.3390/ijms131013414. PMID:23202960.
- Nygren MK, Tekle C, Ingebrigtsen VA, Makela R, Krohn M, Aure MR, Nunes-Xavier CE, Perala M, Tramm T, Alsner J, et al. Identifying microRNAs regulating B7-H3 in breast cancer: the clinical impact of microRNA-29c. Br J Cancer. 2014;110(8):2072–80. doi:10.1038/bjc.2014.113. PMID:24577056.
- Kalniete D, Nakazawa-Miklasevica M, Strumfa I, Abolins A, Irmejs A, Gardovskis J, Miklasevics E. High expression of miR-214 is associated with a worse disease-specific survival of the triple-negative breast cancer patients. Hered Cancer Clin Pract. 2015;13(1):7. doi:10.1186/s13053-015-0028-z. PMID:25705321.
- Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2011;3(3):151–8. doi:10.1093/jmcb/mjq042. PMID:21183529.
- Keller A, Leidinger P, Vogel B, Backes C, ElSharawy A, Galata V, Mueller SC, Marquart S, Schrauder MG, Strick R, et al. miRNAs can be generally associated with human pathologies as exemplified for miR-144. BMC Med. 2014;12:224. doi:10.1186/s12916-014-0224-0. PMID:25465851.
- Bao H, Li X, Li H, Xing H, Xu B, Zhang X, Liu Z. MicroRNA-144 inhibits hepatocellular carcinoma cell proliferation, invasion and migration by targeting ZFX. J Biosci. 2017;42(1):103–11. PMID:28229969
- Pan HL, Wen ZS, Huang YC, Cheng X, Wang GZ, Zhou YC, Wang ZY, Guo YQ, Cao Y, Zhou GB. Down-regulation of microRNA-144 in air pollution-related lung cancer. Sci Rep. 2015;5:14331. doi:10.1038/srep14331. PMID:26395400
- Sun J, Shi R, Zhao S, Li X, Lu S, Bu H, Ma X, Su C. E2F8, a direct target of miR-144, promotes papillary thyroid cancer progression via regulating cell cycle. J Exp Clin Cancer Res. 2017;36(1):40. doi:10.1186/s13046-017-0504-6. PMID:28270228.
- Chang YC, Wu CH, Yen TC, Ouyang P. Centrosomal protein 55 (Cep55) stability is negatively regulated by p53 protein through Polo-like kinase 1 (Plk1). J Biol Chem. 2012;287(6):4376–85. doi:10.1074/jbc.M111.289108. PMID:22184120.
- Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, Gabrielli BG, Robinson PJ, Nigg EA, Ono Y, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9(4):477–88. doi:10.1016/j.devcel.2005.09.003. PMID:16198290.
- Jiang W, Wang Z, Chen G, Jia Y. Prognostic significance of centrosomal protein 55 in stage I pulmonary adenocarcinoma after radical resection. Thorac Cancer. 2016;7(3):316–22. doi:10.1111/1759-7714.12330. PMID:27148417.
- Jiang W, Wang Z, Jia Y. CEP55 overexpression predicts poor prognosis in patients with locally advanced esophageal squamous cell carcinoma. Oncol Lett. 2017;13(1):236–42. doi:10.3892/ol.2016.5414. PMID:28123547.
- Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, Araiza-Chavez J, Trevino V, Rodriguez-Padilla C, Resendez-Perez D. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9(1):53–9. doi:10.1111/j.1743-7563.2012.01548.x. PMID:22898264.
- Orso F, Quirico L, Virga F, Penna E, Dettori D, Cimino D, Coppo R, Grassi E, Elia AR, Brusa D, et al. miR-214 and miR-148b Targeting inhibits dissemination of Melanoma and Breast Cancer. Cancer Res. 2016;76(17):5151–62. doi:10.1158/0008-5472.CAN-15-1322. PMID:27328731.
- Shimono Y, Mukohyama J, Nakamura S, Minami H. MicroRNA Regulation of Human Breast cancer stem cells. J Clin Med. 2015;5(1):2. doi:10.3390/jcm5010002. PMID:26712794.
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. doi:10.1038/nrd.2016.246. PMID:28209991.
- Guo L, Yuan J, Xie N, Wu H, Chen W, Song S, Wang X. miRNA-411 acts as a potential tumor suppressor miRNA via the downregulation of specificity protein 1 in breast cancer. Mol Med Rep. 2016;14(4):2975–82. doi:10.3892/mmr.2016.5645. PMID:27572271.
- Madhavan D, Peng C, Wallwiener M, Zucknick M, Nees J, Schott S, Rudolph A, Riethdorf S, Trumpp A, Pantel K, et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016;37(5):461–70. doi:10.1093/carcin/bgw008. PMID:26785733.
- Peng T, Zhou W, Guo F, Wu HS, Wang CY, Wang L, Yang ZY. Centrosomal protein 55 activates NF-kappaB signalling and promotes pancreatic cancer cells aggressiveness. Sci Rep. 2017;7(1):5925. doi:10.1038/s41598-017-06132-z. PMID:28724890.
- Chen CH, Lu PJ, Chen YC, Fu SL, Wu KJ, Tsou AP, Lee YC, Lin TC, Hsu SL, Lin WJ, et al. FLJ10540-elicited cell transformation is through the activation of PI3-kinase/AKT pathway. Oncogene. 2007;26(29):4272–83. doi:10.1038/sj.onc.1210207. PMID:17237822.
- Sakai M, Shimokawa T, Kobayashi T, Matsushima S, Yamada Y, Nakamura Y, Furukawa Y. Elevated expression of C10orf3 (chromosome 10 open reading frame 3) is involved in the growth of human colon tumor. Oncogene. 2006;25(3):480–6. doi:10.1038/sj.onc.1209051. PMID:16170351.
- Tao J, Zhi X, Tian Y, Li Z, Zhu Y, Wang W, Xie K, Tang J, Zhang X, Wang L, et al. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour Biol. 2014;35(5):4389–99. doi:10.1007/s13277-013-1578-1. PMID:24390615.
- Chen S, Li P, Li J, Wang Y, Du Y, Chen X, Zang W, Wang H, Chu H, Zhao G, et al. MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cell Physiol Biochem. 2015;35(3):997–1007. doi:10.1159/000369755. PMID:25660220.
- Pan Y, Zhang J, Fu H, Shen L. miR-144 functions as a tumor suppressor in breast cancer through inhibiting ZEB1/2-mediated epithelial mesenchymal transition process. Onco Targets Ther. 2016;9:6247–55. doi:10.2147/OTT.S103650. PMID:27785072.
