ABSTRACT
Phosphoinositide-3-kinase regulatory subunit 3(PIK3R3) is overexpressed in different types of human cancer. We previously reported the important role of PIK3R3 in colorectal cancer (CRC). However, the prognosis effect of PIK3R3 in CRC is still remaining unclear. In this study, we explored online clinical databases to analyze the prognosis differences between higher and lower expression of PIK3R3 in CRC patients. Interestingly, we found that better disease-free survival (DFS) were occurred in patients with higher expression of PIK3R3, but there is no significant difference in overall survival (OS). For further, we showed that PIK3R3 could enhance 5-FU induced apoptosis by regulating the expression of thymmidine phosphorylase (TP). In conclusion, PIK3R3 could be considered as a predictor of 5-FU sensitivity for personalized treatment, and a therapeutic target for colorectal cancer.
1. Introduction
Colorectal cancer (CRC) is the most common cancer in the world, with its incidence and mortality rising rapidly in Asia.Citation1,Citation2 In the past decades, surgical resection and chemotherapy have been used as the main treatment for patients with CRC. 5-fluorouracil (5-FU) based chemotherapy plays a critical role in prolonged patient survival after surgery. However, drug resistance occurs frequently and results in treatment failure.Citation3,Citation4 Overcoming drug resistance and selecting appropriate treatment programs for patients are the key concerns that arise in 5-FU based chemotherapy.
PIK3R3 is a regulatory subunit of phosphatidylinositol kinase, encoded by PIK3R3.Citation5 It is well known that PIK3R3 is overexpressed in different types of cancer.Citation6,Citation7 Our previous study found that PIK3R3 is overexpressed in CRC and promoted cancer cell proliferation.Citation8,Citation9 However, there is no clinical study has reported the relationship between the expression level of PIK3R3 and patients' outcome in CRC. In this study, we first reported the different prognoses in patients with higher and lower expression of PIK3R3. All the data used in this study were derived from open online databases.
Thymidine phosphorylase (TP), also known as platelet derived endothelial cell growth factor (PD-ECGF), was upregulated in various malignancies compared to that in normal tissues.Citation10 Interestingly, TP has dual effect on tumor development and chemotherapy.Citation10,Citation11 On one hand, TP could stimulate cancer cell migration and proliferation.Citation10 On the other hand, some chemotherapeutic agents (5-fluorouracil [5-FU], capecitabine [Xeloda], Ftorafur, etc.] were converted to their active forms through TP enzymes.Citation10-12
In present study, we showed that the overexpression of PIK3R3 could enhance 5-FU based chemo-sensitivity and subsequently promote apoptosis in vitro and in vivo. Database analysis indicates that high level of PIK3R3 showed better outcome after surgery. In conclusion, our study indicates further information of PIK3R3 as a target to improve the chemotherapeutic efficacy of 5-FU based treatment for CRC.
2. Results
2.1. Overexpression of PIK3R3 predicts better DFS in patients with CRC
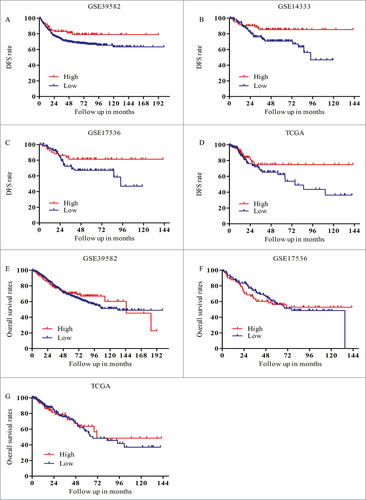
To better understand the prognosis of PIK3R3 in patients with CRC, we downloaded and analyzed the clinical data from TCGA and three other clinical datasets from GEO (as described in methods). CRC patients with metastasis and lack of follow up information were excluded, TCGA and three other CRC cohorts were available (TCGA n = 328, GSE39582 n = 557, GSE17536 n = 145, GSE14333 n = 226). DFS were analyzed in each group by using Kaplan-Meier analysis. As a result, we found that patients with elevated expression of PIK3R3 showed better DFS than those with lower expression of PIK3R3 (). However there is no significant difference in OS ().
Figure 1. Overexpression of PIK3R3 predicts better DFS in CRC patient. Three independent microarray data sets and TCGA microarray expression were used for this study. (A-D) DFS analysis A. P = 0.0565. B. P = 0.0127. C. P = 0.0769. D. P = 0.1760. (E-G) OS analysis E. P = 0.5277. G. P = 0.7728. F. P = 0.8671.
2.2. Relationship between the expression levels of PIK3R3 and TP
Surgical resection and chemotherapy are two major therapeutic schedules for patients with CRC. Chemotherapy is commonly used after surgery for the improvement of clinical prognosis. 5-FU and its prodrugs are the most commonly used clinical chemotherapeutic agents. All of these drugs are converted by TP to their active therapeutic form of 5-fluoro-2-deoxyuridine.Citation11,Citation12 We previously reported that PIK3R3 activated the NF-kB pathway, which is the main transcription factor of TP.Citation13,Citation14
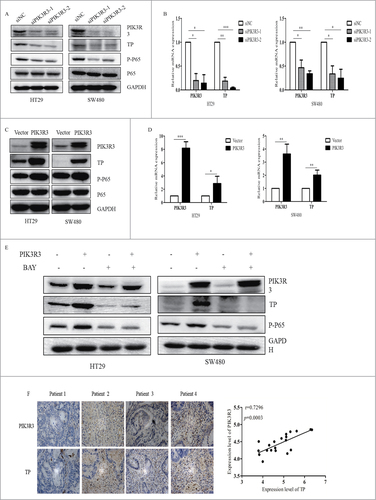
To confirm the relationship of PIK3R3 and TP, we used two CRC cell lines (HT-29 and SW480 derived from primary tumor). Our data showed that knockdown of PIK3R3 could induce the decreasing of TP (). Moreover, overexpression of PIK3R3 significantly increased the expression level of TP, and this process was accomplished by activation of the NF-kB pathway (). To confirm whether PIK3R3 regulation of TP were depending on NF-kB pathway, we established a rescue experiment. Indeed, we found that PIK3R3 couldn't increase TP expression at the present of NF-kB inhibitor (). We then analyzed the expression level of TP and PIK3R3 in primary tumors from 20 patients by IHC. Data showed that upregulated TP was observed in patients with overexpressed PIK3R3. We also noticed that the expression of PIK3R3 was positively correlated with TP expression (). Collectively, we suggest that PIK3R3 regulates the transcription of TP.
Figure 2. Relationship between the expression levels of PIK3R3 and TP. A-C. Protein expression of PIK3R3, TP, p-P65, P65 and GAPDH after transfection with Vector and PIK3R3 in HT29 and SW480 cells. B-D. Real-time PCR analysis of PIK3R3 and TP mRNA expression in HT29 and SW480 after transfected with Vector and PIK3R3. E. Western blot analysis of PIK3R3, TP, p-P65, P65 expression after transfected with PIK3R3 at the presence of BAY11-7082 (NF-Kb specific inhibitor). F. IHC analyses indicated that PIK3R3 levels were positively correlated TP expression in primary human CRC (n = 20, magnified × 400). Data are presented as mean ± SE. #p < 0.05; ##p < 0.01; ###p < 0.001, as compared with control group.
2.3. PIK3R3 enhances cell survival and chemosensitivity of CRC cells
PIK3R3 has been shown to play an important role in CRC cell proliferation and metastasis.Citation13 For further study of the function of PIK3R3 in CRC cells, we established stabilized PIK3R3 overexpressing cells as described in methods (, ). Cells were seeded at a density of 5000 cells/well, and cell survival was measured after 72 h. We observed that PIK3R3 promoted survival of CRC cells (, ). This result is consistent with our previous study.Citation8,Citation13
Figure 3. PIK3R3 enhances cell survival and chemosensitivity of CRC cells. (A, D). Overexpression of PIK3R3 in stable cell lines was confirmed by Western blotting. (B, D). Effects of PIK3R3 overexpression on cell growth were evaluated by CCK-8. (C, F). Cell viability was estimated after the cells treated with different dose of 5-FU for 72 h. Data shown are representative of 3 independent sets of experiments. Data are presented as the mean ± SE of three independent experiments. #p < 0.05; ##p < 0.01; ###p < 0.001, as compared with control group.
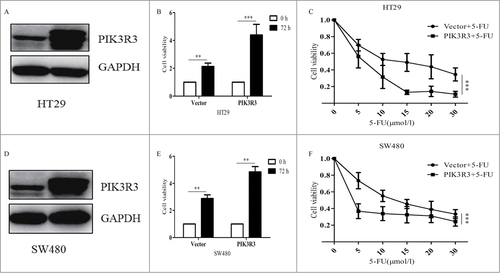
5-FU is the most commonly used anticancer agent in clinics; hence, it was used in the treatment group in this study. 50% inhibitory concentration (IC50) values of HT-29 and SW480 were 11.246 and 16.217 µmol/l, respectively. We then measured cell viability in both cells with empty vectors and cells with overexpressed PIK3R3, at different drug concentration. In line with our assumption, when exposed to 5-FU, proliferation in cells with overexpressed PIK3R3 significantly decreased as compared to that in cells bearing empty vectors (, ). These results strongly affirm our hypothesis and findings of our previous study. In conclusion, we noticed that PIK3R3 could enhance 5-fu toxicity in CRC cell lines.
2.4. Effect of PIK3R3 on cell cycle and apoptosis induced by 5-FU
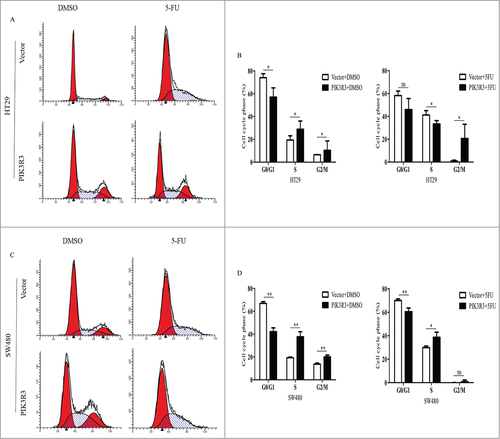
To better understand the effect of PIK3R3 on 5-FU-induced cytotoxicity in vitro, cells were treated with DMSO and 5-FU (2 µmol/l) for each group. Data showed that the percentage of cells in the S phase slightly increased in PIK3R3-overexpressed cells when treated with DMSO (). These results were consistent with our previous study. However, when the cells were treated with 5-FU, we observed a significant S phase blockage in vector cells than DMSO, but not in PIK3R3 overexpressed cells ().Citation15
Figure 4. Effect of PIK3R3 on cell cycle induced by 5-FU. HT29 and SW480 cells were treated with DMSO and 5-FU (2 µmol/l) for 72 h. (A-D). Cell cycle was determined by flow cytometric analysis using PI staining. Data are presented as the mean ± SE of three independent experiments. #p < 0.05; ##p < 0.01; ###p < 0.001, as compared with control group.
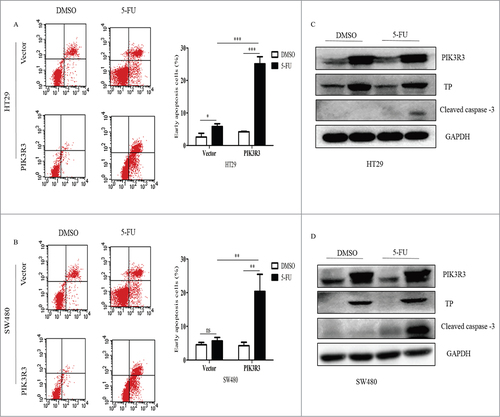
Additionally, overexpressed PIK3R3 had no effect on the apoptosis in cells treated with DMSO, but it did significantly increase the early apoptosis in cells exposed to 5-FU (). We also found increased cleaved caspase-3 level in PIK3R3 over expressed cells when the cells were treated with 5-FU (). These data suggest that PIK3R3 has no effect on 5-FU induced cell cycle arrest; however, it could increase 5-FU induced apoptosis in CRC cells.
Figure 5. Effect of PIK3R3 on apoptosis induced by 5-FU. HT29 and SW480 cells were treated with DMSO and 5-FU (2 µmol/l) for 72 h. (A-B). Early apoptosis was determined by flow cytometric analysis using Annexin V/PE staining. Annexin V was set as the horizontal axis and PE was set as the vertical axis. Mechanically damaged cells are located in the upper left quadrant, apoptotic or necrotic cells in the upper right quadrant, dual negative andnormal cells in the lower left quadrant, and early apoptotic cells in the lower right quadrant of the flow cytometric dot plot. (C-D). protein level of PIK3R3, TP and cleaved caspase-3 were analyzed by western blot. Data are presented as the mean ± SE of three independent experiments. #p < 0.05; ##p < 0.01; ###p < 0.001, as compared with control group.
2.5. PIK3R3 promotes CRC chemosensitivity in vivo
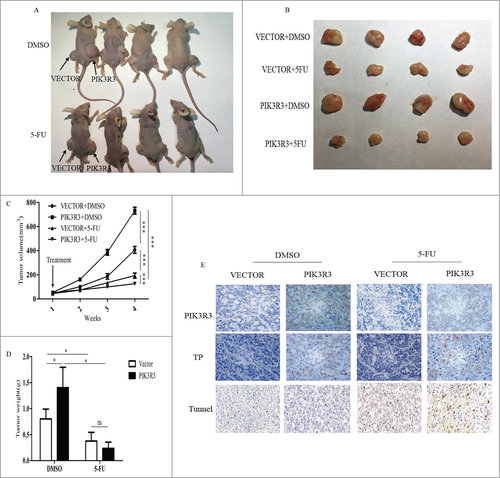
To determine the in vivo effects of PIK3R3 on tumor development and 5-FU chemosensitivity, the HT-29 cell line was used for animal studies, and cells overexpressing PIK3R3 were subcutaneously injected on the right side and empty vector on the left side on the back of each mouse (). After one week, the mice were randomly separated into two groups and treated as described in methods. Tumor volume and tumor weight in each group were measured and the P value was calculated (). Experimental data revealed that tumors with overexpressed PIK3R3 showed sharp increases in tumor size and weight when compared with those carrying vectors without 5-FU treatment (). This result was consistent with our previous study, which suggested that PIK3R3 contributed to tumor progression.Citation18 Interestingly, we found obviously low tumor progression in the PIK3R3-overexpressing group, when mice treated with 5-FU (). IHC results showed that more TP-positive cells were found in the PIK3R3-overexpressing group, with or without treatment with 5-FU (). TUNEL staining showed that 5-FU was able to induce apoptosis in both vector and PIK3R3-overexpressing groups (). Importantly, this effect was significantly increased in PIK3R3-overexpressing tumors. Therefore, consist with our in vitro study, overexpressed PIK3R3 enhances 5-FU induced apoptosis in mice xenograft tumors.
Figure 6. PIK3R3 promotes CRC chemosensitivity in vivo. HT29 cells were cultured, collected, washed and resuspended in culture medium (about 2 × 107/ml) and injected into subcutaneous of athymic nude mice (100 μl/tumor). Mice were treated as described in methods. A-B. After 3 weeks of treatment, all the mice were killed and the tumors were collected. C. The tumor growth curves of four groups during 4 weeks, the tumor volumes were monitored before they are killed. The length, width and depth of tumors are measured every weeks to determine tumor volume (mean±s.d, n = 6). D. The mean tumor weight of each group was calculated when mice were sacrificed. E. The expression of PIK3R3 and TP was detected by immunohistochemistry in each group magnified x 400. Data are presented as the mean ± SE of three independent experiments. #p < 0.05; ##p < 0.01; ###p < 0.001, as compared with control group.
3. Discussion
CRC is one of the commonly diagnosed cancers in the world, with high rates of morbidity and mortality.Citation1,Citation16,Citation17 Chemotherapy could improve patients' outcome, however, the response rates are not satisfied, mainly because the resistance during treatment process.Citation4,Citation18 Treatment failure has directly increased the recurrence of malignancy, which is the major threat to a patient's survival. Therefore, it is important to select more effective and comprehensive treatments, which is individualized treatment, for improve patients' clinical outcome. In the present study, we used database analysis and discovered that PIK3R3 predicts better outcome in CRC. For further study we found that PIK3R3 could accelerate tumor development, additionally it could enhance 5-FU sensitivity in vivo and in vitro. This is similar to the dual effect of TP in cancer development.Citation10-12
5-FU based chemotherapy has been used as first-line treatment in various malignant tumors. After its absorption, 5-FU is converted to its active form of FUDR through TP, and the toxic activity of 5-FU is enhanced.Citation10,Citation19 Moreover, 5-FU prodrugs, such as capecitabine, are transformed to their final active form by three steps. The key step in the process, which is the conversion of 5-deoxy-5-fluorouridine (5-DFUR) to active 5-FU, is catalyzed by TP.Citation20 Therefore, elevated levels of TP contribute to higher local concentration of the active drugs and increased sensitivity of the cells to chemotherapy of 5-FU.Citation21,Citation22 Studies have reported that high level of TP expression promotes chemosensitivity and predicts a better outcome in different cancer types.Citation12,Citation20,Citation22-24 Increased level of TP was observed in different pathways, among which activation of NF-kB pathway is one of the main reason for the regulation of TP expression.Citation11,Citation22 In this study, we observed that PIK3R3 could regulate the transcription level of TP by activation of the NF-kB pathway.
It is well established that PIK3R3 is related to tumor malignancy.Citation25 Several studies have reported that PIK3R3 was upregulated in different human malignancy.Citation6,Citation7 We previously reported that overexpression of PIK3R3 in CRC tissue than normal colorectal mucosa, and has important role on cell proliferation, tumor growth, and metastasis.Citation8,Citation9,Citation13 Blocking PIK3R3 by its specific inhibitor peptide Ad-N24 could prevent cancer metastasis in animals.Citation9 In this study we found the same phenomenon of PIK3R3 in CRC. Moreover, as a new function of PIK3R3 in CRC, we observed that PIK3R3 promotes 5-fu induced apoptosis by regulating the expression of TP.
In conclusion, we demonstrated that PIK3R3 has a dual effect on tumor development and chemotherapy. On one hand, it can stimulate tumor progression and metastasis. On the other hand, it can promote chemosensitivity to 5-FU and its prodrugs. Thus, this study indicates that CRC patients with higher expression of PIK3R3 was more suitable for 5-FU based chemotherapy, and have an improved prognosis after surgery.
4. Materials and methods
4.1. Ethics statement
All experiments were performed according to the guidelines and approved protocols of the Huazhong University of Science and Technology Animal Care and Use Committee. All experiments involving human participants have been approved by the Huazhong University of Science and Technology Ethics Committee, and we have obtained informed consent. All the methods were performed in accordance with relevant guidelines.
4.2. Bioinformatics analysis
We conducted bioinformatics analysis on CRC mRNA datasets to identify the expression of PIK3R3 in CRC. Clinical data was downloaded from The Cancer Genome Atlas (TCGA) website (http://www.cbioportal.org/) for 328 CRC samples. In addition, three different microarray datasets (GSE14333 Sieber OM et al. 2010, GSE39582 Marisa L et al. 2013, GSE17536 Smith JJ et al. 2009), were downloaded from the Gene Expression Omnibus (GEO) website. These datasets following criteria: obtained using a similar chip platform (Affymetrix, GPL570) with raw data in recent ten years, and survival information was available, obtaining no less than 150 patients. Overall survival (OS) and disease free survival (DFS) was measured by Kaplan-Meier method. R package survival was used to analyze the relationship between PIK3R3 expression and survival in this study.
4.3. Cell culture and antibodies
Human colon adenocarcinoma cell lines, HT29 and SW480, were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured at 37°C, 5% CO2 in Dulbecco's Modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclon, Thermo Scientific, Rockford, IL, USA). The primary antibodies PIK3R3, TP, cleave caspase-3, P65 and p-P65 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). NF-kB inhibitor BAY 11−7082 purchased from MedChem Express (MCE, USA).
4.4. Transfection and lentivirus infection
Cell transfection and lentivirus infection of stabilized PIK3R3 overexpressing cells were performed as described.Citation13 Short hairpin RNAs (shRNA-1, 5′-GGACUUGCUUUAUGGGAAA-3′, shRNA-2 5′-AAGCTTTGGACAACCGAGAAA-3′), targeting the p55PIK, and a non-targeting
RNA sequence, serving as negative control (Scr-shRNA), Cells were incubated at 37°C, 5% CO2 for 48–72 h before for further study, following which protein expression levels were determined by western blotting.
4.5. Cell viability assay
Cells were seeded in a 96-well plate at a density of 5000 cells/well. The next day, cells were treated with different doses of 5-FU for 72 h, then the culture medium was removed and replaced with CCK-8 solution for 2h at 37°C, then cells absorbance was measured at 450 nm using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
4.6. Cell cycle and apoptosis analysis
After treated with 5-FU and DMSO (2 µmol/l) for 72 h, the cells were harvested and fixed in 75% ice-cold ethanol at 4°C overnight. The next day, these cells were washed twice in PBS, then incubated with propidium iodide staining buffer (BD Pharmingen, San Diego, CA, USA) for 30 min and analyzed using FACSCalibur from BD Biosciences (Franklin Lakes, NJ, USA).
For apoptosis analysis, the cells were treated with 5-FU and DMSO (2 µmol/l) for 72 h. The cells were measured using FACSAria flow cytometer (BD Biosciences, USA), and apoptotic cells were detected using a PE Annexin V Apoptosis Detection Kit from BD Pharmingen (BD Pharmingen, San Diego, CA, USA). The experiment was performed in triplicate.
4.7. Western blot analysis
The cells were lysed in NP-40 lysis buffer containing 10 nmol/l of phenylmethylsulfonylfluoride and protease inhibitor. The lysate contents were measured using a bicinchoninic acid assay kit (Thermo). We added an appropriate volume loading buffer to each sample to its final concentration of 1X. Samples were boiled for 5 minutes at 95°C and then subjected to SDS-PAGE. Following SDS-PAGE, proteins were transferred to polyvinylidene difluoride membranes for immunoblotting. Blots were incubated overnight with primary antibodies at 4°C. The appropriate secondary antibodies were then used to determine the presence of various proteins in the samples.
4.8. RNA isolation and quantitative PCR assays
RNA was extracted using Trizol reagent (Invitrogen), and cDNA was synthesized from isolated RNA with a Superscript Reverse Transcriptase Kit (Transgene) following the manufacturer's instructions. We conducted quantitative PCR (qPCR) assays on an ABI7300 instrument using the Super SYBR Green kit (Transgene). Specific primer pairs were synthesized for the amplification and quantitation of the following gene expression products: PIK3R3 (5′-ATGTACAATACGGTGTGGAGTATG-3′ and 5′-GCTGGAGGATCCATTTCAAT-3′), TP (5′-TGGAAACATTCTGCTGGGAAG-3′ and 5′-CTGTGGAGGTGACAACTGCG-3′), and GAPDH (5′-GAGAGACCCTCACTGCTG-3′ and 5′-GATGGTACATGACAAGGTGC-3′). Light Cycler Data Analysis Version 3.1.102 (Roche) was used to analyze the obtained qPCR data. The fold change in mRNA was calculated through relative quantification (2-ΔΔCt).
4.9. TUNEL
Terminal deoxyribonucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay was performed to assess apoptosis. Tumor tissue sections were stained according to the manufacturer's instructions (In situ Cell Death Detection kit, fluorescein; Roche Diagnostics, Indianapolis, IN, USA; cat. no. 11684795910). Sections were observed using a fluorescent microscope (Olympus Corp.) and apoptotic cell nuclei were stained in green.
4.10. Immunohistochemical (IHC) analysis
Sections of paraffin-embedded tissues were dewaxed and heated in a pressure cooker. Endogenous peroxidase activity was blocked using H2O2. Sections were incubated with normal goat serum buffer for 30 minutes and then with a polyclonal goat anti-PIK IgG (diluted 1:100) for 1 h at room temperature. Sections were then incubated with rabbit anti-goat horseradish peroxidase–conjugated secondary antibody before aDABkit (BosterBio) was used to develop the color reaction. Sections were counter-stained with H&E and dehydrated through xylene and alcohol before mounting. Images were acquired with a Leica2000 fluorescence microscope.
4.11. Animal study
Four-week-old female athymic nude mice were housed under controlled conditions of light, and were fed for one week. Tumors were then generated by subcutaneous injection of 2 × 106 HT-29 cells. The tumor volume was calculated by the formula: volume = (length × widthCitation2)/2 and a tumor growth curve was drawn. After one week, the animals were randomly divided into two groups. DMSO was used for the control and 5-FU (100 mg/kg per week) was used for the treatment groups. All treatments were injected intraperitoneally weekly for 3 weeks before the animals were euthanized. Tumor samples were dissected and stored in 4% paraformaldehyde for further analysis.
4.12. Statistical analysis
GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) was used for all statistical analysis. Data are expressed as mean ± standard error (SE). Differences between groups were examined for statistical significance by t-test. Two-sided P-values were calculated, and a value of P < 0.05 was considered to be statistically significant.
Abbreviations
| CRC | = | colorectal cancer |
| DFS | = | disease-free survival |
| DMSO | = | Dimethyl sulfoxide |
| OS | = | overall survival |
| GEO | = | Gene Expression Omnibus |
| IHC | = | Immunohistochemical |
| PD-ECGF | = | platelet derived endothelial cell growth factor |
| PIK3R3 | = | Phosphoinositide-3-kinase regulatory subunit 3 |
| TP | = | thymmidine phosphorylase |
| TCGA | = | The Cancer Genome Atlas |
| 5-FU | = | 5-fluorouracil |
Acknowledgments
Supported by National Natural Science foundation (No. 81372323, No. 81372662).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262.
- Liu W, Zhang Z, Zhang Y, Chen X, Guo S, Lei Y, Xu Y, Ji C, Bi Z, Wang K. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biology & Therapy. 2015;16(4):511–517. doi:10.1080/15384047.2015.1017691.
- Peters GJ, Backus HH, Freemantle S, van Triest B, Codacci-Pisanelli G, van der Wilt CL, Smid K, Lunec J, Calvert AH, Marsh S and others. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta. 2002;1587(2–3):194–205. doi:10.1016/S0925-4439(02)00082-0.
- Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmuller C, Kahl C, Seipelt G and others. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15(10):1065–75. doi:10.1016/S1470-2045(14)70330-4.
- Pons S, Asano T, Glasheen E, Miralpeix M, Zhang Y, Fisher TL, Myers MG, Jr., Sun XJ, White MF. The structure and function of p55PIK reveal a new regulatory subunit for phosphatidylinositol 3-kinase. Mol Cell Biol 1995;15(8):4453–65. doi:10.1128/MCB.15.8.4453.
- Zhang L, Huang J, Yang N, Greshock J, Liang S, Hasegawa K, Giannakakis A, Poulos N, O'Brien-Jenkins A, Katsaros D and others. Integrative genomic analysis of phosphatidylinositol 3'-kinase family identifies PIK3R3 as a potential therapeutic target in epithelial ovarian cancer. Clin Cancer Res. 2007;13(18 Pt 1):5314–21. doi:10.1158/1078-0432.CCR-06-2660.
- Zhou J, Chen GB, Tang YC, Sinha RA, Wu Y, Yap CS, Wang G, Hu J, Xia X, Tan P and others. Genetic and bioinformatic analyses of the expression and function of PI3K regulatory subunit PIK3R3 in an Asian patient gastric cancer library. BMC Med Genomics. 2012;5:34. doi:10.1186/1755-8794-5-34.
- Hu J, Xia X, Cheng A, Wang G, Luo X, Reed MF, Fojo T, Oetting A, Gong J, Yen PM. A peptide inhibitor derived from p55PIK phosphatidylinositol 3-kinase regulatory subunit: a novel cancer therapy. Mol Cancer Ther. 2008;7(12):3719–28. doi:10.1158/1535-7163.MCT-08-0499.
- Xia X, Cheng A, Akinmade D, Hamburger AW. The N-Terminal 24 Amino Acids of the p55 Gamma Regulatory Subunit of Phosphoinositide 3-Kinase Binds Rb and Induces Cell Cycle Arrest. Mol Cell Biol. 2003;23(5):1717–1725. doi:10.1128/MCB.23.5.1717-1725.2003.
- Bronckaers A, Gago F, Balzarini J, Liekens S. The dual role of thymidine phosphorylase in cancer development and chemotherapy. Med Res Rev. 2009;29(6):903–53. doi:10.1002/med.20159.
- Elamin YY, Rafee S, Osman N, KJ OB, Gately K. Thymidine Phosphorylase in Cancer; Enemy or Friend? Cancer Microenviron. 2016;9(1):33–43. doi:10.1007/s12307-015-0173-y.
- Kobashi N, Matsumoto H, Zhao S, Meike S, Okumura Y, Abe T, Akizawa H, Ohkura K, Nishijima K, Tamaki N and others. The Thymidine Phosphorylase Imaging Agent 123I-IIMU Predicts the Efficacy of Capecitabine. J Nucl Med. 2016;57(8):1276–81. doi:10.2967/jnumed.115.165811.
- Wang G, Yang X, Li C, Cao X, Luo X, Hu J. PIK3R3 induces epithelial-to-mesenchymal transition and promotes metastasis in colorectal cancer. Mol Cancer Ther. 2014;13(7):1837–47. doi:10.1158/1535-7163.MCT-14-0049.
- Wang G, Chen C, Yang R, Cao X, Lai S, Luo X, Feng Y, Xia X, Gong J, Hu J. p55PIK-PI3K stimulates angiogenesis in colorectal cancer cell by activating NF-kappaB pathway. Angiogenesis. 2013;16(3):561–73. doi:10.1007/s10456-013-9336-y.
- Wang G, Cao X, Lai S, Luo X, Feng Y, Xia X, Yen PM, Gong J, Hu J. PI3K stimulates DNA synthesis and cell-cycle progression via its p55PIK regulatory subunit interaction with PCNA. Mol Cancer Ther. 2013;12(10):2100–9. doi:10.1158/1535-7163.MCT-12-0920.
- Hammond WA, Swaika A, Mody K. Pharmacologic resistance in colorectal cancer: a review. Ther Adv Med Oncol. 2016;8(1):57–84. doi:10.1177/1758834015614530.
- Li X, Wang S, Ren H, Ma J, Sun X, Li N, Liu C, Huang K, Xu M, Ming L. Molecular correlates and prognostic value of tmTNF-α expression in colorectal cancer of 5-Fluorouracil-Based Adjuvant Therapy. Cancer Biology & Therapy. 2016;17(6):684–692. doi:10.1080/15384047.2016.1187551.
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–26. doi:10.1038/nrc3599.
- Lam SW, Guchelaar HJ, Boven E. The role of pharmacogenetics in capecitabine efficacy and toxicity. Cancer Treat Rev. 2016;50:9–22. doi:10.1016/j.ctrv.2016.08.001.
- Terranova-Barberio M, Roca MS, Zotti AI, Leone A, Bruzzese F, Vitagliano C, Scogliamiglio G, Russo D, D'Angelo G, Franco R and others. Valproic acid potentiates the anticancer activity of capecitabine in vitro and in vivo in breast cancer models via induction of thymidine phosphorylase expression. Oncotarget. 2016;7(7):7715–31. doi:10.18632/oncotarget.6802.
- Daniele G, Gallo M, Piccirillo MC, Giordano P, D'Alessio A, Del Giudice A, La Porta ML, Perrone F, Normanno N, De Luca A. Pharmacokinetic evaluation of capecitabine in breast cancer. Expert Opin Drug Metab Toxicol. 2013;9(2):225–35. doi:10.1517/17425255.2013.759939.
- Chen CC, Chen LC, Liang Y, Tsang NM, Chang YS. Epstein-Barr virus latent membrane protein 1 induces the chemotherapeutic target, thymidine phosphorylase, via NF-kappaB and p38 MAPK pathways. Cell Signal. 2010;22(7):1132–42. doi:10.1016/j.cellsig.2010.03.008.
- Ogawa M, Watanabe M, Mitsuyama Y, Anan T, Ohkuma M, Kobayashi T, Eto K, Yanaga K. Thymidine phosphorylase mRNA expression may be a predictor of response to post-operative adjuvant chemotherapy with S-1 in patients with stage III colorectal cancer. Oncol Lett. 2014;8(6):2463–2468.
- Shigeta K, Ishii Y, Hasegawa H, Okabayashi K, Kitagawa Y. Evaluation of 5-fluorouracil metabolic enzymes as predictors of response to adjuvant chemotherapy outcomes in patients with stage II/III colorectal cancer: a decision-curve analysis. World J Surg. 2014;38(12):3248–56. doi:10.1007/s00268-014-2738-1.
- Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc Natl Acad Sci U S A. 2008;105(46):17724–9. doi:10.1073/pnas.0809844105.
