ABSTRACT
To determine PD1/PDL1 expression status in triple-negative breast cancer (TNBC) at both protein and mRNA levels, and to analyze the relationship between their expression and clinical parameters of the TNBC patients.
Immunohistochemistry and RNAscope were used to semi quantitively evaluate PD1/PDL1 protein and mRNA expression in 195 TNBC cases on tissue microarrays. Tumor infiltrating lymphocyte (TILs) abundance was assessed using hematoxylin-eosin staining. Both tumor cells and TILs expressed PDL1. PDL1 protein and mRNA positivity was 6.7% and 74.4% respectively in tumor cells, and 31.3% and 50.9% respectively in TILs. PDL1 protein and mRNA expressions had no significant association with patient prognosis. PD1 protein was only detected in TILs (70.3% positivity). PD1 protein expression was significantly related to PDL1 expression, higher TIL abundance, Ki-67 index, basal-like subtypes, and distant metastasis. Furthermore, it was significantly associated with longer disease free survival (P<0.001) and overall survival (P = 0.004). There was no significant association between PD1 mRNA expression and clinicopathological characteristics. PD1/PDL1 protein and mRNA expressions were inconsistent (kappa = 0.705 and 0.061, respectively). PD1 protein expression in TILs, but not PDL1 in tumor cells, was a favorable prognostic factor in TNBC. PD1/PDL1 mRNA and protein expressions were inconsistent.
Introduction
Breast cancer lacking the expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) is termed triple-negative breast cancer (TNBC). It is a clinically and molecularly heterogeneous disease that encompasses more than one entity.Citation1,Citation2 In total, 70–80% of the TNBCs belong to the basal-like subtype.Citation3,Citation4 Basal-like breast cancers express at least one basal marker, such as cytokeratin(CK) 5/6, CK14, or epidermal growth factor receptor (EGFR).Citation5,Citation6 TNBC, especially the basal-like subtype, does not benefit from any routinely available targeted therapy. And it brings about the poorest disease-free survival (DFS) and overall survival (OS) of any breast cancer.Citation7,Citation8 Lymphocytes were often seen in the tumor stroma of TNBC, namely tumor-infiltrating lymphocytes (TILs). Fridman et al reviewed the impact of TILs on the clinical outcomes of various epithelial cancers. In breast cancer, some reports showed TILs were associated better outcome.Citation9
As is well known, programmed cell death protein 1/programmed cell death ligand 1 (PD1/PDL1) are important immune checkpoint components that mainly regulate the function of tumor cells and TILs.Citation10 PD1/PDL1 activation in tumor cells and TILs attenuates local immune responses, thus shielding the tumor from T cell-mediated killing.Citation11-13 Clinical trials of agents targeting PD1/PDL1in these tumors have displayed obvious tumor regression and extended stabilization of disease in some patients.Citation14-16
Most research focuses on the relationship between PD1/PDL1 expression and targeted therapeutic effectiveness. The prognosis value of PDL1 expression mostly were based on mRNA level and the result were controversial in breast cancer.Citation17,Citation18 The correlations between PD1/PDL1 protein expression and clinicopathological factors such as TIL abundance, basal-like markers and prognosis are seldom researched.
In light of previous researches, and with the interest in finding an association between PD1/PDL1 expression and TNBC clinicopathological factors, we analyzed PD1/PDL1 expression status in TNBC on both the protein and mRNA level in tumor cells and TILs. We then analyzed the expression data in relation to clinicopathological parameters and prognosis of TNBC. Finally, the consistency of mRNA and protein of PD1/PDL1 expression were assessed.
Results
Clinicopathological characteristics of TNBC patients
This study enrolled altogether195 consecutive TNBC cases. The average age of the patients was 50.8 years old at diagnosis (range: 24–90). Clinicopathological characteristics of the patients are presented in . 177 patients (90.8%) were histologically non-specific type and the majorities had a high-grade lesion. 175 (89.7%) patients underwent modified radical mastectomy. 82 (42.1%) had positive lymph node metastases in surgical specimen; 103 patients (69.2%) received adjuvant chemotherapy only, and 32 (16.4%) patients received both adjuvant chemotherapy and radiotherapy. For the 135 patients who received chemotherapy, recurrence was observed in 67 patients (34.4%). 57 of them presented distant metastases and 10 (5.1%) had local recurrence. 35 (17.9%) patients died of the disease during the follow-up.
Table 1. Patient characteristics.
Protein expression of PDL1 in tumor cells and PD1 in TILs and their relationships with clinicopathological features
PD1 or PDL1 protein expression assays were successful in 195 cases. PDL1 protein expression could be detected in both tumor cells and TILs. There were 13 PDL1 positive cases as evaluated by tumor cells and 61 by TILs (namely tumor cells PDL1 protein positivity and TILs PDL1 protein positivity thereafter, respectively). Tumor cells PDL1 protein positivity was significantly related to TILs PDL1 protein positivity (P = 0.001) (). However, no significant relationship was found between tumor cells PDL1 protein positivity and assessed clinicopathological characteristics. Kaplan–Meier and Cox analysis showed that PDL1 positivity in neither tumor cells nor TILs was correlated with recurrence or survival.
Table 2. Tumor cells PDL1 protein and mRNA expression relationship with clinicopathological features.
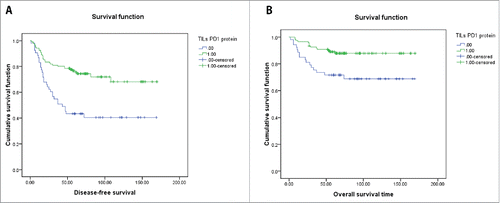
No tumor cells PD1 protein positivity was identified in the present cohort. On the other hand, TILs PD1 protein positivity was found in 70.3% of the cases. TILs PD1 protein positivity was significantly related to basal-like subtype, TILs abundance, TILs PDL1 protein positivity and distant metastasis(P < 0.05) (). Kaplan–Meier tests revealed that TILs PD1 protein positivity was significantly related to DFS and OS (P = 0.000 and 0.004, respectively) (A and B). Of all factors, stage, lymph node or distant metastases, tumor diameter>5cm, TILs abundance ≥30%, and TILs PD1 protein positivity were significantly correlated with DFS and OS in the univariate test. Multivariate analyses showed that TILs PD1 positivity was an independent prognostic factor.
Table 3. TILs PD1 protein and mRNA expression and their relationship with clinicopathological features.
PD1/PDL1 mRNA expression and its relationship with clinicopathological features
101 cases were subjected to in situ PD1 and PDL1 mRNA expression analyses with RNAscope. The successful rates for PD1 and PDL1 assays were 73.3% (74 out of 101 cases) and 82.2% (83 out of 101 cases), respectively.
Both tumor cells and TILs expressed PDL1 mRNA. There were 61 tumor cells PDL1 positive cases (74.4%), far higher than the rate of PDL1 protein expression (8.4%). Tumor cells PDL1 mRNA expression was also significantly related to TILs PDL1 mRNA positivity (P = 0.006), which is similar to the corresponding protein expression. Tumor cells PDL1 mRNA positive cases are more likely to appear in basal-like subtypes, tumors with higher Ki-67 index (≥20%), and high grade cases (P = 0.033, 0.025 and 0.034, respectively) (). PDL1 mRNA positivity in neither tumor cells nor TIL was significantly related to recurrence or survival by Kaplan-Meier analysis.
PD1 mRNA positivity was mainly found in TILs. Tumor cells PD1 mRNA expression was only detected in two cases. PD1 mRNA expression did not correlate with any characteristics (), except for tumor cells and TILs PDL1 mRNA expression.
Consistencies of PD1/PDL1 mRNA and protein expression in tumor cells and lymphocytes
The tumor cells PDL1 mRNA positive rate was 74.7%, figure was much lower when evaluated at protein level (8.4%). Tumor cells PDL1 protein positive case was 100% positive for mRNA, but not vice versa (). Some cases with high mRNA expression did not have corresponding high protein expression. The TILs PDL1 mRNA and protein positive rate was 50.6% and 21.7%, respectively. mRNA expression was not consistent with protein expression in either tumor cells or TILs. The Kappa value of PDL1 mRNA and protein consistence was 0.061 and 0.234 for tumor cells and TILs, respectively (). PDL1 positivity in tumor cells was also significantly correlated with TILs PDL1 expression at mRNA level.
Figure 2. PDL1 protein and mRNA staining in triple negative breast cancer. (A) IHC shows PDL1 protein expression in the tumor cells (200x). (B) RNAscope shows PDL1 mRNA expression in the same case as in A (400x). (C) IHC shows negative PDL1 protein staining in the tumor cells (200x). (D) RNAscope shows positive PDL1 mRNA expression in the same case as in C (400x).
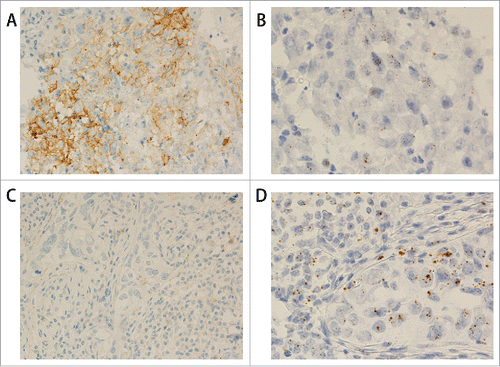
Table 4. PD1/PDL1 mRNA and their corresponding protein comparison.
The TILs PD1 mRNA positive rate was 36.5% and the protein positive rate was 51.4%. There were 11 PD1 protein positive cases that were PD1 mRNA negative. We didn't find high consistence in PD1 mRNA and protein expression, too (with kappa value = 0.705) (, ).
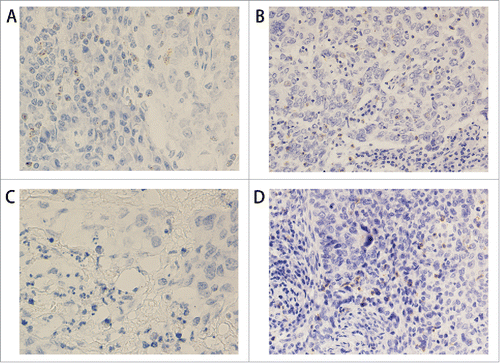
Figure 3. PD1 protein and mRNA staining in triple negative breast cancer. (A) RNAscope shows PD1 mRNA expression in TILs (400x). (B) IHC shows PD1 protein expression in the same case as in A (200x). (C) RNAscope shows negative PD1 mRNA expression in TILs (400x). (D) IHC shows PD1 protein expression in the same case as in C (200x).
Discussion
Blockade of the PD1/PDL1 pathway is a new and promising therapeutic approach in oncology. Research has focused primarily on the relationship between PDL1 expression level and response to PDL1 targeted therapy.Citation14,Citation19 However, the expression and prognostic value of PD1/PDL1expression in TNBC has been insufficiently investigated. To our knowledge, this is the first large cohort study (n = 195) that analyzed PD1 and PDL1 expression at both the mRNA and protein levels at the same time and analyzed the relationship between expression and clinicopathological characteristics.
In this study, there was a low tumor cells PDL1 protein positivity in the 195 enrolled cases (n = 13, 6.7%). PDL1 positivity was reported to be between19% and 64% in existing literatures.Citation20-22 However, the antibodies were from different companies and the positive cut off value was different. The low positivity in the present study might be attributable to the strict criteria recommended by the antibody producer, which required ≥25% tumor cell membrane staining, instead of 1% or 5% membrane or cytoplasm staining being sufficient for a case to be classified as PDL1 positive, as was the case in previous studies.
The results in the literature regarding the relationship between PDL1 expression and prognosis are conflicting. One study showed that tumor cells PDL1 expression was associated with poor prognosis in breast cancer.Citation23 However, other groups showed that PDL1 mRNA was associated with better prognosis.Citation24,Citation25 Our mRNA expression analysis found similar results to those described by Muenst et al, who found that PDL1 upregulation was associated with poor-prognostic features.Citation25 Tumor cell PDL1 mRNA expression tended to appear in cases with higher grade (P = 0.034), larger size (P = 0.094), higher Ki-67 index (≥20%, P = 0.025), and basal-like subtypes (P = 0.033). In our study, cases that were determined to be tumor cells PDL1 mRNA positive had shorter DFS and OS on average, which was consistent with their relation to poor-prognostic features, although this prognostic relationship was not significant. But we were unable to establish an association between tumor cells PDL1 protein positivity and clinical outcome, which was different to the result of Beckers et al.Citation22 They also noticed that PDL1 expression in stromal immune cells was associated with a lower mortality rate. Our findings in TILs also showed that PDL1 expression cases had longer DFS and OS, but this was not statistically significant.
This is the first study reporting that PD1 expression in triple negative breast cancer could be of prognostic significance. PD1 protein was only detected in lymphocytes. It was positively related to basal-like subtype, TIL abundance, and PDL1 protein expression, but negatively related to distant metastasis. More importantly, survival analysis showed PD1 protein positivity was an independent prognostic factor relating to TIL abundance, stage, large tumor size, and lymph node metastasis. A higher abundance of TILs was considered a good prognosticcator.Citation26-28 Our data also showed that higher numbers of TILs could significantly prolong DFS and OS. Previous research has verified that TILs in breast cancer were related to the expression of CD8+T-cell–related genes.Citation29 PD1 itself was reported to be expressed in activated T cells.Citation30 Therefore, it is possible that PD1 may act through a TIL-mediated anti-tumor inflammatory response rather than in tumor immune evasion. PD1 expression was significantly related to TILs PDL1 expression. This might explain why the PDL1 expression in TILs tended to be associated with better prognosis. Vassiliki et al pointed out that the effectiveness of anti-PD1 therapy requires the existence of both PD1+CD8+ T cells and PDL1expressing cells in the tumor microenvironment.Citation31 Although tumor cells PDL1 expression was low; there was a subset of TILs that did show PDL1 expression (31.3%). This may indicate that anti-PD1 or PDL1 therapy could still be effective even if tumors have low expression levels overall.
As PDL1 expression was analyzed simultaneously at mRNA and protein levels, we checked the concordance between the two methodologies. PDL1 protein and mRNA expression were inconsistent. For PDL1, there were cases with positive mRNA expression but negative protein expression. PDL1 mRNA expression in both tumor cells and TILs was not significantly related to survival. PD1 mRNA and protein expression were also not having a high concordance with a kappa value of 0.705. For PD1, positivity evaluated by mRNA was lower than protein and this may be due to mRNA degradation after long-term storage of the tissue. The oldest cases in our study has been stored for 10 years; some scientists proposed that analyzing the relationship between expression and clinical outcomes by protein was more reasonable because mRNA is not always translated into protein and protein is more stable than mRNA over long periods of time.Citation32 There was no significant association between PD1 mRNA expression and clinicopathological features, indicating that protein detection is more reliable than PD1 mRNA detection.
Our research had some limitations. Firstly, the use of TMA may not detect true protein or mRNA status due to intra-tumor heterogeneity. We tried to avoid this heterogeneity by sampling 3 from representative regions in every tumor. Another limitation is that we could not obtain mRNA expression data in all cases. When we performed statistics, we included a subset cohort that contained cases with both RNA and protein detection.
In conclusion, no significant association was established between PDL1 expression and patient survival. However, higher TIL PD1protein expression is associated with longer DFS and OS. The protein and mRNA expression levels of PD1/PDL1 were not consistent and protein detection may better reflect the true status of PD1/PDL1 gene expression.
Materials and methods
Case enrollment, Tissue Microarrays (TMAs) Preparations
Breast cancer cases that underwent curative operations between May 2002 and May 2012 and showed negative ER, PR, and HER-2 immunohistochemistry (IHC) were retrospectively enrolled from the pathology department of Peking Union Medical College Hospital (PUMCH). Cases with insufficient paraffin-embedded tumor tissue or those with preoperational neoadjuvant treatment were excluded from the study. Altogether, 195 cases with complete medical records were enrolled. Hematoxylin and Eosin (H&E) slides of all enrolled cases were reviewed by two experienced pathologists (XY Ren and HW Wu) to confirm the diagnosis and select representative tumor areas. Pathological features, including tumor size and nodal metastasis status were obtained. The follow-up time began from the date of surgery until June 30, 2016. The primary endpoint was the progression or relapsing of disease and the secondary endpoint was death. DFS and OS were defined as the period between surgery and the date of breast cancer-related relapse and death, respectively. A tissue microarray construction machine (Quick-Ray UT-06, UNITMA) was used. Three 1mm cores per case were obtained using a needle and arrayed in a recipient block for representativeness. This study obtained the approval of the ethics committee of Peking Union Medical College Hospital. Written informed consent was obtained from each patient.
Immunohistochemistry (IHC) staining and result interpretation
4μm thick sections were mounted on adhesion slides from tissue microarray blocks. The immunohistochemistry staining was done on a Ventana Benchmark XT autostainer (Ventana Medical Systems Inc., Tucson, AZ) according to the manufacturer's protocols. Antibodies against ER, PR, Her-2, EGFR, CK14, CK5/6, P53, Ki-67 and their respective interpretation criteria were the same as previously described, except that we changed the cutoff value for Ki67 index to 20%.Citation33 Briefly, all antigens were subjected to thermal remediation by 1 mM EDTA in 10 mM Tris buffer (pH 8.5). Antibodies against PD1, PDL1 and CK14 and their respective optimizations are listed in . Positive control and negative control slide were included in each run for each antibody. IHC slides were independently evaluated by two experienced pathologists. PD1, PDL1, and CK14 positivity was defined by criteria listed in . ER, PR, Her-2, EGFR, CK5/6, P53, Ki-67 HER-2 were interpreted as previously described.Citation33 Of note, Her-2 staining was evaluated according to the HER-2 test guide for breast cancer.Citation34 For HER-2 (2+) cases by IHC, fluorescent in situ hybridization (FISH) were conducted. HER-2–negative cases confirmed by FISH assay were enrolled in the study. Basal-like phenotype was defined by positivity for any of the following markers: CK5/6, EGFR, and CK14 in the present study.
Table 5. Antibodies used for the immunohistochemical analysis.
In Situ evaluation of PD1/PDL1 RNA expression
In situ assays for PD1/PDL1 transcripts in formalin-fixed paraffin embedded (FFPE) TMA samples was performed according to the manual of the RNAscope® FFPE 2.0 HD detection kit (Brown) (Advanced Cell Diagnostics, Hayward, CA, USA, Catalog Number 310035).Citation35-37 Before running the RNAscope® Assay on our samples for the first time, we ran positive and negative control probes on our samples to assess sample RNA quality and determine the optimal permeabilization conditions. Briefly, 5-μm sections were deparaffinized, boiled with preamplification reagent for 15min, subjected to protease digestion, followed by hybridization for 2h with target probes to human PD1/PDL1 mRNA (CAT #602021 and 600861). Hybridization signals were visualized by 3,3'-diaminobenzidine tetrahydrochloride (DAB) substrate. Positive staining was indicated by brown punctuate dots in the cytoplasm or nucleus. PD1/PDL1 mRNA expression levels were categorized into 5 grades according to the manufacture's scoring guidelines: score 0, no staining or <1 dot per 10 cells; score 1, 1–3 dots per cell (visible at 20–40x); score 2, 4–9 dots per cell (visible at 20–40x); score 3, 10–15 dots per cell and <10% positive cells have dot clusters (visible at 20–40x); score 4,>15 dots per cell and >10% positive cells have dot clusters (visible at 20–40x). Cases that scored higher than 0 were considered positive.
Evaluation of tumor infiltrating lymphocytes
The scoring of TILs was carried out on Hematoxylin&Eosin-stained TMA slides according to methods described previously by 2 experienced pathologists (XY Ren and HW Wu).Citation25 A score of 0 indicated virtual absence of TILs, 1+ = low TILs (<30%), 2+ = moderate (30–60%), and 3+ = marked increase in the lymphocytic infiltrate (>60%). Cases that could not be appropriately evaluated for technical reasons (e.g. bad staining, low tumor area, etc.) were designated as not evaluable. In the case of inter-observer discordance in TIL abundance, the particular slide was reviewed jointly and a agreeable score was assigned.
Statistical analysis
Statistical analysis was performed by SPSS 17.0 (SPSS, Chicago, IL, USA). Qualitative variables were compared using the chi-square test. The analysis of DFS and OS were performed firstly by a Kaplan–Meier plot and log-rank test and later with Cox regression to adjust for covariates. P-values were two sides and a p < 0.05 was considered statistically significant.
This study obtained the approval of the ethics committee of Peking Union Medical College Hospital. Written informed consent was obtained from each patient.
(Reagents listed in the Materials and Methods: PDL1 antibody were purchased from Ventana Medical Sstems,Inc,ready-to-use,code:263; PD1 antibody were from OriGene, MRQ-22; CK14 antibody were from NovocatraTM,Leica Biosystem, LL002. RNAscope® FFPE 2.0 HD detection kit (Brown) were from (Advanced Cell Diagnostics, Hayward, CA, USA, Catalog Number 310035; human PD1/PDL1 mRNA probes were from CAT #602021 and 600861.).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jefferey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature 2000; 406:747–752. doi:10.1038/35021093. PMID:10963602.
- Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med 2009; 360:790–800. doi:10.1056/NEJMra0801289. PMID:19228622.
- Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi:10.1002/cncr.22381. PMID:17146782.
- Tischkowitz M, Brunet JS, Begin LR, Huntsman DG, Cheang MC, Akslen LA, Nielsen TO, Foulkes WD. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;24:134. doi:10.1186/1471-2407-7-134.
- Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi:10.1158/1078-0432.CCR-07-1658. PMID:18316557.
- Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA Evans AJ, et al. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 2009; 15:2302–2310. doi:10.1158/1078-0432.CCR-08-2132. PMID:19318481.
- Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–167. doi:10.1038/modpathol.2010.200. PMID:21076464.
- Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–5657. doi:10.1200/JCO.2006.06.5664. PMID:17116942.
- Kurozumi S, Fujii T, Mastsumoto H, Inoue K, Kurosumi M, Horiguchi J, Kuwano H. Significance of evaluating tumor-infiltrating lymphocytes (TILs) and programmed cell death-ligand 1(PD-L1) expression in breast cancer. Med Mol Morphol. 2017;50(4):185–194. [ Epub ahead of print] doi:10.1007/s00795-017-0170-y. PMID:28936553.
- Dong H, Strome SE, Salomao DR, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi:10.1038/nm730. PMID:12091876.
- Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, Rosenberg SA. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114:1537–1544. doi:10.1182/blood-2008-12-195792. PMID:19423728.
- Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi:10.1084/jem.20090847. PMID:20008522.
- Hamel KM, Cao Y, Wang Y, Rodeghero R, Kobezda T, Chen L, Finnegan A. B7-H1 expression on non-B and non-T cells promotes distinct effects on T- and B-cell responses in autoimmune arthritis. Eur J Immunol. 2010;40:3117–3127. doi:10.1002/eji.201040690. PMID:21061440.
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi:10.1056/NEJMoa1200694. PMID:22658128.
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi:10.1056/NEJMoa1200690. PMID:22658127.
- Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi:10.1038/nature13904. PMID:25428503.
- Ghebeh H, Barhoush E, Tulbah A, Elkum N, Al-Tweigeri T, Dermime S. FOXP3+ Tregs and B7-H1+/PD-1+ T lymphocytes co-infiltrate the tumor tissues of high-risk breast cancer patients: Implication for immunotherapy. BMC Cancer. 2008;8:57. doi:10.1186/1471-2407-8-57. PMID:18294387.
- Sabatier R, Finetti P, Mamessier E, Adelaide J, Chaffanet M, Ali HR, Viens P, Caldas C, Birnbaum D, Bertucci F. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget. 2015;6:5449–5464. doi:10.18632/oncotarget.3216. PMID:25669979.
- Sznol M, Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res. 2013;19:1021–1034. doi:10.1158/1078-0432.CCR-12-2063. PMID:23460533.
- Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM, Akcakanat A, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2:361–370. doi:10.1158/2326-6066.CIR-13-0127. PMID:24764583.
- Gatalica Z, Snyder C, Maney T, Ghazalpour A, Holterman DA, Xiao N, Overberg P, Rose I, Basu GD, Vranic S, et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev. 2014;23:2965–2970. doi:10.1158/1055-9965.EPI-14-0654. PMID:25392179.
- Beckers RK, Selinger CI, Vilain R, Madore J, Wilmott JS, Harvey K, Holliday A, Cooper CL, Robbins E, Gillett D, et al. Programmed death ligand 1 expression in triple-negative breast cancer is associated with tumour-infiltrating lymphocytes and improved outcome. Histopathology. 2016;69:25–34. doi:10.1111/his.12904. PMID:26588661.
- Muenst S, Schaerli AR, Gao F, Däster S, Trella E, Droeser RA, Muraro MG, Zajac P, Zanetti R, Gillanders WE, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2014;146:15–24. doi:10.1007/s10549-014-2988-5. PMID:24842267.
- Bertucci F, Finetti P, Birnbaum D, Mamessier E. The PD1/PDL1 axis, a promising therapeutic target in aggressive breast cancers. Oncoimmunology. 2016;5:e1085148. doi:10.1080/2162402X.2015.1085148. PMID:27141340.
- Schalper KA, Velcheti V, Carvajal D, Wimberly H, Brown J, Pusztai L, Rimm DL. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res. 2014;20:2773–2782. doi:10.1158/1078-0432.CCR-13-2702. PMID:24647569.
- Mahmoud SM, Lee AH, Paish EC, Macmillan RD, Ellis IO, Green AR: The prognostic significance of B lymphocytes in invasive carcinoma of the breast. Breast Cancer Res Treat. 2012;132:545–553. doi:10.1007/s10549-011-1620-1. PMID:21671016.
- Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, Green AR. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–1955. doi:10.1200/JCO.2010.30.5037. PMID:21483002.
- Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J Clin Oncol. 2013;31:860–867. doi:10.1200/JCO.2011.41.0902. PMID:23341518.
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi:10.1038/nm1764. PMID:18438415.
- Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol. 2007;19:813–824. doi:10.1093/intimm/dxm057. PMID:17606980.
- Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. 2016;375:1767–1778 doi:10.1056/NEJMra1514296. PMID:27806234.
- Wang X, Teng F, Kong L, Yu J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Therapy. 2016;9:5023–5039. doi:10.2147/OTT.S105862.
- Ren X, Yuan L, Shen S, Wu H, Lu J, Liang Z. c-Met and ERbeta expression differences in basal-like and non-basal-like triple-negative breast cancer. Tumor Biol. 2016;37:11385–11395. doi:10.1007/s13277-016-5010-5.
- Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, et al. American Society of Clinical Oncology; College of American Pathologists: Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi:10.1200/JCO.2013.50.9984. PMID:24101045.
- Bordeaux JM, Cheng H, Welsh AW, Haffty BG, Lannin DR, Wu X, Su N, Ma XJ, Luo Y, Rimm DL. Quantitative in situ measurement of estrogen receptor mRNA predicts response to tamoxifen. PLoS One. 2012;7:e36559. doi:10.1371/journal.pone.0036559. PMID:22606272.
- Rajbhandari P, Schalper KA, Solodin NM, Ellison-Zelski SJ, Ping Lu K, Rimm DL, Alarid ET. Pin1 modulates ERalpha levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene. 2014;33:1438–1447. doi:10.1038/onc.2013.78. PMID:23542176.
- Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, Herbst RS, Gettinger SN, Chen L, Rimm DL. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94:107–116. doi:10.1038/labinvest.2013.130. PMID:24217091.