 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Objective: To explore the relationship between miR-122-5p and DUSP4 and their effects on gastric cancer (GC) cell mobility and invasiveness.
Methods: Abnormally expressed miRNAs and mRNAs were analyzed using microarrays. The miR-122-5p and DUSP4 mRNA expression levels in GC tissues and cells were determined by RT-qPCR. The target relationship between miR-122-5p and DUSP4 was validated by dual luciferase reporter assay. GC cell mobility and invasiveness were respectively observed by wound healing assay and transwell invasion assay. Western blot and immunohistochemistry were used for detection of the expressions of DUSP4 protein and MMP2 and MMP9 proteins related to cell invasion and migration. The migration and invasion abilities of gastric cancer cells in vivo were evaluated according to the number of lung metastatic nodules in mice.
Results: The expression of miR-122-5p in GC tissues and cells was significantly down-regulated, whereas DUSP4 expression was up-regulated. Bioinformatics prediction strategies and dual luciferase reporter assay verified the binding sites of miR-122-5p on 3′UTR of DUSP4 and the target relationship between miR-122-5p and DUSP4. Overexpression of miR-122-5p and knockdown of DUSP4 in BGC-823 cells observantly suppressed GC cell mobility and invasiveness, whereas downregulation of miR-122-5p expression promoted cell metastasis. MiR-122-5p inhibited GC cell mobility and invasiveness and pulmonary tumor metastasis via downregulation of DUSP4.
Conclusion: MiR-122-5p restrained migration and invasion abilities of GC cells by repressing DUSP4.
Introduction
Gastric cancer (GC) is identified as one of the most prevalent malignancies, with over 1 million death cases annually worldwide.Citation1 Up to now, smoking, alcohol consumption and helicobacter pylori infection have been found to be main culprits of GC.Citation2,Citation3 Due to its high metastasis allowing the neoplasm to rapidly progress into advanced stages, early diagnosis of GC remains difficult.Citation4 Although enormous advancement has been achieved in the treatment of GC, multiple therapeutic strategies still cannot cure such a malignant cancer.Citation5 Moreover, despite traditional TNM staging system is widely applied to clinical treatment, fewer biomarkers can accurately predict the metastasis and recurrence of GC.Citation6 However, more and more scientists start to focus on the potential mechanisms of GC and explore new therapeutic targets for the treatment of GC.Citation7
MicroRNAs (miRNAs), as a class of small, single stranded and non-coding RNAs, play critical parts in post-transcriptional regulation and function as either tumor inhibitor or facilitator in multiple cancers.Citation8,Citation9 It have been reported that these small molecules usually repress mRNAs and prevent transcription by combining with 3′-UTR.Citation10 Previous studies substantiated that dysregulation of miRNAs often occurs in multiple cancers.Citation11 Furthermore, aberrant expressions of miRNAs could interfere with tumorgenesis and development.Citation12,Citation13 In a large family of miRNAs, miR-122 has been thought to be down-regulated in several types of cancers, including GC,Citation14 gallbladder carcinoma,Citation15 bladder cancerCitation16 and so on. Furthermore, Ergün et al. has demonstrated the suppressive function of miR-122-5p on human breast cancer by targeting ADAM10, indicating that miR-122-5p may act as a therapeutic target in breast cancer.Citation17 Bai et al. revealed that miR-122-5p is commonly repressed in primary hepatocellular carcinomas (HCCs) and can inhibit somatotropin-releasing factor (SRF) activities and tumorgenesis,Citation18 suggesting that miR-122-5p can act as a tumor inhibitor through different molecular pathways. In the present study, we delve deeper into the molecular mechanisms of miR-122-5p in GC.
Dual-specificity phosphatases (DUSPs), as a heterogeneous group of protein phosphatases, can dephosphorylate tyrosine and serine or threonine residues.Citation19 Currently, 25 human DUSPs genes have already been explored, of which approximately 11 genes are found to result in MAPK signaling inactivation and others play a regulatory role in the dephosphorylation of diverse targets.Citation20 Several DUSPs involved in the development of specific human cancers have been reported over the past decade. Zhang et al. has revealed the inhibitory effects on sanguinarine on growth and invasiveness of human gastric cancer cells via down-regulation of DUSP4/ERK pathway.Citation21 De et al. verified that higher expression of DUSP4 was associated with a worse overall survival and with clinical characteristics of colorectal cancer patients.Citation22 Wei et al. substantiated that miR-101 inhibited macrophage-induced cell growth of hepatocellular carcinoma through targeting DUSP1.Citation23 Samsonov et al. demonstrated that miR-145 restrained papillary thyroid cancer cell reproduction by targeting DUSP6.Citation24 However, there are fewer studies on the relationship between DUSP6 and miR-122-5p and their underlying mechanism on GC.
The current study probed into the impacts of miR-122-5p and DUSP4 on GC and the association between them. We first conjectured that there was a target relationship between miR-122-5p and DUSP4 and miR-122-5p exerted a certain influence on GC cell by modulating DUSP4. Next, the target relationship between miR-122-5p and DUSP4 was validated by bioinformatics prediction strategies and dual luciferase reporter. The impacts of miR-122-5p on GC cells were detected by Immunohistochemistry, HE staining, wound healing assay, transwell assay and pulmonary metastasis model assay.
Materials and methods
Clinical specimens
GC tissues and the adjacent tissues (at least 3 cm away from the tumor) were collected from 30 patients undergoing surgical treatment at Jingjiang People's Hospital during the period between April 2015 and April 2017. All samples were assessed by clinicopathological examination and all subjects had been confirmed to receive no radiotherapy or chemotherapy. The samples were conserved in liquid nitrogen at −80°C for subsequent experiments. All experiments were ratified by the Ethics Committee of Jingjiang People's Hospital, and informed consents were provided by all subjects.
Microarray analysis
Differentially expressed miRNAs were screened out through the GSE78091 gene microarray hybridization, which contains 3 random pairs of GC tissues and adjacent tissues. Totally there were 3554 probes and 2095 human genes and some genes were measured by multiple probes. Abnormally expressed mRNAs were screened out through the GSE2685 gene microarray hybridization, which contains 22 primary human advanced gastric cancer tissues samples and 8 adjacent gastric tissues samples. Totally there were 7129 probes and 7129 human genes and each gene was measured by one probe. Fold change value greater than 2 and P less than 0.05 were regarded as the screening criteria for aberrantly expressed miRNAs and mRNAs.
Cell culture and cell transfection
HEK-293T cells were acquired from the stem cell bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). Human normal gastric mucosal cell line GES1 and human gastric cancer cell lines BCG-823, SGC-7901, MGC-803, HGC-27 were both procured from BeNa Culture Collection Company (Beijing China). Cells were cultured in RPMI-1640 medium (Gibco-Invitrogen, USA) with 10% FBS (Hyclone, USA) at 37°C and dissolved using 0.25% trypsin (Invitrogen, USA). Transfection was carried out through Lipofectamine 2000 following the instruction of manufacturer (Invitrogen). GC cells were assigned to five groups: (1) negative control (NC) group: GC cells transfected with scrambles and siRNA control. (2) miR-122-5p mimics group: GC cells transfected with miR-122-5p mimics. (3) miR-122-5p inhibitor group: GC cells transfected with miR-122-5p inhibitor. (4) si-DUSP4 group: GC cells transfected with DUSP4 siRNA (si-DUSP4). (5) MIX group: GC cells co-transfected with miR-122-5p inhibitor and si-DUSP4. The transfection efficiency was detected b under an inverted fluorescence microscope.
qRT-PCR
The extraction of total RNA was implemented with TRIzol reagent (Invitrogen) following the protocol of the manufacturer. Reverse transcription of extracted RNA and real-time quantitative PCR were successively performed according to instruction of RT-PCR kit (Invitrogen) and the fluorescence quantitative PCR kit (Invitrogen). Primer sequences used were presented at with GAPDH as an internal reference for mRNAs and U6 for miRNAs, and the relative expressions of miR-122-5p and DUSP4 were quantified by method.
Table 1. Primer sequences for RT-qPCR.
Western blot
Total proteins were isolated by using 1 ml radioimmunoprecipitation assay buffer with 10 l phenylmethanesulfonyl fluoride. Then protein concentration was examined by a bicinchoninic acid protein assay kit. Proteins were separated using 8% SDS-PAGE, then electro-transferred onto PVDF membranes, which were sealed with 5% non-fat milk for 2 h and incubated in the primary antibodies rabbit polyclonal anti-DUSP4 (ab72593, 1:500, Abcam Trading, Shanghai, China), rabbit polyclonal anti-MMP2 (ab37150, 1:1000, Abcam Trading), rabbit polyclonal anti-MMP9 (1:1000, ab38898, Abcam Trading), rabbit polyclonal anti-β-actin (ab8227, 1:1000, Abcam Trading) and rabbit polyclonal anti-GAPDH (ab9485, 1:2500, Abcam Trading). After rinsed by tris buffer saline containing 20% Tween twice, the PVDF membrane was incubated for 2 h at 37°C in the diluted HRP-labeled goat anti-rabbit IgG H&L secondary antibodies (1:2000). Then the membranes were rinsed using TBST for 20 min and visualized using an ECL Plus kit (Life Technology Inc., USA). GAPDH and
-actin were respectively considered as internal references for DUSP4 and MMP2/ MMP9.
Immunohistochemistry
The primary and secondary antibodies used in this assay were same as those in western blot. The tissues were fixed in 4% formaldehyde buffered with phosphate-buffered saline, then dehydrated, embedded in paraffin and serial section cut (4 μm thick). DUSP4 protein was detected after incubation with the rabbit polyclonal primary antibody (1:100) and HRP-conjugated anti-rabbit secondary antibody (1:2000). Afterwards, immunoreactivity in the sections was detected using a horseradish peroxidase (3,3′-diaminobenzidine substrate) kit (BioGenex, Fremont, CA, USA). The slides were then counterstained with hematoxylin (Beyotime, Shanghai, China), dehydrated and mounted. The sections were evaluated by an optical microscopy.
Hematoxylin-Eosin (HE) staining
The tissues embedded in paraffin were first deparaffinized in xylene, and then soaked in alcohol with gradient concentration (100%, 95%, 90%, 80%, 70%) for 10 min each. Hematoxylin was instilled to stain the tissues for 15 min after washed with distilled water for 2 min. The stained tissues were disposed in 1% hydrochloric acid ethanol to separate color for 20 s, and then soaked in warm water (50°C) for 5 min. The 0.25% eosin dye solution was instilled for counterstaining for 2 min, and the stained tissues were dehydrated by being soaked in gradient concentration of alcohol (70%, 80%, 90%, 95%, 100% ethyl alcohol) for 10 min each. The samples were observed by using an optical microscope (200 ×).
Wound healing assay
GC cells BCG-823 were inoculated on 6-well plates, and the germfree pipette was utilized to create a linear scratch in cell monolayer when cell confluence was up to 80%. The cells were then washed with PBS for 2∼3 times and cultivated in the serum-free medium. Lastly, we observed the wound distance at 0 h and 24 h by using image-Pro Plus 6.0 (Media Cybernetics, Inc., Rockville, MD, USA) and measure the cell mobility.
Transwell assay
The transfected cells were seeded onto the upper chamber along with serum-free medium. The medium containing 10% FBS was instilled into the lower chamber and incubated for 24 h. The cells on the upper chamber were gently removed by cotton swabs. The migrating or invading cells in the lower chamber were fixed with alcohol for 15 min and stained using crystal violet for 10 min. Lastly, the cells were detected under an optical microscope (200 ×).
Dual luciferase reporter assay
The sequence 3′UTR of DUSP4 was cloned into pMIR eukaryotic expression vectors (Promega, Madison, USA) including luciferase genes to construct pMIR-DUSP4-wt plasmids after recovery, purification and enzyme digestion. Likewise, the pMIR-DUSP4-mut plasmids were also constructed based on this method. The recombinant plasmids were identified through PCR, double-restrict-enzyme digestion of EarI (CTCTTC) and PspOMI (GGGCCC) and DNA sequence. The HEK-293T cells were first carefully seeded into 24-well plates. Following transfection, RIPA lysis buffer (Thermo Fisher Scientific) was added into the 24-well plates (100 μL / well). Then the mixture was centrifuged and the supernatant was collected into a 96-well plate. Renilla luciferase activity was properly normalized to firefly luciferase activity. Finally, the relative luciferase activities were was determined following the instructions of Dual-Luciferase Reporter Assay kit (Promega).
Murine pulmonary metastasis models
C57 BL/6 nude mice (4-5 weeks old, 18–20 g, male and female) were bought from Shanghai Experimental Animal Center (Shanghai, China) and maintained in specific pathogen free (SPF) level environment. All the experimental procedures were approved by Animal Ethics Committee in Jingjiang People's Hospital. 12 C57 BL/6 mice were randomly divided into two groups and labeled. 5 × 105 transfected cells in 100 μl of PBS were first injected into the tail vein of the mice. All mice were sacrificed at 24th day after injection. Next, the lung, liver and brain tissues were isolated and the number of lung metastatic nodules was counted under anatomical microscope. At last, 4% paraformaldehyde was used to fix lung, liver and brain tissues for HE staining and weighting.
Statistical analysis
Statistical software SPSS version 19.0 (SPSS Inc., USA) and Graphpad 6.0 (GraphPad Software, Inc., USA) were applied to data analysis. Results were presented in form of mean ± standard deviation. The differences between two groups were compared by Student's t-test, while that among three or more groups was analyzed using one-way ANOVA method. All experiments were conducted in triplicate and P < 0.05 was considered as an indicator of statistical significance.
Results
MiR-122-5p was low-expressed in gastric cancer tissues and cells
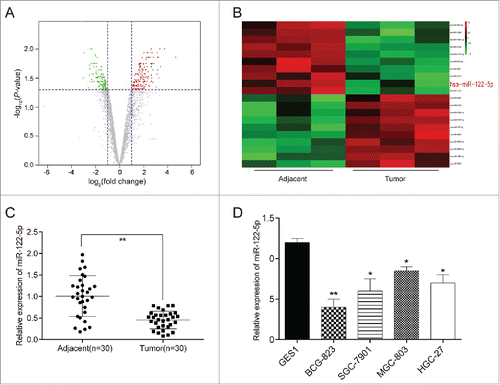
We took fold change over 2 and P < 0.05 as the filtration standard to screen out abnormally expressed miRNAs () and found that there were 111 up-regulated miRNAs and 76 down-regulated miRNAs in GC tissues in comparison with the adjacent tissues. Among these miRNAs, miR-122-5p was found to be down-regulated and miR-122-5p expression in GC tissues was averagely 2.99 times lower than that in the adjacent tissues (P = 0.001249428). The top 10 highly expressed miRNAs and the first 10 lowly expressed miRNAs were selected for the heat map (). Furthermore, qRT-PCR results exhibited that miR-122-5p expression was observantly lower in GC tissues than in adjacent tissues (P < 0.01, ). Beyond that, the expressions of miR-122-5p in human gastric cancer cell lines (BGC-823, SGC-7901, MGC-803 and HGC-27) were conspicuously lower compared with human normal gastric mucosal cell line GES1 (P < 0.05, ). BCG-823 cell line was chosen for subsequent experiments as presented the most significant difference of expression in contrast to GES1 cell line (P < 0.01).
Figure 1. MiR-122-5p was low-expressed in gastric cancer tissues and cells (A) Aberrant expression miRNAs in gastric cancer tissues were reflected by the volcano plot. (B) MiR-122-5p was a lowly expressed miRNA in gastric cancer tissues than in adjacent tissues shown in the heat map. (C) MiR-122-5p expression was significantly lower expression in GC tissues than in adjacent tissues determined by qRT-PCR. (D) The expressions of miR-122-5p in human gastric cancer cell lines (BGC-823, SGC-7901, MGC-803 and HGC-27) were conspicuously lower compared with human normal gastric mucosal cell line GES1 determined by qRT-PCR. *P < 0.05, **P < 0.01, compared with normal tissues or GES1 cell line.
DUSP4 was high-expressed in gastric cancer tissues and cells
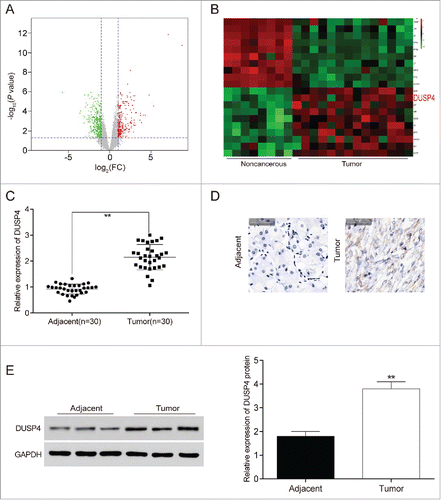
Likewise, we screened out aberrantly expressed mRNAs based on fold change over 2 and P < 0.05 (), finding that 245 mRNAs were up-regulated and 336 mRNAs were down-regulated in GC tissues. DUSP4 was found in the up-regulated mRNAs, and the DUSP4 expression level was averagely 1.01 times higher in gastric carcinoma tissues than in noncancerous tissues (P = 0.003334112). The top 10 highly or lowly expressed mRNAs were selected for the heat map (). In addition, qRT-PCR also exhibited that DUSP4 expression in GC tissues was dramatically higher compared with adjacent tissues (P < 0.01, ). In addition, the results from immunohistochemistry () and western blot () manifested that the expression of DUSP4 protein displayed a considerable increase in GC tissues (P < 0.01).
Figure 2. DUSP4 was high-expressed in gastric cancer tissues and cells (A) Abnormal expression mRNAs in gastric carcinoma tissues were reflected by the volcano plot. (B) DUSP4 was a highly expressed mRNA in gastric carcinoma tissues than in noncancerous gastric tissues shown in the heat map. (C) The expression of DUSP4 in GC tissues was dramatically higher than that in adjacent tissues determined by qRT-PCR. (D-E) The protein expression of DUSP4 experienced a significant increase in GC tissues detected by immunohistochemistry and western blot. **P < 0.01, compared with noncancerous or adjacent tissues.
DUSP4 was a potential target of miR-122-5p
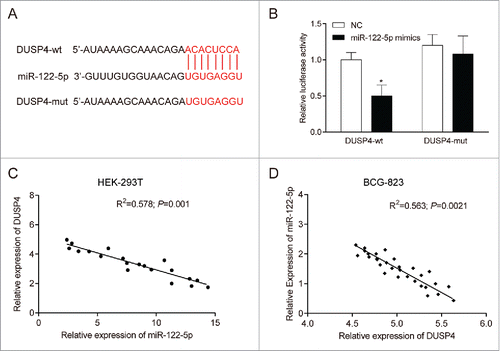
The candidate target gene of miR-122-5p was predicted by bioinformatics software, of which the results suggested that there was a potential binding site of miR-122-5p on the 3'-UTR of DUSP4 (). Meanwhile, dual luciferase reporter assay also illustrated that after transfected with pMIR-DUSP4-wild type plasmids, the luciferase activity of BGC-823 cells decreased by about 50% in miR-122-5p mimics group compared with NC group (P < 0.05, ). However, no significant difference was found between miR-122-5p mimics group and NC group in terms of luciferase activity of the cells after transfection with pMIR-DUSP4-mutated type plasmids (P > 0.05), suggesting that miR-122-5p could directly bind to 3'-UTR of DUSP4 and hinder the transcriptional activity of DUSP4. Furthermore, the results of qRT-PCR indicated that the relative expression of DUSP4 mRNA was negatively correlated to that of miR-122-5p both in HEK-293T cells (P < 0.05, ) and BCG-823 cells (P < 0.05, ), which further validated the above results.
Figure 3. DUSP4 was a potential target of miR-122-5p (A) There was a potential binding site of miR-122-5p on the 3'-UTR of DUSP4 predicted by bioinformatics software. (B) MiR-122-5p could inhibit the luciferase activity of the cells transfected wild-type 3′UTR of DUSP4, but had little influence on that of those transfected with mutated-type 3′UTR of DUSP4. (C-D) There was a negative correlation between DUSP4 and miR-122-5p in HEK-293T cells and BCG-823 cells. *P < 0.05, compared with NC group.
MiR-122-5p suppressed gastric cancer cell mobility and invasiveness by targeting DUSP4
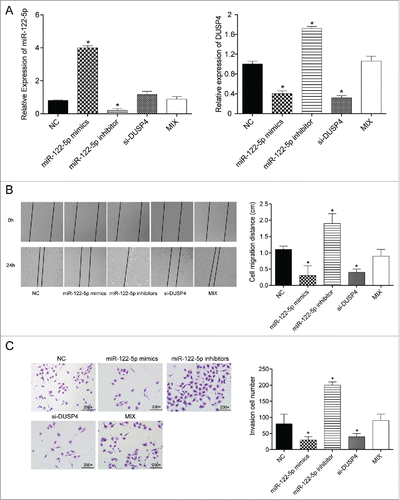
The mRNA expressions of miR-122-5p and DUSP4 after cell transfection were also examined by qRT-PCR. As shown in , the expression of miR-122-5p was dramatically up-regulated in miR-122-5p mimics group, whereas that in miR-122-5p inhibitor group was remarkably down-regulated (both P < 0.05). Conversely, DUSP4 expression levels in miR-122-5p mimics group and si-DUSP4 group drastically decreased, while those in miR-122-5p inhibitor group observantly increased (all P < 0.05). There was no conspicuous distinction between Mix group and NC group in terms of the mRNA expressions of miR-122-5p and DUSP4 (both P > 0.05). Additionally, wound healing assay results exhibited that the cell mobility ability was conspicuously attenuated after transfection with miR-122-5p mimics or DUSP4 siRNA, while the migration ability of the cells transfected with miR-122-5p inhibitor was significantly enhanced (all P < 0.05, ). No significant difference of cell mobility was detected between Mix group and NC group (P > 0.05). Furthermore, the GC cell invasion was determined by transwell invasion assay, of which the results suggested that the number of invasion cells in miR-122-5p mimics group and si-DUSP4 group was remarkably fewer compared with NC group, whereas that in miR-122-5p inhibitor group was considerably more than that in NC group (all P < 0.05, ). Little difference could be observed between NC group and Mix group (P > 0.05). Overall, miR-122-5p inhibited GC cell mobility and invasiveness by down-regulating DUSP4.
Figure 4. MiR-122-5p inhibited gastric cancer cell migration and invasion ability by targeting DUSP4 (A) The transfection efficiency was confirmed by qRT-PCR. (B) The wound distance and cell mobility of the experimental groups at different time points were measured by wound healing assay. The migration ability of the cells in miR-122-5p group or si-DUSP4 group was significantly lower than in NC group. The NC group and the Mix group could not be significantly distinguished (× 200). (C) The number of invasion cells in miR-122-5p mimics group and si-DUSP4 group was remarkably fewer compared with NC group, whereas that in miR-122-5p inhibitor group was considerably more than that in NC group detected by transwell invasion assay. Little difference could be found between NC group and the Mix group. *P < 0.05, compared with NC group.
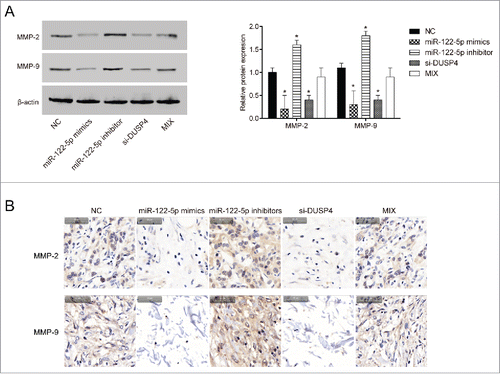
MiR-122-5p influenced the expression of MMP2 and MMP9 proteins by regulating DUSP4
Western blot and immunohistochemistry were both employed to identify the expressions of MMP2 and MMP9 proteins related to cell metastasis. Results from western blot () immunohistochemistry () both indicated that the expressions of MMP2 and MMP9 proteins were observantly down-regulated in BGC-823 cells transfected with miR-122-5p mimics or si-DUSP (P < 0.05). Conversely, the expressions of MMP2 and MMP9 were significantly up-regulated in BGC-823 cells transfected with miR-122-5p inhibitor (P < 0.05). The expressions of MMP2 and MMP9 in MIX group were equivalent to those in NC group (P > 0.05), suggesting that miR-122-5p affected the expression of MMP2 and MMP9 proteins related to cell migration and invasiveness by regulating DUSP4.
Figure 5. MiR-122-5p influenced the expression of MMP2 and MMP9 proteins in gastric cancer by regulating DUSP4 (A-B) The protein expressions of MMP2 and MMP9 were observantly down-regulated in BGC-823 cells transfected with miR-122-5p mimics or si-DUSP detected by western blot and immunohistochemistry. Conversely, the expressions of MMP2 and MMP9 were significantly up-regulated in BGC-823 cells transfected with miR-122-5p inhibitor. The expressions of MMP2 and MMP9 in MIX group were equivalent to those in NC group. *P < 0.05, compared with NC group.
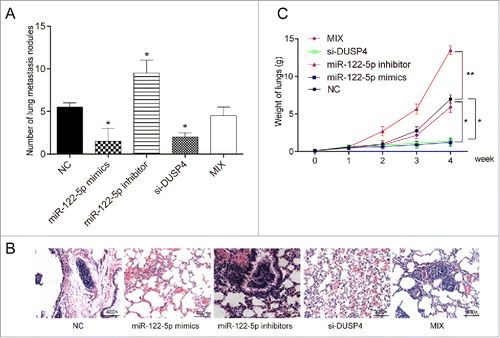
MiR-122-5p inhibited pulmonary tumor metastasis by repressing DUSP4 in vivo
To further confirm the effects of miR-122-5p and DUSP4 on GC cell metastasis, murine pulmonary metastasis model assay was performed, from which the results displayed that the number of mice lung metastasis nodules in miR-122-5p mimics group or si-DUSP4 group remarkably decreased, while that in miR-122-5p inhibitor group conspicuously increased (, P < 0.05). The number of mice lung metastasis nodules in Mix group was equivalent to that in NC group (P > 0.05). HE staining assay also indicated that the lungs of mice injected with transfected cells in miR-122-5p mimics group or si-DUSP4 group had almost no visible metastatic nodules, while that of mice in miR-122-5p inhibitor group was fully covered with nodules (, P < 0.05). The number of mice lung nodules in Mix group was equivalent to that in NC group (P > 0.05). Additionally, the lung weight of mice in miR-122-5p mimics group or si-DUSP4 group decreased significantly (P < 0.05), while that of mice in miR-122-5p inhibitor group increased sharply (, P < 0.01). There was no significant difference between Mix group and NC group in terms of lung weight (P > 0.05). Taken together, miR-122-5p restrained pulmonary metastasis by repressing DUSP4 in vivo.
Figure 6. MiR-122-5p inhibited pulmonary tumor metastasis by repressing DUSP4 in vivo (A) The number of murine pulmonary nodules determined by dissecting microscope. The number of mice lung metastasis nodules in miR-122-5p mimics group or si-DUSP4 group remarkably decreased, while that in miR-122-5p inhibitor group conspicuously increased. The number of mice lung metastasis nodules in Mix group was equivalent to that in NC group. (B) HE staining was conducted to detect the number of lung metastatic nodules after tail vein injection. The lungs of mice injected with transfected cells in miR-122-5p mimics group or si-DUSP4 group had almost no visible metastatic nodules, while that of mice in miR-122-5p inhibitor group was fully covered with nodules. (C) The lung weight of mice in miR-122-5p mimics group or si-DUSP4 group decreased significantly, whereas that of mice in miR-122-5p inhibitor increased sharply. There was no significant difference between Mix group and NC group in terms of lung weight. *P < 0.05, **P < 0.01, compared with NC group.
Discussion
The current experiments have already identified that miR-122-5p was low-expressed, while DUSP4 was high-expressed in GC tissues and cell lines. Over-expression of miR-122-5p or knockdown of DUSP4 could inhibit the metastasis of GC cells. We disclosed that miR-122-5p played a tumor-suppressive role in cell mobility and invasiveness by targeting DUSP4 in GC cells. Meanwhile, we also validated that miR-122-5p impeded pulmonary tumor metastasis by repressing DUSP4 in vivo.
The molecular mechanism of GC was a process involving multiple factor, including epigenetic factors and genetic factors. Up to now, considerable researches have substantiated that miRNAs play a crucial function in the gene regulation in multiple cancers. Zhang et al. unraveled that miR-200c suppressed GC cell reproduction and metastasis through directly targeting FN1.Citation25 Wang et al. demonstrated that miR-190b enhanced the radio-sensitivity of GC cells through negatively regulated Bcl-2.Citation26 Ding et al. revealed that miR-27a stimulated the tumorgenesis of GC by activating the AKT/GSK3β pathway.Citation27 Mi et al. validated that miR-181a-5p facilitated the development of GC via RASSF6-modulated MAPK signaling pathway.Citation28 Recently, miR-122-5p has been identified as a potential novel biomarker. For instance, miR-122-5p could play key roles in diagnosis and specific early prognosis of acute myocardial infarction.Citation29, Citation30 In the present research, we substantiated that miR-122-5p could exerted inhibitory influence in GC cell metastasis through down-regulating DUSP4, suggesting that miR-122-5p could function as a tumor inhibitor in GC.
Additionally, the regulation function of DUSP4 was reported in many previous study reports,Citation31–33 which indicated that DUSP4 had been thought to be a critical signaling regulator of multiple cellular processes, such as cell invasion, migration, differentiation and so forth. Zhang et al. found that sanguinarine exerted a inhibitive influence on GC cell growth and invasion via regulation of the DUSP4/ERK pathway.Citation21 Lai et al. reported that the increased phosphorylation of ERK, correlated with decreased abundance of the phosphatases DUSP4 and DUSP6, might counteract resistance to c-Met inhibitors in GC.Citation34 Our study verified that DUSP4 could be repressed by miR-122-5p, hence impeding the progression of GC. Besides, a few signaling pathways have been found to be involved in DUSP-mediated regulation like ERK1/2/JNK pathwayCitation35 and MAPK pathways.Citation36 However, a lack of study on signaling pathway was one of the limitations in this study. Apart from it, the small number of tissue samples might make our results not completely convincing.
In conclusion, over-expression of miR-122-5p could inhibit the cell mobility and invasiveness in GC through targeting DUSP4. The finding not only laid a foundation for an intensive study on GC, but also provided two promising therapeutic targets in GC treatment.
Conflict of interest
The authors declare no potential conflicts of interest.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64. doi:10.1016/S0140-6736(16)30354-3.
- Li L, Ying XJ, Sun TT, Yi K, Tian HL, Sun R, Tian JH, Yang KH. Overview of methodological quality of systematic reviews about gastric cancer risk and protective factors. Asian Pac J Cancer Prev: APJCP. 2012;13:2069–79. doi:10.7314/APJCP.2012.13.5.2069.
- Kato M, Asaka M. Recent knowledge of the relationship between Helicobacter pylori and gastric cancer and recent progress of gastroendoscopic diagnosis and treatment for gastric cancer. Jpn J Clin Oncol. 2010;40:828–37. doi:10.1093/jjco/hyq119.
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115–32.
- Zheng L, Jiao W, Song H, Qu H, Li D, Mei H, Chen Y, Yang F, Li H, Huang K, et al. miRNA-558 promotes gastric cancer progression through attenuating Smad4-mediated repression of heparanase expression. Cell Death Dis. 2016;7:e2382. doi:10.1038/cddis.2016.293.
- Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol: official journal of the European Society for Medical Oncology. 2016;27:763–9. doi:10.1093/annonc/mdw040.
- Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH, Lu PH, Lin KH. Potential Diagnostic, Prognostic and Therapeutic Targets of MicroRNAs in Human Gastric Cancer. Int J Mol Sci. 2016;17. doi:10.3390/ijms17060945.
- Sundaram P, Hultine S, Smith LM, Dews M, Fox JL, Biyashev D, Schelter JM, Huang Q, Cleary MA, Volpert OV, et al. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res. 2011;71:7490–501. doi:10.1158/0008-5472.CAN-11-1124.
- Bar-Eli M. Searching for the ‘melano-miRs’: miR-214 drives melanoma metastasis. EMBO J. 2011;30:1880–1. doi:10.1038/emboj.2011.132.
- An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, Nie Y, Zhao Q. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015;6:e1766. doi:10.1038/cddis.2015.123.
- Zhang X, Zeng J, Zhou M, Li B, Zhang Y, Huang T, Wang L, Jia J, Chen C. The tumor suppressive role of miRNA-370 by targeting FoxM1 in acute myeloid leukemia. Mol Cancer. 2012;11:56. doi:10.1186/1476-4598-11-56.
- Zhu M, Zhang N, He S, Lui Y, Lu G, Zhao L. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014;588:600–7. doi:10.1016/j.febslet.2013.12.028.
- Miko E, Margitai Z, Czimmerer Z, Varkonyi I, Dezso B, Lanyi A, Bacsó Z, Scholtz B. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585:1191–6. doi:10.1016/j.febslet.2011.03.039.
- Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang Q, Chen L, Pang X, Leng W, Bi F. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep. 2014;31:1863–70. doi:10.3892/or.2014.3004.
- Lu W, Zhang Y, Zhou L, Wang X, Mu J, Jiang L, Hu Y, Dong P, Liu Y. miR-122 inhibits cancer cell malignancy by targeting PKM2 in gallbladder carcinoma. Tumour Biol. 2015. doi:10.1007/s13277-015-4308-z.
- Wang Y, Xing QF, Liu XQ, Guo ZJ, Li CY, Sun G. MiR-122 targets VEGFC in bladder cancer to inhibit tumor growth and angiogenesis. Am J Transl Res. 2016;8:3056–66.
- Ergun S, Ulasli M, Igci YZ, Igci M, Kirkbes S, Borazan E, Balik A, Yumrutaş Ö, Camci C, Cakmak EA, et al. The association of the expression of miR-122-5p and its target ADAM10 with human breast cancer. Mol Biol Rep. 2015;42:497–505. doi:10.1007/s11033-014-3793-2.
- Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–27. doi:10.1074/jbc.M109.016774.
- Patterson KI, Brummer T, O'Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–89. doi:10.1042/BJ20082234.
- Huang CY, Tan TH. DUSPs, to MAP kinases and beyond. Cell Biosci. 2012;2:24. doi:10.1186/2045-3701-2-24.
- Zhang R, Wang G, Zhang PF, Zhang J, Huang YX, Lu YM, Da W, Sun Q, Zhu JS. Sanguinarine inhibits growth and invasion of gastric cancer cells via regulation of the DUSP4/ERK pathway. J Cell Mol Med. 2017;21:1117–27. doi:10.1111/jcmm.13043.
- De Vriendt V, De Roock W, Di Narzo AF, Tian S, Biesmans B, Jacobs B, Budinska E, Sagaert X, Rossi S, D'Ario G, et al. DUSP 4 expression identifies a subset of colorectal cancer tumors that differ in MAPK activation, regardless of the genotype. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2013;18:516–24. doi:10.3109/1354750X.2013.819038.
- Wei X, Tang C, Lu X, Liu R, Zhou M, He D, Zheng D, Sun C, Wu Z. MiR-101 targets DUSP1 to regulate the TGF-beta secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget. 2015;6:18389–405. doi:10.18632/oncotarget.4089.
- Samsonov R, Burdakov V, Shtam T, Radzhabovsmall a CZ, Vasilyev D, Tsyrlina E, Titov S, Ivanov M, Berstein L, Filatov M, et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:12011–21. doi:10.1007/s13277-016-5065-3.
- Zhang H, Sun Z, Li Y, Fan D, Jiang H. MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomed Pharmacother = Biomedecine & pharmacotherapie. 2017;88:285–92.
- Wang C, Qiao C. MicroRNA-190b confers radio-sensitivity through negative regulation of Bcl-2 in gastric cancer cells. Biotechnol Lett. 2017;39:485–90. doi:10.1007/s10529-016-2273-2.
- Ding L, Zhang S, Xu M, Zhang R, Sui P, Yang Q. MicroRNA-27a contributes to the malignant behavior of gastric cancer cells by directly targeting PH domain and leucine-rich repeat protein phosphatase 2. J Exp Clin Cancer Res: CR. 2017;36:45. doi:10.1186/s13046-017-0516-2.
- Mi Y, Zhang D, Jiang W, Weng J, Zhou C, Huang K, Tang H, Yu Y, Liu X, Cui W, et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017;389:11–22. doi:10.1016/j.canlet.2016.12.033.
- Cortez-Dias N, Costa MC, Carrilho-Ferreira P, Silva D, Jorge C, Calisto C, Pessoa T, Robalo Martins S, de Sousa JC, da Silva PC, et al. Circulating miR-122-5p/miR-133b Ratio Is a Specific Early Prognostic Biomarker in Acute Myocardial Infarction. Circ J: official journal of the Japanese Circulation Society. 2016;80:2183–91. doi:10.1253/circj.CJ-16-0568.
- Yao XL, Lu XL, Yan CY, Wan QL, Cheng GC, Li YM. Circulating miR-122-5p as a potential novel biomarker for diagnosis of acute myocardial infarction. Int J Clin Exp Pathol. 2015;8:16014–9.
- Barajas-Espinosa A, Basye A, Angelos MG, Chen CA. Modulation of p38 kinase by DUSP4 is important in regulating cardiovascular function under oxidative stress. Free Radic Biol Med. 2015;89:170–81. doi:10.1016/j.freeradbiomed.2015.07.013.
- Schmid CA, Robinson MD, Scheifinger NA, Muller S, Cogliatti S, Tzankov A, Müller A. DUSP4 deficiency caused by promoter hypermethylation drives JNK signaling and tumor cell survival in diffuse large B cell lymphoma. J Exp Med. 2015;212:775–92. doi:10.1084/jem.20141957.
- Kim SY, Han YM, Oh M, Kim WK, Oh KJ, Lee SC, Bae KH, Han BS. DUSP4 regulates neuronal differentiation and calcium homeostasis by modulating ERK1/2 phosphorylation. Stem Cells Dev. 2015;24:686–700. doi:10.1089/scd.2014.0434.
- Lai AZ, Cory S, Zhao H, Gigoux M, Monast A, Guiot MC, Huang S, Tofigh A, Thompson C, Naujokas M, et al. Dynamic reprogramming of signaling upon met inhibition reveals a mechanism of drug resistance in gastric cancer. Sci Signal. 2014;7:ra38. doi:10.1126/scisignal.2004839.
- Xu T, Wu X, Chen Q, Zhu S, Liu Y, Pan D, Chen X, Li D. The anti-apoptotic and cardioprotective effects of salvianolic acid a on rat cardiomyocytes following ischemia/reperfusion by DUSP-mediated regulation of the ERK1/2/JNK pathway. PloS One. 2014;9:e102292. doi:10.1371/journal.pone.0102292.
- Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW, Gómez H, Cook RS, Arteaga CL. Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer Res. 2013;73:6346–58. doi:10.1158/0008-5472.CAN-13-1385.
