ABSTRACT
Objective: This study investigated miR-422a and PLP2 expressions in breast cancer cells and breast cancer stem cells (BCSCs). Besides, their influences on polymorphism changes were observed.
Methods: Flow cytometry and fluorescence-activated cell sorting was performed and CD24−/CD44+ cells were sorted from breast cancer cells and recognized as BCSCs. Microarray was applied to search for the differentially expressed miRNAs and mRNAs between MCF7 and BCSCs. The aberrant expression of miR-422a and PLP2 was further confirmed by RT-qPCR and the direct targeted relationship was verified by dual-luciferase reporter assay. After in vitro transfection, the expression of miR-422a and PLP2 were manipulated and biological functions of BMSCs were compared with CCK-8, colony formation and sphere formation assay. The tumorigenesis ability of transfected BMSCs was also investigated in NOD/SCID tumor mice models.
Results: BMSCs were successfully established from MCF7 cells and miR-422a expression was downregulated while PLP2 level decreased in BMSCs. MiR-422a directly targets the 3′UTR of PLP2 and suppressed its expression. Besides, the up-regulation of miR-422a contributed to weakened ability of proliferation and microsphere formation of BMSCs, while PLP2 overexpression facilitated those biological abilities. Tumorigenesis of BMSCs in mice models was impaired by either overexpression of miR-442a or silencing of PLP2.
Conclusion: Up-regulation of miR-422a attenuated microsphere formation, proliferation and tumor formation of breast cancer stem cells via suppressing the PLP2 expression.
KEYWORDS:
Introduction
Breast cancer was the primary cause of cancer-related deaths worldwide.Citation1 Sustained proliferation, activated invasion, migration and resistance to cell death contributed to its rapid recurrence and low survival rate. Cancer stem cells are potential origins of breast cancer and may dictate tumor phenotype.Citation2 They share properties including self-renewal, tumor initiation, indefinite replicative potential, and the ability to generate differentiated progeny. Breast cancer stem cells (BCSCs) could be recognized by activities of CD44+/CD24− and enzyme aldehyde dehydrogenase (ALDH+).Citation3 Besides, BCSCs possessing both the CSC markers (CD44+CD24− and ALDH+) have shown the greatest tumor-initiating capacity in previous studies.Citation4 SOX6 has previously reported to be important for the maintenance of stem cells and OCT4 as well a weighted regulator of stemness in various cancers, both as representative tumor markers.Citation5,Citation6 Therefore, stemness properties downregulation is of great significance in breast cancer studies.
Recently, miRNAs (small non-coding RNAs) have been reported to function as oncogenes or tumor suppressors. They are capable of regulating breast cancer at the post-transcriptional level.Citation7 MiR-422a has been proved to suppress cell proliferation in non-small cell lung cancer by targeting at KLK4.Citation8 MiR-422a downregulation could aggravate cell proliferation, migration and invasion of osteosarcoma.Citation9 The aberrant expression of miRNAs may also regulate the stemness of cancer stem cells and contribute to the tumorigenicity.Citation10,Citation11 Via the control of DNMT3b protein, miR-221 facilitates the self-renewal and differentiation in breast cancer stem cells and promotes the pathogenesis of breast cancer.Citation12 However, the potential role of miR-422a in cell stemness has not been explored, which would possibly span miRNA studies in stem properties of breast cancer.
Proteolipid protein 2 (PLP2), also known as A4 protein, has been shown to be upregulated in several cancers, including melanoma cancer, alzheimer's disease, renal cell carcinoma and so on.Citation13,Citation14 Its overexpression enhances cell proliferation, adhesion, and invasion in melanoma cells.Citation13 It is also involved in CCR1 signaling pathway and stimulates the migration of human osteosarcoma cells.Citation15 However, except from the study of Longo A et.al, no correlation between PLP2 and breast cancer cells have been reported before.Citation16 Our microarray results found the aberrant expression of PLP2 in breast cancer stem cells compared with the origin MCF-7 cells, the influence of PLP2 on stemness was then explored.
Herein, we investigated influences of miR-422a and PLP2 on breast cancer stem cells. By changing miR-422a and PLP2 expressions, polymorphism changes in vitro and tumor growth in vivo were recorded, which suggested BCSCs properties fluctuations. These discoveries may provide a novel insight into breast cancer studies from an aspect of reducing stemness properties.
Methods
Cell culture and transfection
MCF-10A and MCF-7 cells were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) and maintained in DMEM (Gibco, Grand Island, NY, USA) supplemented with 10% FBS, 100 ml penicillin, 100 ml streptomycin and 2 mM L-glutamine. Isolated BCSCs were cultured in serum-free RPMI1640 medium containing bFGF、EGF、B27 (Shanghai Novoprotein technology co. LTD, China). When cells in culture had grown to 80% confluence, the tissue culture medium was replaced with medium containing 1.6 mM (24 wells) or 3 mM (6 wells) of the plasmid per well. MiR-422a mimics, negative mimics, PLP2-pcDNA3.1, pcDNA3.1 plasmid vector and PLP2 shRNA were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China) and transfected into cells using Lipofectamine 2000 (Life Technologies, USA) according to manufacturer's instructions. Groups were set as follows: NC group included cells transfected with negative mimics; miR-422a mimics group included cells transfected with miR-422a mimics; PLP2 group included cells transfected with PLP2-pcDNA3.1; miR-422a mimics+PLP2 group included cells transfected with miR-422a mimics and PLP2-pcDNA3.1.
Bioinformatics analysis
GSE68271 and GSE99394 were used to screen out differentially expressed miRNAs and mRNAs. R language was applied in the formation of heat map and volcano plot. MiRanda (http://www.microrna.org/microrna/getMirnaForm.do) was used to predict the target site of miRNA. A higher mirSVR score indicated a more stable complementarity and a higher PhastCons score indicated a greater conservation.
BCSCs isolation
These cells were sorted into two sub-populations based on surface markers CD44+ and CD24-: cells with CD24-/CD44+ (BCSCs) and non CD24-/CD44+ (non BCSCs). Breast cancer cells were trypsinized, suspended into single-cell mixtures and washed with PBS. After incubation on ice for 30 min with monoclonal antibodies specific for human cell surface markers including CD44 and CD24 (PE conjugated) (eBioscience, San Diego, CA, USA). After washing, cells were analyzed using a FACS Calibur flow cytometer (BD Biosciences, Franklin Lakes, New Jersey) using Cell Quest Pro software (BD Biosciences).
RT-qRCR
Total RNA was extracted with Trizol reagent (Life Technologies, USA) and reverse-transcribed with PrimeScript II 1st Strand cDNA Synthesis Kit. RT-qPCR was then performed using SYBR Premix Ex Taq according to the manufacturer's protocol to detect the expression level of miR-422a and PLP2. The expression of RNA was calculated using the 2-ΔΔCt method. The primers were provided by Sangon (shanghai, China). All primers were detailed in .
Table 1. Primers for qRT-PCR.
Western blot
Total protein was separated on by SDS-PAGE polyacrylamide gel electrophoresis polyacrylamide and then electro-transferred onto PVDF membranes at constant voltage 80 V. After 1 h blocking with BSA, the membrane were incubated with antibodies. Primary antibodies included mouse anti-PLP2 (1:5000; BD Biosciences, San Jose, CA, USA), mouse anti-OCT4 (1:1000, Abcam, Cambridge, MA, USA) and mouse anti-SOX2 (1:2000, Abcam). Secondary antibodies included rabbit anti-mouse IgG-HRP (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Mouse anti-GAPDH (Kangchen, Shanghai, China) was used as an internal parameter. All antibodies were diluted with 5% milk in phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBS-T) and incubated for either 1 h at room temperature or overnight at 4°C. Protein bands were visualized with ECL Western blotting substrate (Pierce, Rockford, IL, USA).
Dual-Luciferase reporter assay
For luciferase reporter experiments, the 3′UTR sequence of PLP2 predicted to interact with miR-422a or a mutated sequence within the predicted target sites was synthesized and inserted into the Mlu I and Hind III sites of a pGL3 vector (Promega, Madison, WI, USA) (wt forward 5′-GAATTCGCGAACTTCCCTCA-3′, wt reverse 5′- GGATCCTTTGATGAAAGGATTACT -3′, mut forward: 5′-AGTTAGATTTCAGAGTCCAGGCCCTAGGTTGG-3′, mut reverse: 5′-ACCTAGGGCCTGGACTCTGAAATCTAACTCC-3′). These constructs were known as pGL3-PLP2-3′UTR-wt or pGL3-PLP2-3′UTR-mut, respectively. For the reporter assay, the HEK 293T cells (Cell library of the Chinese academy of sciences, Shanghai, China)\ cells were plated onto 12-well plates, and then co-transfected with the above-mentioned constructs and 5 ng of pRL-TK (Promega), with or without miR-214 or miR-con using Lipofectamine 2000 reagent (Invitrogen). Approximately 48 h later, the cells were harvested. The luciferase and Renilla signals were determined using the Dual-Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions.
Spheroid formation assay
Breast cancer stem cells were transfected with miRNAs or plasmids for 24 h in six-well plates. Single–cell suspensions were prepared and cells were plated on six-well ultralow attachment plates (Corning-Costar Inc., Corning, NY, USA) at a density of 1,000 cells/ml. Cells were grown in DMEM supplemented with 1% N2 Supplement (Invitrogen), 2% B27 Supplement (Invitrogen), 20 ng/ml human platelet growth factor (Sigma-Aldrich), 100 ng/ml epidermal growth factor (Invitrogen) and 1% antibiotic-antimycotic (Invitrogen). After 7 ‘d’ of culturing, the mammospheres with no less than 50. 0 μm diameter were counted.
Colony formation assay
MCF-7 cells were trypsinized, counted, and seeded for colony formation assay in 6-well plates at 300 cells per well. During colony growth, the culture medium was replaced every 3 ‘d’. At 7 ‘d’ after seeding, the colonies were stained with 0.02% crystal for 1 h and then were counted in 5 random chosen fields under an inverted phase-contrast microscope (Olympus IX73; Olympus, Tokyo, Japan). The colony was counted only if it contains more than 50 cells. Each treatment was carried out in triplicate.
CCK-8 assay
Cells were plated in 96-well plates at a density of 3 × 104 per well. At 24, 48, 72 and 96 h post-plating, 10 μl Cell Counting Assay Kit-8 solution (Sigma, St. Louis, MO) was added to each well and incubated for 2 h, and the absorbance at 490 nm was measured using a microplate reader. Results represented the average of samples from three independent studies per well.
NOD/SCID Xenograft model
Cells were cultured in antibiotic-free normal growth medium supplemented with FBS till 80% confluency. PLP2 shRNA and pre-miR-422a were sub-cloned into pCMV-Red-Firefly-Luc plasmid and the lentivirus was packed in 293T cells. PCMV-Red-Firefly-Luc vector was utilized as negative control. BCSCs were infected with the harvested lentivirus (MOI = 10). After the selection with Doxycycline (2μg/ml) for two weeks, stably expressing miR-442a or PPL2-knockdown BCSCs were injected into female non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice (6 weeks old) fat pad at a concentration of 2 × 106 /ml and 5 × 106/ml. Tumor growth in vivo was measured by Bioluminescent IVIS imaging system 200 (Xenogen CA, USA). The animals were sacrificed after 80 ‘d’. Animal maintenance and experiments were performed in accordance with the animal care guidelines of the Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All animal experiments were approved by the Animal Care and Use Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology.
Statistical analyses
Each experiment was done in triplicate. Data were expressed as the mean ± standard deviation (x±s). When two groups were compared, the differences between groups were analyzed using Student's t-test and when more than two groups were compared, a one-way analysis of variance (ANOVA) was used. Statistical analyses were performed in GraphPad Prism 6.0 (GraphPad Softwares Inc, San Diego, CA, USA). A probability level of 0.05 indicated statistical significance.
Results
MiR-422a was lowly expressed while PLP2 was highly expressed in BCSCs
Flow cytometry were applied to selected breast stem cells in breast cancer cells with stem properties marker CD24-CD44+. Besides, OCT4 and SOX6 protein expression were tested to validate the stemness of isolated BCSCs (Figure S1A-B). MiRNA expressions in breast cancer cell line MCF-7 and BCSCs were evaluated by microarray analysis (Table S1). Among 33 down-regulated miRNAs, miR-422a barely been investigated before and was selected for further study (). GSE99394 also indicated twenty most significantly overexpressed genes, including PLP2 (). As suggested in Figure S1C, miR-422a was significantly correlated with TLR2 signaling pathway. MRNA expressions in breast cancer cell line MCF-7 and BCSCs were evaluated by microarray analysis at dataset (Table S2). Besides, differentially expressed mRNAs shown in Table S2 indicated that they were significantly related to RNA binding and B cell homeostasis (Figure S1C). Then, qRT-PCR validated that miR-422a expression was significantly higher in CD24-CD44+ marking BCSCs than in normal epithelial cells (P = 0.0038) and breast cancer cells (P = 0.0027). Besides, no obvious difference was observed between non-BCSCs group and breast cancer group (). QRT-PCR and western blot also confirmed the prediction: PLP2 had a higher mRNA and protein expression in CD24-CD44+ marking BCSCs compared with breast cancer cells (P = 0.001, P = 0.0048) and normal epithelial cells (P = 0.004, P = 0.003). Therefore, PLP2 was high expressed in BCSCs compared with normal breast cancer cells (all P < 0.05, -).
Figure 1. MiR-422a was lowly expressed while PLP2 was highly expressed in BCSCs. (A) Microarray analysis indicated that miR-422a was lower expressed in BCSCs than in breast cancer cell line MCF-7. (B) Microarray analysis indicated that PLP2 was higher expressed in BCSCs than in breast cancer cells. (C) qRT-PCR of miR-422a expression verified that miR-422a was lower expressed in BCSCs than in breast cancer cells. (D-E) qPCR and western blot analysis verified that PLP2 was higher expressed in BCSCs than in breast cancer cells. **P < 0.01, ***P < 0.001 indicated significant difference compared with normal epithelial cells. ##P < 0.01, ###P < 0.001 indicated significant difference compared with breast cancer cells.
MiR-422a down-regulated PLP2 expression in BCSCs
MiRanda predicted SOX6 as well as PLP2 as miR-422a targets. However, SOX6 expression didn't show significant differences in BCSCs and PLP2 was therefore the only suitable target (Figure S2C-2D). MiR-422a might bind to PLP2 wt at position 282 (). A mirSVR score of −0.2454 which was lower than −0.1 indicated that the binding was stable and a PhastCons score of 0.5865 suggested a relatively high conservation. To confirm the direct target relationship between miR-422a and PLP2, dual luciferase reporter assay was performed. It turned out that miR-422a mimics significantly decreased the luciferase activity in pGL3-PLP2 wt group but caused minor changes in pGL3-PLP2 mut group (P < 0.05, ), indicating that miR-422a directly targeted at PLP2 and inhibited its expression. Their regulation relationship was further confirmed in -. MiR-422a mimics increased its expression in miR-422a mimics group and PLP2-pcDNA3.1 up-regulated PLP2 expression. Meanwhile, miR-422a mimics significantly down-regulated PLP2 expression while PLP2 overexpression exerted little influence on miR-422a expression. However, miR-422a mimics+ PLP2-pcDNA3.1 group showed no much changes of PLP2. Therefore, miR-422a could directly target at PLP2 and inhibit the expression of PLP2 in BCSCs.
Figure 2. MiR-422a downregulated PLP2 expression in BCSCs. (A) MiRanda predicted site 285 as binding sites. (B) Dual-Luciferase reporter assay verified that miR-422a directly targeted at PLP2 wild type and downregulated luciferase activity (P < 0.05). (C-F) MiR-422a expression was increased in miR-422a mimics group and miR-422a mimics+PLP2 group. PLP2 expression was decreased in miR-422a mimics group and increased in PLP2 group, but was not influenced significantly in miR-422a mimics+PLP2 group. **P< 0.01 indicated significant difference compared with NC group.
MiR-422a suppressed BCSCs properties by PLP2 down-regulation
As shown in , the transfection of miR-422a mimics and PLP2 overexpression vector significantly changed stem cell population. MiR-422a overexpression greatly reduced CD44+CD24- cell population in isolated BCSCs, indicating a decrease on BCSCs stemness. On the contrary, PLP2 overexpression increased stem cell population in isolated BCSCs which meant an enhancement on BCSCs stemness (). Altered protein expressions of OCT4 and SOX2 were also recorded. MiR-422a could elevate OCT4 and SOX2 expressions and overexpression of PLP2 would suppress expressions of stemness markers (). However, the combination of miR-422a and PLP2 failed to either change stem cells population or influence expressions of those proteins.
Figure 3. MiR-422a suppressed BCSCs properties by PLP2 downregulation. (A) BCSCs population decreased in miR-422a mimics group and increased in PLP2 overexpression group. (B) Stemness markers OCT4 and SOX2 expression were significantly suppressed in miR-422a mimics group and elevated in PLP2 overexpression group.
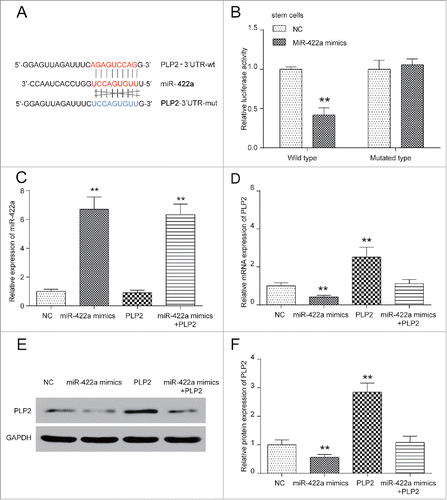
MiR-422a inhibited while PLP2 promoted BCSCs proliferation
To investigate the role of miR-422 and PLP2 in BCSCs, the cell proliferation rate of co-transfected compounds was determined using tumorsphere formation assay, CCK-8 and colony formation assay. Influences of miR-422a and PLP2 on cell proliferation in normal breast cancer cells were also proved to show similar trend in BCSCs. MiR-422a mimics significantly inhibited colony formation and cell viability and PLP2 greatly improved colony formation and cell viability (Figure S2A-2B). Since this study focused on BCSCs, we only study changes in stem cells. In tumorsphere formation assay, the number of glomus cells with no less than 50 μm diameter was counted. Knockdown efficiency of shRNAs was firstly validated in Figure S2E-2F which indicated shRNA1 as the most efficient inhibitor of PLP2 expression and therefore was chosen in following PLP2 knockdown experiments (Figure S2E-2F). As shown in , the number decreased in miR-422a mimics group, increased in PLP2 group, but showed no marked changes in the miR-422a mimics+PLP2 group. Therefore, miR-422a mimics suppressed the sphere formation efficiency in BCSCs, while PLP2 exerted the opposite effect. Similarly, fewer colonies were observed in miR-422a mimics group while more colonies were detected in PLP2 group, which indicated that miR-422a inhibited cell proliferation, PLP2 promoted cell proliferation, but miR-422a and PLP2 together exert no obvious effect on cell proliferation (P>0.05, ). CCK-8 at the same time proved the effect of miR-422a and PLP2: OD value in miR-422a mimics group was significantly lower and that in PLP2 group was remarkably higher than NC and mix group, suggesting effectiveness on stemness suppression of miR-422a overexpression and PLP2 suppression (P>0.05, ).
Figure 4. MiR-422a inhibited while PLP2 promoted BCSCs proliferation. (A) Bright field images of sphere formation assays displayed that miR-422a mimics led to a decrease while PLP2 overexpression led to an increase in tumorsphere formation. (B) MiR-422a inhibited cell proliferation and PLP2 accelerated the proliferation while joint expression of miR-422a and PLP2 revealed no significant changes in cell formation assay. (C) Cell proliferation was significantly repressed in miR-422a mimics group while upregulated in PLP2 group in CCK8 assay. Similar proliferation rate was detected in the mix group compared with NC group. **P < 0.01 indicated significant difference compared with NC group. Scale bar: 50 μm.
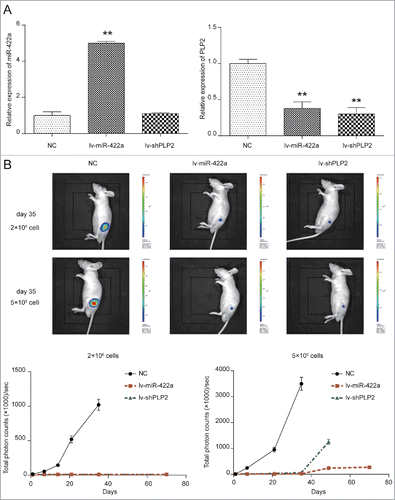
Upregulation of miR-422a or knockdown of PLP2 inhibited tumorigenesis of BMSCs in vivo
For investigating the impact of miR-442a and PLP2 expression on the tumor formation ability of BMSCs, we first established the stably miR-422a overexpressing and PLP2-knockout BCSCs. The successful manipulating of miR-422a and PLP2 expression in BCSCs was confirmed in . IVIS image of mice at ‘d’ 35 were shown in . NOD/SCID mice in NC group inoculated with 2 × 106 /ml cells into fat pad formed 1.5 cm diameter tumors in 35 ‘d’, while those in lv-miR-422a group and lv-shPLP2 group failed to form palpable tumors (both P < 0.001). However, NOD/SCID mice in lv-NC group inoculated with 5 × 106 /ml cells formed tumors of 1.5cm diameter in only 28 ‘d’, but tumors in lv-shPLP2 grew more slowly (both P < 0.01). Total photon counts in lv-shPLP2 group reached 1000 after 49 ‘d’ while those in miR-422a overexpressing group were barely zero in total 70 ‘d’. Therefore, increased level of miR-422a or downregulation of PLP2 could attenuate the tumor formation of BCSCs.
Figure 5. MiR-422a mimics and PLP2 shRNA attenuated BCSCs tumorigenesis in vivo. (A) MiR-422a expression was increased in miR-422a overexpressing group and PLP2 expression was decreased in lv-miR-422a group and lv-shPLP2 group. (B) Mice in lv-NC group inoculated with 2 × 106 /ml cells into fat pad formed 1.5 µm tumors in 35 ‘d’, while those in lv-miR-422a group and lv-shPLP2 group failed to form palpable tumors. Mice in lv-NC group inoculated with 5 × 106 /ml cells into fat pad formed 1.5 µm tumors in 28 ‘d’ but lv-shPLP2 group as well as lv-shPLP2 group displayed no significant changes. **P < 0.01 indicated significant difference compared with NC group.
Discussion
In the present study, we have discovered that miR-422a suppressed BCSCs properties by suppressing PLP2. After overexpressing miR-422, microsphere formation, cell proliferation abilities in vitro and tumor initiation rate in vivo were significantly down-regulated in BCSCs. Therefore, miR-422a might function as a suppressor in tumorigenesis of breast cancer. This discovery would not only contribute to miRNA investigation in breast cancer but also facilitate the development of treatment strategies for breast cancer.
BCSCs could cause treatment relapse as they have higher migratory potential than differentiated, non-tumorigenic, breast cancer cells. They are of great capacity for self-renewal and differentiation, which lead to initiation, progression, metastasis and recurrence of tumor.Citation17 BCSCs could enable the re-establishment of tumors, therefore targets silencing stemness properties are needed. Instead of stemness correlated proteins or EMT involved proteins, this study focused on PLP2, an integral membrane protein that localizes to the endoplasmic reticulum in colonic epithelial cells.
Downregulated miR-422a expression has been found in several types of cancers, including osteosarcoma (OS), hepatocellular carcinoma and colorectal cancer.Citation18 MiR-422a was revealed as a valuable prognostic marker for osteosarcoma patients since it was correlated with large tumor size, advanced TNM stage, distant metastasis and grade of tumor.Citation18 Consistently, Downregulated miR-422a expression in BCSCs might be of great value in breast cancer treatment.
Then we further investigated how miR-422a functioned in BCSCs. MiRanda predicted various mRNAs that could be regulated by miR-422a, including PLP2, a differentiation correlated gene. Besides, their combination was relatively stable and highly conserved, which was why the relationship between miR-422a and PLP2 was further investigated in BCSCs and mice models. PLP2 had been shown to involve in various cancers, including breast cancer, hepatocellular carcinoma, osteosarcoma, and melanoma.Citation13 Zhu et al. reported that miR-664 directly targeted the 3′UTR region of PLP2 in T-cell acute lymphoblastic leukemia.Citation19 Ding et al. suggested that PLP2 was a bona fide target of miR-664 in cutaneous malignant melanoma.Citation13 This study proved that PLP2, which was downregulated by miR-422a, exhibit tumor initiating potential in BCSCs.
In vitro experiments proved that miR-422a mimics weakened whereas PLP2 enhanced the tumorsphere formation ability and cell proliferation. In vivo experiments confirmed functions of miR-422a and PLP2 in tumor outgrowth in mice models, which suggested that miR-422a overexpression could inhibit the tumorigenesis of BCSCs via downregulating the expression of PLP2.
Limitations have to be taken into consideration in this study. BCSCs sorted out in this study might not be replicable in other researches. Besides, some other targets except PLP2 could be regulated by miR-422a and other pathways affected by miR-442a and PLP2 was not explored in this study. Moreover, the underlying mechanism responsible for the aberrant miR-442a was attractive but not attended in this manuscript due to the limited financial support. However, those could be involved in our further study.
Conclusion
In conclusion, we have proved that miR-422a directly targeted at PLP2 in BCSCs and suppressed stemness properties. MiR-422a mimics weakened ability of tumorsphere formation, cell proliferation in vitro and tumor growth in vivo in BCSCs by downregulating PLP2. Therefore, miR-422a may be a potential therapeutic target for breast cancer treatment.
Author contribution
Yanmei Zou, Yuandong Chen and Shuo Yao contributed to research conception and design as well as manuscript drafting; Guangrui Deng, Dian Liu, Yihua Wang and Xun Yuan analysed and interpreted data; Shunfang Liu, Jie Rao and Huihua Xiong made statistical analysis; Xianglin Yuan, Shiying Yu, Feng Zhu and Hua Xiongrevised the manuscript and took the role of funding collectors. In addition, all authors approved final manuscript.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Maramba-Ceballos L. Philippines empirical dental remedies against pain. Dent Mirror (Quezon City). 1966;3(1):13–6. PMID:5228385.
- Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, Chen H, Dong C, Yang R, Liu S, et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6(4):533–44. doi:10.7150/thno.14315. PMID:26941846.
- Bilek SL, Lay T. Variation of interplate fault zone properties with depth in the japan subduction zone. Science. 1998;281(5380):1175–8. doi:10.1126/science.281.5380.1175. PMID:9712578.
- Xie G, Ji A, Yuan Q, Jin Z, Yuan Y, Ren C, Guo Z, Yao Q, Yang K, Lin X, et al. Tumour-initiating capacity is independent of epithelial-mesenchymal transition status in breast cancer cell lines. Br J Cancer. 2014;110(10):2514–23. doi:10.1038/bjc.2014.153. PMID:24755887.
- Ohta S, Misawa A, Lefebvre V, Okano H, Kawakami Y, Toda M. Sox6 up-regulation by macrophage migration inhibitory factor promotes survival and maintenance of mouse neural stem/progenitor cells. PLoS One. 2013;8(9):e74315. doi:10.1371/journal.pone.0074315. PMID:24066135.
- Kuo KK, Lee KT, Chen KK, Yang YH, Lin YC, Tsai MH, Wuputra K, Lee YL, Ku CC, Miyoshi H, et al. Positive feedback loop of OCT4 and c-JUN expedites cancer stemness in liver cancer. Stem Cells. 2016;34(11):2613–24. doi:10.1002/stem.2447. PMID:27341307.
- Adler CP, Riede UN, Wurdak W, Zugmaier G. Cytophotometric measurements of the DNA in Hodgkin lymphomas and non-Hodgkin lymphomas. Pathol Res Pract. 1984;178(6):579–89. doi:10.1016/S0344-0338(84)80091-6. PMID:6207516.
- Zhu SP, Wang JY, Wang XG, Zhao JP. Long intergenic non-protein coding RNA 00858 functions as a competing endogenous RNA for miR-422a to facilitate the cell growth in non-small cell lung cancer. Aging (Albany NY). 2017;9(2):475–86 PMID:28177876.
- Wang H, Tang C, Na M, Ma W, Jiang Z, Gu Y, Ma G, Ge H, Shen H, Lin Z. miR-422a inhibits glioma proliferation and invasion by targeting IGF1 and IGF1R. Oncol Res. 2017;25(2):187–94. doi:10.3727/096504016X14732772150389. PMID:28277190.
- Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, Miao N, Shen J, Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi:10.1016/j.canlet.2017.06.009. PMID:28642170.
- Tung SL, Huang WC, Hsu FC, Yang ZP, Jang TH, Chang JW, Chuang CM, Lai CR, Wang LH. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis. 2017;6(5):e326. doi:10.1038/oncsis.2017.25. PMID:28459431.
- Roscigno G, Quintavalle C, Donnarumma E, Puoti I, Diaz-Lagares A, Iaboni M, Fiore D, Russo V, Todaro M, Romano G, et al. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget. 2016;7(1):580–92. doi:10.18632/oncotarget.5979. PMID:26556862.
- Ding Z, Jian S, Peng X, Liu Y, Wang J, Zheng L, Ou C, Wang Y, Zeng W, Zhou M. Loss of MiR-664 expression enhances cutaneous malignant melanoma proliferation by upregulating PLP2. Medicine (Baltimore). 2015;94(33):e1327. doi:10.1097/MD.0000000000001327. PMID:26287415.
- Ozawa H, Sonoda Y, Suzuki T, Yoshida-Hoshina N, Funakoshi-Tago M, Kasahara T. Knockdown of proteolipid protein 2 or focal adhesion kinase with an artificial microRNA reduces growth and metastasis of B16BL6 melanoma cells. Oncol Lett. 2012;3(1):19–24. doi:10.3892/ol.2011.422. PMID:22740849.
- Lee SM, Shin H, Jang SW, Shim JJ, Song IS, Son KN, Hwang J, Shin YH, Kim HH, Lee CK, et al. PLP2/A4 interacts with CCR1 and stimulates migration of CCR1-expressing HOS cells. Biochem Biophys Res Commun. 2004;324(2):768–72. doi:10.1016/j.bbrc.2004.09.118. PMID:15474493.
- Longo A, Librizzi M, Luparello C. Effect of transfection with PLP2 antisense oligonucleotides on gene expression of cadmium-treated MDA-MB231 breast cancer cells. Anal Bioanal Chem. 2013;405(6):1893–901. doi:10.1007/s00216-012-6182-5. PMID:22729357.
- Li W, Ma H, Zhang J, Zhu L, Wang C, Yang Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci Rep. 2017;7(1):13856. doi:10.1038/s41598-017-14364-2. PMID:29062075.
- Bahador R, Taheriazam A, Mirghasemi A, Torkaman A, Shakeri M, Yahaghi E, Goudarzi PK. Tissue expression levels of miR-29b and miR-422a in children, adolescents, and young adults' age groups and their association with prediction of poor prognosis in human osteosarcoma. Tumour Biol. 2016;37(3):3091–5. doi:10.1007/s13277-015-4140-5. PMID:26423405.
- Zhu H, Miao MH, Ji XQ, Xue J, Shao XJ. miR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia. Biochem Biophys Res Commun. 2015;459(2):340–5. doi:10.1016/j.bbrc.2015.02.116. PMID:25735976.
