ABSTRACT
Background: LncRNA PTCSC3 is a tumor suppressor in thyroid cancer, and its role in drug resistance of anaplastic thyroid cancer (ATC) to chemotherapy drug doxorubicin was investigated in this study.
Methods: Expression of RNA and protein was analyzed by qRT-PCR and western blot, respectively. Flow cytometry was used to analyze the expression rate of CD133+ cells. The endogenous expression of related genes was modulated by recombinant plasmids and cell transfection. Combination condition and interaction between PTCSC3 and STAT3 were determined by RIP and RNA pull-down assay, respectively. MTT assay was performed to detect cytotoxicity. Chromatin immunoprecipitation was conducted to identify interactions between STAT3 and DNA promoter of INO80.
Results: LncRNA PTCSC3 was low-expressed in ATC tissues and cells. Over-expressed PTCSC3 inhibited the drug resistance of ATC to doxorubicin. PTCSC3 negatively regulated STAT3, and STAT3 promoted expression of INO80. PTCSC3 regulated INO80 through STAT3. PTCSC3 suppressed stem cells properties and drug resistance of ATC to doxorubicin.
Conclusion: LncRNA PTCSC3 inhibits INO80 expression by negatively regulating STAT3, and thereby attenuating drug resistance of ATC to chemotherapy drug doxorubicin.
KEYWORDS:
Introduction
Thyroid cancer is the most common endocrine malignancy and accounts for the major deaths from it, with increasing incidence.Citation1 Apart from parafollicular cells, over 95% of thyroid cancers originate from follicular cells and can be classified into four major histologic groups, including papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), medullary thyroid carcinoma (MTC), and anaplastic thyroid carcinoma (ATC).Citation2 ATC is an aggressive, lethal and recurrent solid tumor that usually treated with conventional therapies and chemotherapy including doxorubicin,Citation3 but the effects were heavily affected by drug resistance attributed to the over-expression of multidrug resistance gene (MDR).Citation4 The expression of CD133 is frequently associated with poor outcome of cancer and it has been used for cancer stem cell (CSC) identification in various cancers, and ATC-CD133+ cells have been proved with CSC features and were involved in tumor progression, aggressiveness and chemotherapeutic drug resistance of ATC.Citation5
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor that regulates cellular functions via responding to cytokine and growth factor signaling.Citation6 STAT3 is defined as an oncogene for its role in tumorigenesis, by promoting inflammation, tumour cell proliferation, survival, invasion and immunosuppression in cancers.Citation7 Microarray bioinformatics analysis suggested that STAT3 plays a pivotal role in modulating CSC properties of ATC-CD133+ cells; as an inhibitor of STAT3, the cucurbitacin I increased sensitivity of ATC-CD133+ to radiation and chemotherapeutic drugs.Citation8 It suggested the adverse impact of STAT3 pathway for evoking resistance in radiation and chemotherapies of ATC. The ATP-dependent chromatin remodeling complex INO80 shows capability of regulating chromatin structure and as a novel self-renewal factor.Citation9 INO80 also participates in many nuclear transactions and implicates in organization of the pluripotency network and regulation of the pluripotent state.Citation10 Moreover, INO80 was up-regulated in ATC-CSCs, and knockdown of INO80 not only decreased the expression of stem cell marker genes (including ALDH1) but also attenuated stem cell-specific properties for forming tumors.Citation11 It suggested the positive role of INO80 in maintaining cell-specific properties of ATC. The binding of STAT3 and promoter of INO80 was predicted by JASPAR software, implying the potential interaction between them.
Long noncoding RNAs (lncRNAs) are non-coding transcripts longer than 200 nucleotides and involved in many disorders. Papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) is one of the lncRNAs that was initially identified in thyroid,Citation12 and to be a tumor suppressor in thyroid cancer.Citation13 It has been manifested that PTCSC3 inhibited proliferation and migration of glioma cells and suppresses Wnt/β-catenin signaling pathway by targeting low density lipoprotein receptor related protein 6 (LRP6) directly.Citation14 Considering the close interaction between Wnt/β-catenin pathway and STAT3,Citation15 we suspected that PTCSC3 may interact with STAT3 and play a role in ATC. This study was undertaken to assess the expression of lncRNA PTCSC3 in ATC and to evaluate whether it play an essential role in drug resistance of ATC by targeting STAT3, which may further have an effect on INO80.
Materials and methods
Patient samples
Twenty FTC patients and twenty ATC patients used in this study were admitted to the First Affiliated Hospital of Zhengzhou University from January 2014 to December 2016, and the FTC tissues (n = 20) and ATC tissues (n = 20) were separated from the corresponding patients. All patients were signed informed consent and the study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University. This study was performed according to the Helsinki Declaration.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from FTC or ATC tissue by using Trizol reagent (Invitrogen) according to the manual. The total RNA was transcribed with a cDNA Reverse Transcription Kit (Applied Biosystems). The cDNA was quantified by qRT-PCR on an ABI 7300-fast Real Time PCR system (Applied Biosystems) with SYBR Select Master Mix (Applied Biosystems). Primers were designed and synthesized by Sangon Biotech (Shanghai, China). GAPDH and U6 served as the internal control and the relative expression was analyzed by 2−ΔΔCt method.
Western blot analysis
For protein extraction, tissues or cells were treated with RIPA lysis buffer (Beyotime), and the concentration of protein was measured with a BCA protein assay kit (Beyotime). Total protein was separated with 12% SDS-PAGE and transferred into the polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The membrane was then blocked with 5% nonfat milk for 1 hour at RT, and then incubated with the following primary antibodies: anti-STAT3 antibody (Abcam, 1:3000), anti-INO80 antibody (1:1000), anti-ALDH antibody (1:500), anti-MDR1 antibody (1:2000), anti-β-actin antibody (Abcam, 1:2000) at 4°C for overnight. The membrane was then incubated with HRP-bounded antibodies for 1 hour at RT and the target proteins were visualized with an ECL Plus Western Blotting Substrate (Thermo Fisher). β-actin served as control.
Cell culture
Two human thyroid carcinoma cell lines were provided by European Collection of Animal Cell Cultures (Salisbury, UK). An undifferentiated thyroid cancer cell line (8505C) was derived from primary culture of a human ATC tumor. Follicular thyroid carcinoma (FTC) cell line 238 (FTC 238) and FTC cell line 133 (FTC 133) were established from a follicular thyroid carcinoma. The thyroid cell lines were cultured in MEM with 1% non-essential amino acids (for 8505C) and in Dulbecco's modified Eagle's medium (DMEM): Ham F12 (1:1) (for FTC 238 and FTC133) obtained from Eurobio (Les Ulis, France). The culture media were supplemented with 10% (for 8505C) or 5% (for FTC 238 and FTC133) heat inactivated fetal calf serum (FCS, Gibco), 10 mM HEPES, 2 mM glutamine, 100 mg/ml streptomycin and 100IU/ml penicillin.
Flow cytometry (FCM)
The CD133+ was analyzed by a flow cytometer FACSCalibur (BD Biosciences). FTC 238, FTC 133 or ATC 8505C cells (1 × 106) were washed with phosphate-buffered saline (PBS), and then incubated with 10 μl of CD133 antibody (Miltenyi Biotec) at 4°C for 10 min. Subsequently, the cells were washed by MACS buffer and centrifuged (300 g, 10 min) for supernatant aspirate, and cells were analyzed on the flow cytometer FACSCalibur (BD Biosciences) and determined by the Flow Jo V10 software (Tree Star Inc.).
Cell transfection
For RNAi-mediated knockdown experiment, lncRNA PTCSC3-specific siRNA (si-PTCSC3), negative control (si-control) and si-STAT3 were purchased from GenePharma Co. Ltd (Shanghai, China) for cell transfection. They were transfected into FTC 238, FTC133 or ATC 8505C cells by using Lipofectamine2000 (Invitrogen) according to the instructions. The efficiency of cell transfection was examined by qRT-PCR and western blot.
Detection of cytotoxicity with MTT assay
Cell cytotoxicity was determined by the MTT assay.Citation16 This assay measures the conversion of MTT to dark blue formazan precipitation by succinate dehydrogenase of the intact mitochondria of living cells. FTC 238, FTC133 or ATC 8505C cells were seeded into 96-well plates at a density of 1 × 104 cells per well in DMEM (10% FCS) in six replicates and incubated for 12 h. The doxorubicin were firstly dissolved in dimethyl sulfoxide (DMSO), and then diluted in culture medium with final DMSO concentration of 0.5% (v/v). After that the cells were exposed to doxorubicin with concentration of 0.1, 0.5, 1.0, and 5.0 (μM) for 48 h. Medium containing 0.5% DMSO was used as negative control. After treatment, the treatment medium was replaced with fresh culture medium without FCS containing MTT at a concentration of 0.5 mg/mL, and the cells were further incubated for 4 h at 37°C. The medium was removed and the formazan crystals were dissolved in DMSO.
RNA pull-down assay
RNA pull-down assay was conducted to determine the interaction between PTCSC3 and STAT3. Briefly, the DNA probe complementary to PRNCR1 was synthesized and biotinylated by GenePharma Co., Ltd (Shanghai, China). RNA pull-down assay was carried out by the Magnetic RNA-Protein Pull-Down Kit (Thermo) according to its specification. The RNA-binding protein complexes were washed and eluted for western blot or qRT-PCR analysis.
RNA immunoprecipitation (RIP)
RIP was performed with the RNA-Binding Protein Immunoprecipitation Kit (Millipore). The ATC 8505C cells were lysed with lysis buffer and the cell lysis solutions were incubated with STAT3 antibody or normal mouse IgG. RNA-protein complexes were immunoprecipitated with protein A agarose beads and RNA was extracted by using Trizol (Invitrogen). The IP-western was used to detected AGO2 protein and qRT-PCR was performed to quantify the PTCSC3.
Chromatin immunoprecipitation (ChIP)
Experiments were performed with the ChIP kit (Upstate Biotechnology) according to the manufacturer's instructions.Citation17 Briefly, FTC 238 or ATC 8505C cells cultured at 37°C for 2 d were cross-linked with 0.37% formaldehyde and the genomic DNA was sheared by sonication. The samples were then immunoprecipated with antibody against STAT3 (Abcam). Normal rabbit IgG (Abcam) was used as a negative control. The immune complex was heated at 65°C for 4 h to revert the crosslinking between DNA and proteins. The purified bound DNA was dissolved in 20 ml of Tris buffer (10 mM Tris, pH 8.5). The input DNA was diluted 100 × prior to PCR. The bound and the input DNA were analyzed by PCR and the PCR products were visualized on a 2% agarose gel stained with ethidium bromide.
Plasmid construction
The pcDNA-PTSCS3 and pcDNA-STAT3 expression plasmids were established by using pcDNA3.1 for endogenous expression. The wild-type 3′-UTR of STAT3 genes were PCR amplified from human genomic DNA and purified with gel extraction. With double enzymes restriction and linkage with T4 DNA ligase (Takara), the purified PCR products of PTSCS3 or STAT3 were linked with pcDNA3.1 to integrate into pcDNA-PTSCS3 or pcDNA-STAT3 recombinant plasmids. Then the recombinant plasmid was inserted into E.coli for amplification and select of positive clones. After identification with restriction analysis and sequence analysis (Sangon Biotech), the recombinant plasmid was extracted from positive clones and transfected into FTC 238 cells. The pcDNA were also transfected as control.
Statistical analysis
Data were expressed as mean ± standard deviation (SD), and statistically analyzed by SPSS 21.0 software (SPSS Inc.) Comparisons between two groups were completed by Student's t-test, and comparisons among multi-groups were performed by One-way ANOVA. P < 0.05 was considered statistically significant.
Results
LncRNA PTCSC3 was low-expressed in ATC tissues
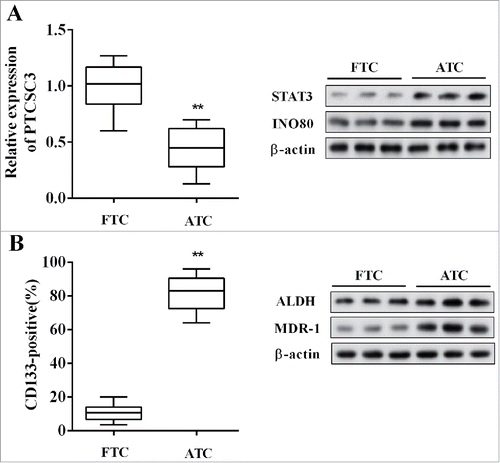
The expression of lncRNA PTCSC3 and the levels of STAT3, INO80, MDR-1, ALDH in ATC (n = 20) and follicular thyroid carcinoma (FTC, n = 20) tissues were detected, and the positive expression rate of CD133 was also examined. Compared with FTC, the PTCSC3 was distinctly decreased in ATC tissues, while the STAT3 and INO80 were significantly up-regulated (); and the MDR-1, stem cell markers including CD133 and ALDH were dramatically increased in ATC tissues ().
Figure 1. LncRNA PTCSC3 was low-expressed in ATC tissues. (A) The expression of PTCSC3 in FTC (n = 20) and ATC tissues (n = 20) was quantified by qRT-PCR. **P < 0.01 vs. FTC; the expression of proteins was analyzed by western blot. (B) The positive expression rate of CD133 was analyzed by flow cytometry. **P < 0.01 vs. FTC; the levels of ALDH and MDR-1 were analyzed by western blot.
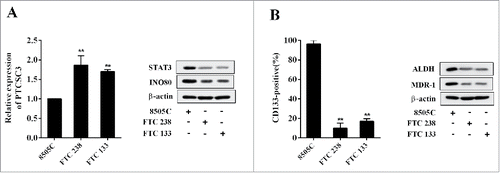
LncRNA PTCSC3 was low-expressed in ATC cells
The expression of PTCSC3 and the levels of STAT3, INO80, MDR-1, ALDH in ATC 8505 cells (8505C) FTC cells (FTC 238), and FTC 133 cells were detected, and the expression rate of CD133 was also examined. Compared with ATC 8505C cells, the PTCSC3 was markedly elevated in FTC 238 and FTC 133, while the STAT3 and INO80 were clearly decreased (); and the MDR-1, stem cell markers including CD133 and ALDH were obviously reduced in FTC 238 and FTC 133 cells ().
Figure 2. LncRNA PTCSC3 was low-expressed in ATC cells. (A) The expression of PTCSC3 in FTC 238, FTC 133 and 8505C cells was quantified by qRT-PCR. **P < 0.01 vs. FTC 238; the expression of proteins was analyzed by western blot. (B) The positive expression rate of CD133 in FTC 238, FTC 133 and 8505C cells was analyzed by flow cytometry. **P < 0.01 vs. FTC 238; the levels of ALDH and MDR-1 were analyzed by western blot.
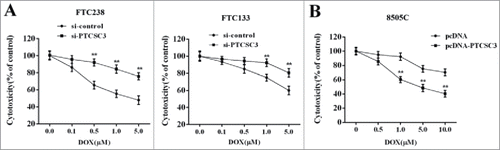
Over-expressed PTCSC3 inhibited the drug resistance of ATC
To investigate the impact of PTCSC3 on drug resistance of ATC, the FTC 238 and FTC 133 cells were divided into two groups and transfected with si-control and si-PTCSC3, independently. 48 h after transfection, cells were treated with doxorubicin (0, 0.1, 0.5, 1.0, 5.0 μM) and the cytotoxicity was determined. As it showed in , compared with si-control group, interference of PTCSC (si-PTCSC3) reduced the sensitivity of FTC 238 and FTC 133 cells to doxorubicin (). The ATC 8050C cells were divided into two groups and transfected with pcDNA and pcDNA-PTCSC3, respectively. 48 h after transfection, cells were treated with doxorubicin (0, 0.1, 0.5, 1.0, 5.0 μM) and the cytotoxicity was determined. Compared with pcDNA group, overexpression of PTCSC3 enhanced the sensitivity of ATC 8050C cells to doxorubicin ().
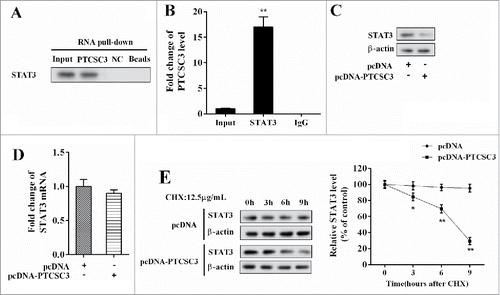
LncRNA PTCSC3 negatively regulated STAT3
In the ATC 8050C cells, the interaction between PTCSC3 and STAT3 was verified by RNA pull-down assay, with NC served as control. STAT3 in the pull-down complex of PTCSC3 was analyzed by western blot. Compared with NC, STAT3 gathered extensively in the pull-down complex of PTCSC3 (). Combination between PTCSC3 and STAT3 was examined by RIP, with IgG as control. Compared with IgG, plenty of PTCSC3 accumulated in the protein deposited by STAT3 (). With PTCSC3 over-expressed, STAT3 protein level was declined (), while the mRNA level of STAT3 remained unchanged (); under treatment of Cycloheximide (CHX), over-expressed PTCSC3facilitated degradation of STAT3 protein ().
Figure 4. LncRNA PTCSC3 negatively regulated STAT3. (A) Combination condition between PTCSC3 and STAT3 was determined by RIP assay. (B) The interaction between PTCSC3 and STAT3 was detected by RNA pull-down assay. **P < 0.01, vs. IgG. (C) The expression of STAT3 was analyzed by western blot. (D) The expression of STAT3 mRNA in 8505C cells was quantified by qRT-PCR. (E) The expression of STAT3 in 8505C cells was analyzed by western blot. *P < 0.05 vs. pcDNA; **P < 0.01 vs. pcDNA.
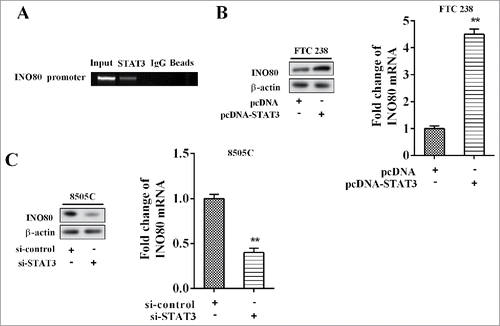
STAT3 promoted expression of INO80
The result of CHIP showed that STAT3 bound with DNA promoter of STAT3 (). With FTC 238 cells transfected with pcDNA-STAT3 and ATC 8505C cells transfected with si-STAT3, and the levels of INO80 mRNA and protein were detected after 48 h. The results suggested that over-expression of STAT3 promoted expression of INO80 mRNA and protein (); while low-expression of STAT3 inhibited expression of INO80 mRNA and protein ().
Figure 5. STAT3 promoted expression of INO80. (A) The chromatin immune-precipitation was used to verify the binding between PTCSC3 and the promoter of STAT3. (B) The expression levels of INO80 mRNA and protein in FTC238 cells were examined by qRT-PCR and western blot, respectively. **P < 0.01 vs. pcDNA. (C) The expression levels of INO80 mRNA and protein in 8505C cells were examined by qRT-PCR and western blot, respectively. **P < 0.01 vs. si-control.
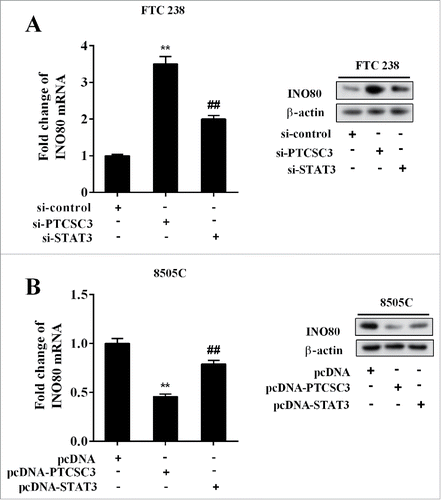
LncRNA PTCSC3 regulated INO80 through STAT3
The FTC 238 cells were divided into three groups and transfected with si-control, si-PTCSC3, and si-PTCSC3+ si-STAT3 separately, and the levels of INO80 mRNA and protein were detected respectively. It showed that interference of PTCSC3 up-regulated INO80 mRNA and protein, which was reversed by si-STAT3 (). The ATC 8505C cells were divided into three groups and transfected with pcDNA, pcDNA-PTCSC3, and pcDNA-PTCSC3+pcDNA-STAT3 respectively, and the levels of INO80 mRNA and protein were detected respectively. It showed that over-expression of PTCSC3 down-regulated INO80 mRNA and protein, which was reversed by pc-STAT3 (). The above results suggested that PTCSC3 regulated INO80 by targeting STAT3.
Figure 6. LncRNA PTCSC3 regulated INO80 through STAT3. (A) The expression levels of INO80 mRNA and protein in FTC238 cells were examined by qRT-PCR and western blot, respectively. **P < 0.01 vs. si-control. ##P < 0.01 vs. si-PTCSC3. (B) The expression levels of INO80 mRNA and protein in 8505C cells were examined by qRT-PCR and western blot, respectively. **P < 0.01 vs. pcDNA. ##P < 0.01 vs. pcDNA-PTCSC3.
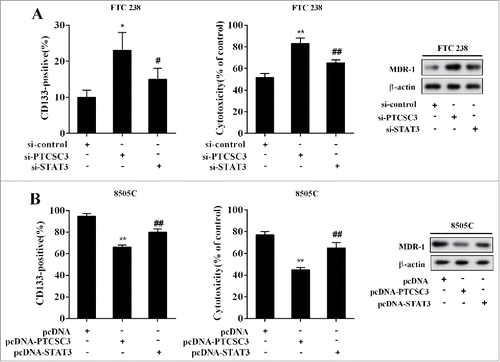
LncRNA PTCSC3 suppressed stem cells characteristics and drug resistance of ATC
The FTC 238 cells were divided into three groups and transfected with si-control, si-PTCSC3, and si-PTCSC3+si-STAT3, separately. 48 h after transfection, cells were treated with doxorubicin (1.0 μM), with the positive expression rate of CD133, cytotoxicity and the level of MDR-1 determined. Compared with si-control, interference of PTCSC3 elevated positive expression rate of CD133 and MDR-1 expression, and decreased the sensitivity of FTC 238 cells to doxorubicin, which were completely reversed by si-STAT3 (). The ATC 8505C cells were divided into three groups and transfected with pcDNA, pcDNA-PTCSC3, and pcDNA-PTCSC3+ pcDNA-STAT3 respectively. 48 h after transfection, cells were treated with doxorubicin (1.0 μM), with the positive expression rate of CD133, cytotoxicity and the level of MDR-1 detected. Compared with pcDNA group, overexpression of PTCSC3 reduced positive expression rate of CD133 and MDR-1 expression, and enhanced the sensitivity of ATC 8050C cells to doxorubicin, which were wholly reversed by pcDNA-STAT3 ().
Figure 7. LncRNA PTCSC3 suppressed stem cells characteristics and drug resistance of ATC. (A) The positive expression rate of CD133 was analyzed by flow cytometry. *P < 0.05 vs. si-control; #P < 0.05 vs. si-PTCSC3; The cytotoxicity of DOX to FTC238 was detected by MTT assay. **P < 0.01 vs. si-control; ##P < 0.01 vs. si-PTCSC3; the levels of MDR-1 was analyzed by western blot. (B) The positive expression rate of CD133 was analyzed by flow cytometry. **P < 0.01 vs. pcDNA; ##P < 0.01 vs. pcDNA-PTCSC3; The cytotoxicity of DOX to FTC238 was detected by MTT assay. **P < 0.01 vs. pcDNA; ##P < 0.01 vs. pcDNA-PTCSC3; the levels of MDR-1 was analyzed by western blot.
Discussion
In the current study, expression of lncRNA PTCSC3, STAT3, INO80 in ATC tissues and cells were investigated, and their influence on drug resistance of ATC to chemotherapy drug doxorubicin were evaluated. All the findings indicated that lncRNA PTCSC3 inhibits INO80 expression by negatively regulating STAT3, and thereby attenuating drug resistance of ATC to chemotherapy drug doxorubicin, providing novel strategies for improving efficiency of chemotherapy for ATC treatment.
As an aggressive, lethal and recurrent solid tumor, ATC is known with the worst prognosis in all types of thyroid cancers for its invasiveness and recurrent resistance to routine therapies. Besides conventional therapies such as surgical resection and radioiodine I (RAI), an additional chemotherapy including doxorubicin is applied in clinical for ATC treatment.Citation18 However, drug resistance caused by mutation of MDR largely impacts the efficiency of chemotherapyCitation4, and the ATC also exhibits significant resistance to chemotherapy for its stem cell differentiation potential.Citation19 In our previous work, we proposed that PTCSC3/miR-574-5p governed cell proliferation and migration of PTC via Wnt/β-catenin signaling, and thereby suppressing tumor growth.Citation20 Similarly, PTCSC3 achieved the same inhibitory effect on PTC by reducing cell motility and invasiveness through down-regulation of the S100A4 pathway,Citation13 while the role of PTCSC3 in other types of thyroid cancers remains uncertain. Here, we revealed that PTCSC3 played a role in ATC for the first time, and it exerted an influence on drug resistance to chemotherapy by modulating STAT3/INO80 pathway, exhibiting its tumor suppressor character in thyroid cancer. This study reconfirmed the role of PTCSC3 in repressing ATC development, and also emphasized its benefit in diminishing thermotherapy drug resistance and suppressing stem cells characteristics, offering a potential strategy in improving effect of thermotherapy in ATC.
STAT3 is a well-known transcription factor that activated in many cancers and participates in evoking resistance in radiation, targeted therapies and chemotherapies of cancers by targeting its downstream targets. For example, feedback activation of STAT3 mediated trastuzumab resistance via up-regulation of MUC1 and MUC4,Citation21 and inhibition of STAT3 sensitizes colorectal cancer to chemoradiotherapy in vitro and in vivo.Citation22 Previous report has also demonstrated that p53 mutations gained antiapoptotic function mediated by STAT3 pathway in ATC cells, implying a facilitative role in tumor progression of STAT3 in ATC.Citation23 In the present study, we identified the STAT3 protein as a direct target of PTCSC3 and demonstrated that STAT3 promoted expression of INO80, involving in drug resistance of ATC. PTCSC3 inhibits INO80 expression by negatively regulating STAT3, and thereby affecting drug resistance of ATC to chemotherapy drug doxorubicin. It suggested the adverse impact of STAT3 signaling pathway in ATC progression, and the STAT3 pathway may be a promising target in designing thyroid cancer therapy.
The INO80 belongs to the INO80 subfamily and functions in many nuclear transactions, such as DNA replication, DNA repair and transcriptional regulation.Citation24 Then its role in regulating stem cell-specific properties was clarified in ATC-CSC, in which INO80 expression was inhibited by miR-148a and the self-renewal of thyroid cancer stem cells was repressed subsequently.Citation11 Our study manifested that INO80 was positively regulated by STAT3, and uniformly exerted an influence on pluripotent state of ATC-CD133+ cells and drug resistance of ATC as well. As important stem cell markers, ATC-CD133+ cells also have been proved to be involved in chemotherapeutic drug and radiotherapy resistance of ATC.Citation5,Citation25 The levels of ALDH and positive expression rate of CD133 determined in this study were used to indicate the pluripotent state of CSCs intuitively.Citation26
Our study illuminated a complicated mechanism network of how lncRNA PTCSC3 influenced drug resistance of ATC to chemotherapeutic drug. As it indicated, PTCSC3 was down-regulated in ATC tissues and cells, and over-expression of PTCSC3 resulted in down-regulation of STAT3 and INO80, and thereby improving drug resistance of ATC to chemotherapy drug doxorubicin. This study initially highlighted the benefit of PTCSC3 in ATC treatment, and identified the STAT3 as a novel target of PTCSC3 to have an impact on drug resistance, bringing about hope for a strategy in enhancing efficiency and reducing resistance of drug treatment in ATC and other cancers.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Carling T, Udelsman R. Thyroid cancer. Annu Rev Med. 2014;65:125–37. doi:10.1146/annurev-med-061512-105739. PMID:24274180.
- Maniakas A, Forest VI, Jozaghi Y, Saliba J, Hier MP, Mlynarek A, Tamilia M, Payne RJ. Tumor classification in well-differentiated thyroid carcinoma and sentinel lymph node biopsy outcomes: a direct correlation. Thyroid. 2014;24(4):671–4. doi:10.1089/thy.2013.0160. PMID:24199963.
- Haddad RI, Lydiatt WM, Ball DW, Busaidy NL, Byrd D, Callender G, Dickson P, Duh QY, Ehya H. Anaplastic Thyroid Carcinoma, Version 2.2015. J Natl Compr Canc Netw. 2015;13(9):1140–50. doi:10.6004/jnccn.2015.0139. PMID:26358798.
- Zheng X, Cui D, Xu S, Brabant G, Derwahl M. Doxorubicin fails to eradicate cancer stem cells derived from anaplastic thyroid carcinoma cells: characterization of resistant cells. Int J Oncol. 2010;37(2):307–15. PMID:20596658.
- Decaussin-Petrucci M, Deladoëy J, Hafdi-Nejjari Z, Sassolas G, Borson-Chazot F, Abu-Khudir R, Fusco A, Descotes F, Cournoyer S, Sartelet H. Expression of CD133 in differentiated thyroid cancer of young patients. J Clin Pathol. 2015;68(6):434–40. doi:10.1136/jclinpath-2014-202625. PMID:25770162.
- Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33(13):1670–9. doi:10.1038/onc.2013.115. PMID:23604114.
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–46. doi:10.1038/nrc3818. PMID:25342631.
- Tseng LM, Huang PI, Chen YR, Chen YC, Chou YC, Chen YW, Chang YL, Hsu HS, Lan YT, Chen KH. Targeting signal transducer and activator of transcription 3 pathway by cucurbitacin I diminishes self-renewing and radiochemoresistant abilities in thyroid cancer-derived CD133+ cells. J Pharmacol Exp Ther. 2012;341(2):410–23. doi:10.1124/jpet.111.188730. PMID:22328572.
- Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B, Papamichos-Chronakis M. INO80 Chromatin Remodeler Facilitates Release of RNA Polymerase II from Chromatin for Ubiquitin-Mediated Proteasomal Degradation. Mol Cell. 2015;60(5):784–96. doi:10.1016/j.molcel.2015.10.028. PMID:26656161.
- Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao YL, Zhou B, Han L. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14(5):575–91. doi:10.1016/j.stem.2014.02.013. PMID:24792115.
- Sheng W, Chen Y, Gong Y, Dong T, Zhang B, Gao W. miR-148a inhibits self-renewal of thyroid cancer stem cells via repressing INO80 expression. Oncol Rep. 2016;36(6):3387–96. doi:10.3892/or.2016.5203. PMID:27779717.
- Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A. 2012;109(22):8646–51. doi:10.1073/pnas.1205654109. PMID:22586128.
- Jendrzejewski J, Thomas A, Liyanarachchi S, Eiterman A, Tomsic J, He H, Radomska HS, Li W, Nagy R, Sworczak K. PTCSC3 Is Involved in Papillary Thyroid Carcinoma Development by Modulating S100A4 Gene Expression. J Clin Endocrinol Metab. 2015;100(10):E1370–7. doi:10.1210/jc.2015-2247. PMID:26274343.
- Xia S, Ji R, Zhan W. Long noncoding RNA papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) inhibits proliferation and invasion of glioma cells by suppressing the Wnt/β-catenin signaling pathway. Bmc Neurology. 2017;17(1):30. doi:10.1186/s12883-017-0813-6. PMID:28187755.
- Fragoso MA, Patel AK, Nakamura RE, Yi H, Surapaneni K, Hackam AS. The Wnt/beta-catenin pathway cross-talks with STAT3 signaling to regulate survival of retinal pigment epithelium cells. PLoS One. 2012;7(10):e46892. doi:10.1371/journal.pone.0046892. PMID:23056515.
- Yang Y, Lu Y, Wu QY, Hu HY, Chen YH, Liu WL. Evidence of ATP assay as an appropriate alternative of MTT assay for cytotoxicity of secondary effluents from WWTPs. Ecotoxicol Environ Saf. 2015;122:490–6. doi:10.1016/j.ecoenv.2015.09.006. PMID:26410194.
- Rodriguez-Ubreva J, Ballestar E. Chromatin immunoprecipitation. Methods Mol Biol. 2014;1094:309–18. doi:10.1007/978-1-62703-706-8_24. PMID:24162998.
- Bisof V, Rakusic Z, Despot M. Treatment of patients with anaplastic thyroid cancer during the last 20 y: whether any progress has been made? Eur Arch Otorhinolaryngol. 2015;272(7):1553–67. doi:10.1007/s00405-014-3108-1. PMID:24890977.
- Giuffrida R, Adamo L, Iannolo G, Vicari L, Giuffrida D, Eramo A, Gulisano M, Memeo L, Conticello C. Resistance of papillary thyroid cancer stem cells to chemotherapy. Oncol Lett. 2016;12(1):687–91. doi:10.3892/ol.2016.4666. PMID:27347201.
- Wang X, Lu X, Geng Z, Yang G, Shi Y. LncRNA PTCSC3/miR-574-5p governs cell proliferation and migration of papillary thyroid carcinoma via Wnt/β-catenin signaling. J of Cellular Bio. 2017;118(12):4745–52. doi:10.1002/jcb.26142. PMID:28513866.
- Li G, Zhao L, Li W, Fan K, Qian W, Hou S, Wang H, Dai J, Wei H, Guo Y. Feedback activation of STAT3 mediates trastuzumab resistance via upregulation of MUC1 and MUC4 expression. Oncotarget. 2014;5(18):8317–29. doi:10.18632/oncotarget.2135. PMID:25327561.
- Spitzner M, Roesler B, Bielfeld C, Emons G, Gaedcke J, Wolff HA, Rave-Fränk M, Kramer F, Beissbarth T, Kitz J. STAT3 inhibition sensitizes colorectal cancer to chemoradiotherapy in vitro and in vivo. Int J Cancer. 2014;134(4):997–1007. doi:10.1002/ijc.28429. PMID:23934972.
- Kim TH, Lee SY, Rho JH, Jeong NY, Soung YH, Jo WS, Kang DY, Kim SH, Yoo YH. Mutant p53 (G199V) gains antiapoptotic function through signal transducer and activator of transcription 3 in anaplastic thyroid cancer cells. Mol Cancer Res. 2009;7(10):1645–54. doi:10.1158/1541-7786.MCR-09-0117. PMID:19825993.
- Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34(2):71–7. doi:10.1016/j.tibs.2008.10.010. PMID:19062292.
- Mei D, Lv B, Chen B, Xiao S, Jiang J, Xie Y, Jiang L. All-trans retinoic acid suppresses malignant characteristics of CD133-positive thyroid cancer stem cells and induces apoptosis. PLoS One. 2017;12(8):e0182835. doi:10.1371/journal.pone.0182835. PMID:28817605.
- Xu X, et al. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015;369(1):50–7. doi:10.1016/j.canlet.2015.08.018. PMID:26319899.