ABSTRACT
Head and neck squamous cell carcinoma is one of the leading cancers in terms of incidence and mortality. However, no reliable marker till date accurately predicts its progression when altered in healthy tissues. The study aims to identify alleles of microsatellites adjacent to important cell cycle regulatory, tumor suppressor genes altered in early head and neck lesions, viz. RBSP3, LIMD1 and CDC25A, which undergo frequent deletion and can be used for population screening and early detection. DNA for tumors and normal tissues was isolated from 143 patients in different stages of head and neck squamous cell carcinoma. The size of microsatellite present in normal tissues and their deletion in the corresponding tumor was identified, along with the correlation of expression in normal epithelium with respect to allele size. The results revealed a range of alleles (CA9 to CA32) for the different microsatellites of the genes in normal tissues. The larger alleles were significantly deleted with differential deletion of alleles observed in tumors, except for LIMD1, in which the smaller allele was significantly deleted. In normal tissues, some alleles represented as stable alleles with high prevalence, while in tumours, specific sizes showed greater propensity for deletion. However, similar expression of the proteins in normal epithelium adjacent to tumors was observed despite variations in allele size, possibly due to the location of the microsatellites. Thus, those alleles when present in normal tissues and undergoing persistent deletion in tumours could be used as markers for screening and early identification of populations at risk of developing head and neck lesions.
Introduction
Head and neck cancer is a global acrimony, ranking sixth in prevalence and the fifth leading cause of cancer- related deaths globally.Citation1 The Indian subcontinent harbors two- third of the global disease burden with predominantly oral cavity and oropharyngeal cancers, accounted for by associated etiological factors such as tobacco, betel nut, alcohol, human papilloma virus infection, etc.Citation2 Notwithstanding the advances in early detection, diagnosis and treatment, the five- year survival of patients has not remarkedly improved over the decades,Citation3 indicating the necessity for more rigorous and effective detection and screening methods.
Microsatellite markers are present ubiquitously in eukaryotic genomes and are one of the most dynamic components of genome, owing to their expansion and contraction and unusual conformations.Citation4 Microsatellite instability has thus been widely used for analysis of genetic status in many pathological conditions including cancer.Citation5-7 Several studies have also indicated the association of microsatellite alterations with pathogenesis of HNSCC.Citation8 However, data regarding the alterations of specific (larger/ smaller) microsatellite alleles in the tumor is grossly lacking. Moreover, it is also not known how variation in microsatellite size affects expression of the corresponding genes, if they are located adjacent or within its promoter region.
Our study thus focuses on three important cell cycle genes, RBSP3, LIMD1 and CDC25A, which were previously shown to be important in the development of head and neck lesions.Citation9,Citation10 These genes showed frequent alterations (deletion, promoter methylation, expression) in dysplasia and HNSCC and showed the presence of several alleles. However, whether certain sized alleles, if present in an individual, have a higher probability of deletion in the tumour is unknown. Moreover, it is also unknown if different- sized alleles correspond to varying levels of expression of the proteins in the normal head and neck epithelium.
To this end, the same large set of HNSCC and paired normal epithelium adjacent to the tumor was used to identify different sized alleles present and frequently deleted alleles, as well as correlation of allelic size in normal epithelium with protein expression. Our data identified the range of alleles present in normal epithelium adjacent to tumors along with the allele sizes which undergo maximum deletion in tumors, indicating their importance as markers for screening and early detection. Additionally, expression of the proteins remained comparable irrespective of variations in allele size, despite their presence in gene promoter (hmLIMD1). Our results suggest the utility of using alleles with high probability of deletion as markers for screening populations at risk of developing HNSCC.
Results
Demographic details
The median age of onset was 50 ± 12.44 years, with buccal mucosa being the most common site affected. A significantly higher number of patients were males and most patients were habitual tobacco users ().
Table 1. Clinico- pathological features of the patients.
Normal epithelium adjacent to tumors showed a range of allele sizes
For marker D3S1298 of RBSP3, a total of 8 different alleles of CA repeats, present at various frequencies were identified in our normal specimens adjacent to HNSCC, ranging from 23 (CA)23 to 32 (CA)32 repeats. Among them, (CA)26 was the most and significantly prevalent (32.18%), followed by (CA)28 (15.33%), (CA)30 (14.18%) and (CA)29 (10.73%). The rest of the repeats were present at less than 10% frequency in the population ().
Table 2. Frequency of alleles present in normal epithelium and their deletion in adjacent tumours. Grey boxes represent stable alleles, bold indicate susceptible alleles.
For LIMD1, the microsatellite marker D3S3582 showed highest prevalence of (CA)16 (29.13%), (CA)19 (25.2%) and (CA)14 (22.83%) (). The marker hmLIMD1 of LIMD1 showed the maximum range of allele sizes, ranging from (CA)9 to (CA)32, with (CA)19 having the highest prevalence ().
Marker D3S3560 for CDC25A presented with the lowest number of alleles in adjacent normal epithelium with (CA)10 and (CA)11 being present in almost equal proportion (43.08%, 43.46% respectively, ). Similarly, for D3S3640, while(CA)21 and (CA)23 were the alleles predominantly present in the samples (31.61%, 32.34% respectively) (). Representative autoradiograph showing allelic deletion can be found in Supp. Fig 1a.
Larger sized alleles were preferentially deleted in tumors
Comparison of the size (smaller, larger) of the allele deleted in the tumor corresponding to the normal epithelium previously studied indicated a preferential deletion of allele size for specific markers. While D3S1298, D3S3582, D3S3640 and D3S3560 showed significantly higher deletion in the larger allele, hmLIMD1 interestingly presented with significant deletion in the smaller sized allele (). There was however no correlation between allele size deleted and stage or grade of tumors (Supp. Fig. 1b, c).
Figure 1. Deletion and expression of the proteins with allele size. a. Allele size (larger/ smaller) undergoing maximum deletion in tumors. Larger allele mostly underwent deletion. b. Immunohistochemical expression of the proteins in normal epithelium with respect to allele size. Similar expression was observed irrespective to size of alleles present. Image magnification 20X, scale bar represents 50µm.
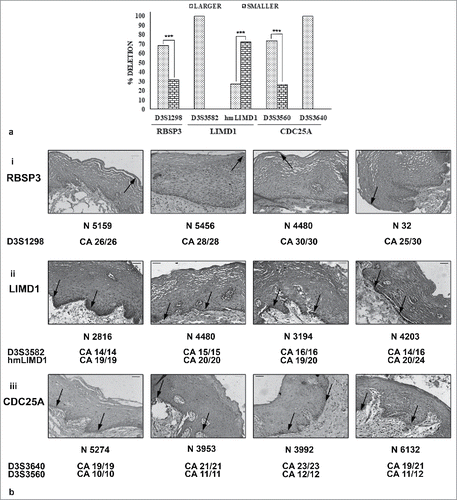
Specifically looking at the length of allele deleted, (CA)29 and (CA)30 showed the highest fraction of deletion in D3S1298 for RBSP3 (64%, 55.56% respectively). However, (CA)26, although most prevalent in adjacent normal epithelium showed nominal deletion in HNSCC tissues, possibly because these alleles were stable and subject to low deletion (). Similarly, for LIMD1, while(CA)19 in D3S3582 showed the highest deletion in tumors (71.88%), (CA)16 showed the highest prevalence in normal tissues, representing its stable nature (). For hmLIMD1, the most deleted alleles were (CA)17, although the stable allele was (CA)19. However, (CA)11, although the highest represented allele in normal for D3S3560 in CDC25A, it underwent the maximum deletion, possibly due to low number of alleles and its fragile status. Interestingly, (CA)10 was observed as the stable allele due to its low deletion despite high prevalence (). Similar phenomenon of highest prevalence/ deletion was observed for (CA)23 in D3S3640 with (CA)21 being the stable allele ().
Expression of the proteins in normal epithelium was not dependent allele size present
Expression of the proteins remained similar in normal epithelial tissues adjacent to tumors, irrespective of allele size or condition (homozygous/ heterozygous) of the adjacent microsatellite markers (, i- iii). The results were comparable for different alleles of hmLIMD1, although the microsatellite marker was located within the promoter region of LIMD1 ( ii).
Discussion
Knowledge on microsatellites regarding the versatility, ubiquitous presence and alterations in pathological conditions including HNSCC is prevalent.Citation8 However, the exact allelic sizes present in normal tissues and those undergoing alterations in corresponding tumors, or the influence of allele size with protein expression in normal head and neck epithelium is unknown.
Microsatellite markers present adjacent to the candidate genes presented with a range of sizes, although some were more prevalent than others. Presence of different allelic sizes at of the genes at variable frequencies in normal tissues has previously been identified in HNSCC.Citation11 Interestingly, tumors mostly showed losses in the larger sizes alleles, probably accounting for their fragility, as previously reported.Citation12,Citation13 Conversely, other researchers have accounted this to either possible normal cell admixtureCitation14 or degradation of DNA.Citation12 We have tried to eliminate both issues by enriching tumours from normal tissues using a high- precision Laser Capture Microdissection, along with checking the integrity of DNA using agarose gel electrophoresis. However, further studies to determine the prognostic significance of larger allele deletion is warranted.
Frequent deletion of these microsatellite markers in HNSCC were previously reported.Citation9-11 However, the allele sizes undergoing maximum deletion is yet unknown, although a study identified the susceptible allele for hmLIMD1 in HNSCC.Citation11 Our study reported the number of alleles present in normal tissues and the fraction of alleles of a particular size undergoing deletion. A similar study indicated the presence of two alleles of D3S3560 in nasopharyngeal carcinoma among Chinese population.Citation15 Differential deletion frequency of the alleles was observed, although highest prevalence in normal tissues not necessarily indicating its highest deletion in the corresponding tumors. In normal tissues, screening of the presence of the alleles which undergo high frequency of deletion in tumors could identify populations at risk of developing HNSCC, similar to the use of microsatellite size instability for detecting different cancers.Citation16,Citation17
Correlation of allele size in normal epithelial tissues with expression of the corresponding genes showed comparable expression, irrespective of allele size, probably due to their distal location from gene promoters. Similar patterns were also observed for hmLIMD1, although it is located within the putative promoter of LIMD1,Citation11 thereby nullifying effects of microsatellite allele size variations on protein expression in normal tissues. Although several reports in HNSCC correlate loss of expression of the proteins with alleleic deletion,Citation18 to the best of our knowledge, no study till date analyzed whether microsatellites play a role in altering protein expression in normal epithelial tissues.
Our results thus indicate the utility of using a panel of microsatellite markers of the genes for screening of populations at risk for developing HNSCC and for its early detection and diagnosis.
Materials and methods
Patients and samples
Tumors and paired normal oral epithelium adjacent to the tumors/ blood were collected from unrelated patients (N = 143) who had visited the outpatient department of Chittaranjan National Cancer Institute, Kolkata, India for diagnosis and/ or treatment. Informed consent was obtained from the patients, the institutional ethical committee and hospital authorities prior to the study. Relevant clinical history including exposure to tobacco and other risk factors and habits was obtained from the patients through an exhaustive oral questionnaire and tumors were classified into TNM stages and histopathologically graded.Citation19,Citation20
Microdissection and genomic DNA isolation
Tumor rich areas were identified in 5µm Hematoxylin and Eosin (H&E) cryosections. After comparing with unstained parallel tumor sections, tumors were enriched using Laser Capture Microdissection (LCM) procedure using PalmRobo software (Palm Microbeam, Zeiss, Germany). DNA was then extracted by standard procedure using Proteinase K digestion, followed by phenol- chloroform extraction.Citation2 Genomic DNA was similarly isolated from normal epithelium and blood.Citation2
Determination of allelic size
Microsatellite markers were selected based on their proximity to the genes. All microsatellites were (CA) repeats. Informative markers were subject to amplification in a standard polymerase chain reaction with [γ-P32] dATP end- labeled forward primer in a 20 μl reaction mixture and electrophoresed in a denaturing acrylamide gel along with pUC19/ HpaII marker end labelled by [αP32] dCTP.Citation10 The autoradiographs were developed and densitometric scanning using Quantity One software (Bio-Rad GS- 800, USA) was used to identify the types of alterations observed and allelic size determined using a specific algorithm pre- installed in the software.Citation20 Size of the alleles was confirmed by sequencing of representative homozygotes using 3130xl Genetic Analyzer (Applied Biosystems, USA). Details of markers used is given in Supp. Tab 1. Number of alleles were counted to determine the total number present in the samples studied. Frequency of deletion of a specific allele was calculated as the percentage among the total alleles of that specific size.
Determination of expression of the genes
Expression of RBSP3, LIMD1 and CDC25A was studied in normal epithelium adjacent to tumors (N = 87) using immunohistochemistry. Primary antibodies RBSP3 (CP-57-09), LIMD1 (CP-30-09) (Imgenix India Pvt. Ltd.) and CDC25A (sc-6947, Santa Cruz Biotechnology, CA, USA) were used at a dilution of 1:100, while HRP conjugated secondary antibodies rabbit anti-goat (sc-2020) and goat anti-rabbit (sc-2004, Santa Cruz Biotechnology, CA, USA) were used as 1:500. Chromogen 3, 3’ diaminobenzidine (DAB) and counterstain hematoxylin was used to develop slides and photograph under a Bright Field microscope (Leica DM1000, Germany).Citation21 Positive cells (<1% = 0, 1–20% = 1, 20–50% = 2, 50–80% = 3 and >80% = 4) and staining intensity (1 = weak, 2 = moderate, 3 = strong) was checked by two independent observers and combined to yield the final expression (0–2 = low, 3–4 = intermediate, 5–7 = high).Citation21 Expression was compared with allelic size of the genes, both in homozygous and heterozygous condition in normal epithelial tissues to determine possible correlation.
Clinico- pathological correlation
Statistical analysis was performed using EpiInfo 7 (CDC, Atlanta). All calculations and plotting of graphs were performed using Microsoft excel (Microsoft Corp., USA).Citation2
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
supp_mat_1449615_KCBT.zip
Download Zip (3.3 MB)Acknowledgments
The authors thank the Director, Chittaranjan National Cancer Institute for his support.
References
- Shi H, Chen X, Lu C, Gu C, Jiang H, Meng R, Niu X, Huang Y, Lu M. Association between P16INK4a promoter methylation and HNSCC: a meta-analysis of 21 published studies. PloS one. 2015;10(4):e0122302. doi:10.1371/journal.pone.0122302. PMID:25835498.
- Sarkar S, Alam N, Chakraborty J, Biswas J, Mandal SS, Roychoudhury S, Panda CK. Human papilloma virus (HPV) infection leads to the development of head and neck lesions but offers better prognosis in malignant Indian patients. Med Microbiol Immunol. 2017;206(3):267–76. doi:10.1007/s00430-017-0502-5. PMID:28343330.
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi:10.1038/nrc2982. PMID:21160525.
- Kejnovsky E, Michalovova M, Steflova P, Kejnovska I, Manzano S, Hobza R, Kubat Z, Kovarik J, Jamilena M, Vyskot B. Expansion of microsatellites on evolutionary young Y chromosome. PloS one. 2013;8(1):e45519. doi:10.1371/journal.pone.0045519. PMID:23341866.
- Mayrhofer M, Kultima HG, Birgisson H, Sundstrom M, Mathot L, Edlund K, Viklund B, Sjoblom T, Botling J, Micke P, et al. 1p36 deletion is a marker for tumour dissemination in microsatellite stable stage II-III colon cancer. BMC cancer. 2014;14:872. doi:10.1186/1471-2407-14-872. PMID:25420937.
- Losso GM, Moraes Rda S, Gentili AC, Messias-Reason IT. Microsatellite instability–MSI markers (BAT26, BAT25, D2S123, D5S346, D17S250) in rectal cancer. Arq Bras Cir Dig. 2012;25(4):240–4. doi:10.1590/S0102-67202012000400006. PMID:23411922.
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–2087 e2073. doi:10.1053/j.gastro.2009.12.064.
- Lin JC, Wang CC, Jiang RS, Wang WY, Liu SA. Microsatellite alteration in head and neck squamous cell carcinoma patients from a betel quid-prevalent region. Sci Rep. 2016;6:22614. doi:10.1038/srep22614. PMID:27009367.
- Ghosh S, Ghosh A, Maiti GP, Alam N, Roy A, Roy B, Roychoudhury S, Panda CK. Alterations of 3p21.31 tumor suppressor genes in head and neck squamous cell carcinoma: Correlation with progression and prognosis. Int J Cancer. 2008;123(11):2594–604. doi:10.1002/ijc.23834. PMID:18792900.
- Ghosh A, Ghosh S, Maiti GP, Sabbir MG, Zabarovsky ER, Roy A, Roychoudhury S, Panda CK. Frequent alterations of the candidate genes hMLH1, ITGA9 and RBSP3 in early dysplastic lesions of head and neck: clinical and prognostic significance. Cancer Sci. 2010;101(6):1511–20. doi:10.1111/j.1349-7006.2010.01551.x. PMID:20412120.
- Ghosh S, Ghosh A, Maiti GP, Mukherjee N, Dutta S, Roy A, Roychoudhury S, Panda CK. LIMD1 is more frequently altered than RB1 in head and neck squamous cell carcinoma: clinical and prognostic implications. Mol cancer. 2010;9:58. doi:10.1186/1476-4598-9-58. PMID:20226061.
- Sieben NL, Kolkman-Uljee SM, Flanagan AM, le Cessie S, Cleton-Jansen AM, Cornelisse CJ, Fleuren GJ. Molecular genetic evidence for monoclonal origin of bilateral ovarian serous borderline tumors. Am J pathol. 2003;162(4):1095–101. doi:10.1016/S0002-9440(10)63906-5. PMID:12651602.
- van Tilborg AA, de Vries A, de Bont M, Groenfeld LE, van der Kwast TH, Zwarthoff EC. Molecular evolution of multiple recurrent cancers of the bladder. Hum Mol Genet. 2000;9(20):2973–80. doi:10.1093/hmg/9.20.2973. PMID:11115841.
- Liu J, Zabarovska VI, Braga E, Alimov A, Klein G, Zabarovsky ER. Loss of heterozygosity in tumor cells requires re-evaluation: the data are biased by the size-dependent differential sensitivity of allele detection. FEBS Lett. 1999;462(1–2):121–8. doi:10.1016/S0014-5793(99)01523-9. PMID:10580104.
- Zeng Z, Zhou Y, Zhang W, Li X, Xiong W, Liu H, Fan S, Qian J, Wang L, Li Z, et al. Family-based association analysis validates chromosome 3p21 as a putative nasopharyngeal carcinoma susceptibility locus. Genet Med. 2006;8(3):156–60. doi:10.1097/01.gim.0000196821.87655.d0. PMID:16540749.
- van Tilborg AA, Kompier LC, Lurkin I, Poort R, El Bouazzaoui S, van der Keur K, Zuiverloon T, Dyrskjot L, Orntoft TF, Roobol MJ, et al. Selection of microsatellite markers for bladder cancer diagnosis without the need for corresponding blood. PloS one. 2012;7(8):e43345. doi:10.1371/journal.pone.0043345. PMID:22927958.
- Zhang X, Li J. Era of universal testing of microsatellite instability in colorectal cancer. World J Gastrointest Oncol. 2013;5(2):12–19. doi:10.4251/wjgo.v5.i2.12. PMID:23556052.
- Jenkins G, O'Byrne KJ, Panizza B, Richard DJ. Genome stability pathways in head and neck cancers. Int J Genomics. 2013;2013:464720. doi:10.1155/2013/464720. PMID:24364026.
- Edge SB, Compton CC. The American Joint Committee on Cancer. the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–74. doi:10.1245/s10434-010-0985-4. PMID:20180029.
- Akhter M, Hossain S, Rahman QB, Molla MR. A study on histological grading of oral squamous cell carcinoma and its co-relationship with regional metastasis. J Oral Maxillofac Pathol. 2011;15(2):168–76. doi:10.4103/0973-029X.84485. PMID:22529575.
- Sarkar S, Maiti GP, Jha J, Biswas J, Roy A, Roychoudhury S, Sharp T, Panda CK. Reduction of proliferation and induction of apoptosis are associated with shrinkage of head and neck squamous cell carcinoma due to neoadjuvant chemotherapy. Asian Pac J Cancer Prev. 2013;14(11):6419–25. doi:10.7314/APJCP.2013.14.11.6419. PMID:24377544.
