ABSTRACT
The growing use of genomic testing presents new treatment options but also new dilemmas. We describe here a heavily-pretreated metastatic triple negative breast cancer patient who failed to respond to conventional treatment. Genomic analyses were performed that discovered several targetable alterations (e.g. FGFR1, CDK6, INSR) and created a clinical challenge – which target to target first? Our solution to this relatively common scenario was using ex-vivo organ culture (EVOC) system to prioritize treatment directed toward the best molecular target. EVOC enabled the trial of several potent targeted agents (Everolimus, Linsitinib, Palbociclib, AZD4547) and allowed semi-quantitative measurement of tumor response. The best response was to FGFR inhibitor, AZD4547. Consequently, the most accessible FGFR inhibiting agents (Pazopanib, then Nintedanib) were administered and some response was achieved. This report provides a potential rationale for utilizing EVOC system to predict tumor response to targeted therapy when multiple targets are proposed.
Introduction
All cancer types are characterized by gene mutations. Recent technological advancements in next-generation sequencing (NGS), together with the steep decrease in their costs lead to widespread use of these methods.Citation1 However, in numerous cancer patients the interpretation of test results is challenging due to multiple gene mutations in various pathways, previously undescribed alterations (with unknown effect on protein function) and non-targetable alterations (without a potent inhibitor in the market).Citation2 When considering the optimistic scenario of multiple targetable mutations, the treating oncologist is facing a difficult dilemma- which target to target? In the largest North American precision medicine studies (TAPUR by American Society of Clinical Oncology and MATCH by the National Cancer Institute), there are no clear algorithms to prioritize targetable mutations to decide ‘which one to target first’.Citation3,Citation4 The most common practice in most institutions is to make decisions using a multi-disciplinary molecular tumor board discussion. Decisions are based upon best available clinical data (known to participants) and biological logic or assumptions.Citation5 One rare example of predefined treatment algorithm was utilized in the randomized phase 2 SHIVA clinical trial (Molecularly targeted therapy based on tumor molecular profiling versus conventional therapy for advanced cancer trialCitation6): first, the hormone receptor with the highest expression was taken into account; second, any mutation, amplification, or deletion was deemed to be of greater importance than was hormone receptor expression; and third, in cases with several mutations, amplifications, or deletions, the tumor board judged alterations to direct targets of a molecularly targeted agent to be of highest priority for the treatment decision—if two molecular alterations that were both direct targets of one of the available molecularly targeted agents were present, the board would make the decision based on which alteration was downstream. For various reasons, the trial failed to improve progression-free-survival using targeted therapies.
However, to our best knowledge, no ex-vivo based trials treated patients prospectively with targeted therapy after performing NGS of tumor tissue. Here we present the use of an ex-vivo organ culture (EVOC) to prioritize targeted treatment in a heavily pretreated metastatic triple negative breast cancer patient.
Case report
58 year old female, previously healthy, diagnosed with metaplastic subtype of triple negative breast cancer (cT3N0M0, stage IIB). Neoadjuvant chemotherapy with Cyclophosphamide and Doxorubicin was initiated. Due to tumor growth during treatment, the patient underwent mastectomy with sentinel lymph node biopsy. Pathologic analysis revealed 5 cm tumor and no axillary involvement (0 out of 5 nodes). Adjuvant chemotherapy with Paclitaxel and Carboplatin was administered. Genetic counseling and testing ruled out germline BRCA1/2 mutations. Two years later, the disease recurred in one ipsilateral axillary lymph node which was retreated with formal dissection (extracapsular extension was detected), radiation therapy (54 Gy to chest wall and axillary basin) and chemotherapeutic protocol comprised of Cyclophosphamide, Methotrexate and 5-Fluorouracil (CMF). A year later, another ipsilateral axillary recurrence was treated with surgery and re-irradiation. Sadly, pulmonary metastases rapidly appeared and were resistant to combination of Carboplatin, Paclitaxel and Bevacizumab. Switch to Eribulin and Bevacizumab did not prevent further tumor growth.
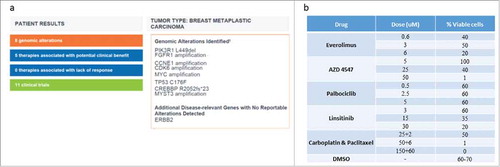
To obtain additional insights into the molecular alterations in the tumor, a sample from the primary tumor was sent for NGS profiling (both our local whole-genome-sequencing (WGS) and commercially available FoundationOne test). Both analyses revealed major molecular alterations (several targetable with available agents); amplifications of FGFR1, CCNE1, CDK6, MYC, MYST3, INSR and mutations in PIK3R1, TP53, CREBBP and (). FGFR1 and CCNE1 amplifications were found on both tests. Detailed WGS methods appear in Supplementary 1.
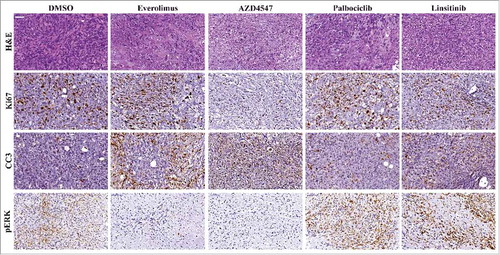
Figure 1. Ex Vivo Organ Culture (EVOC) Results. 1a) Results of tumor sequencing by FoundationOne test. 1b) various drugs in several doses which were used to treat the tumor in the EVOC. Percentage of viable cells as assessed by an expert pathologist examination.
Then, a lung biopsy was performed and small pieces of the tumor were implanted into immune-deficient mice (NSG mice) as patient-derived xenografts (PDX). After 2 passages of tumor between mice to allow expansion (12 weeks period), a tumor was harvested, sliced into 250uM slices and cultured in medium containing antimicrobials. Tissue was incubated in a moist chamber with 80% oxygen and 5% CO2, on titanium mesh inserts, on a rotating platform. After overnight acclimatization, tissue was treated with targeted therapy at 3 concentrations (or DMSO control) for an additional 96 hours (), afterwhich the tumor slices were fixed in paraformaldehyde and embedded in paraffin to allow morphological analysis.
A specialized cancer pathologist examined Hematoxylin & Eosin stained slides and scored the percentage of viable cells (). As presented, best tumor response was to FGFR inhibitors (), manifested also by less proliferation, increased apoptosis and decreased downstream signaling. Multi-disciplinary molecular tumor board decided to start treatment with Pazopanib, a multiple tyrosine kinase inhibitor (TKI) that can also block FGFR1. After two months of treatment with severe side effects (proteinuria and hypertension, partially controlled with intensive therapy and dose reductions), PET-CT showed mixed response to treatment. A second molecular tumor board discussion offered the patient a switch to a less neprotoxic TKI, Nintedanib which continued for 2 months with stable disease. Sadly, a brief time after the imaging the patient suddenly died in her sleep. Autopsy to define the cause of death was not performed.
Discussion
Cancer genomics studies are expanding our knowledge of the molecular disturbances in cancer. More and more alterations are found with increasing accuracy and depth of sequencing. However, the process of translating the molecular findings found by NGS testing to treatment plan is still frustratingly difficult.Citation2 There are major obstacles in clinical practice which make it a daily challenge to decide which molecule or alteration is clinically significant and which to target. Many genetic changes are not described in the major databases (ClinVar, VarSome etc.) and therefore regarded as Variants of Unknown Significance (VUS). To complicate the ‘pure’ bio-medical considerations, the clinician must account for financial, regulatory and bureaucratic matters. One important issue is the accessibility of patients to phase 1 clinical trials; a cumbersome mission outside the largest academic cancer centers of North America or Western Europe.Citation7
Clinicians are enthusiastic to define and use predictive biomarker for every targeted therapy agent. An excellent biomarker will enable better patient selection and better outcomes with lower costs. A great example is the recent U.S. Food & Drug Administration approval of an histology-agnostic biomarker to guide treatment with pembrolizumab in high microsatellite instability (MSI-H) or mismatch repair deficiency (dMMR). However, due to tumor heterogeneity, microenvironment and other factors, many biomarkers for targeted agents fail to perform at a high prediction level. In order to overcome some of those barriers, ex vivo platforms were developed.Citation8 Ex-vivo experiments are cheaper and significantly faster than PDX in mice. They enable maintenance of the tumor microenvironment and permit the trial of multiple different drugs in an individual tumor. Ex-vivo experiments lack the advantages of in-vivo experiments regarding parameters that cannot be recapitulated ex-vivo (e.g. drugs' doses, pharmacodynamics, tumor-organism interactions, and long term response).
Accumulating evidence suggests that ex-vivo systems may serve as a potent and robust platform in various tumor models; breast, head and neck, colorectal etc.Citation9-Citation15 Furthermore, ex-vivo platforms may reflect patients' response to treatment and allow for the study of clinically important molecular processes in the tumor following drug exposure with additional clinical implications.Citation16
In the present report, we show the successful use of an established platform (EVOC) exploited to answer a novel treatment challenge: prioritization of genomic targets found by NGS, to guide the treatment of an aggressive subtype of triple negative breast cancer. Our patient's metastatic disease had progressed on every chemotherapeutic regimen and achieved a stable disease of several months using treatment prioritized using EVOC. Until future functional assays or standardized clinical algorithms become available, this approach can be used to treat selected patients with several targetable genomic alterations to prioritize the most likely effective therapy. Worldwide, initial reports on the outcome of NGS-based targeted therapy approach in pretreated metastatic patients show initial promising results.Citation17,Citation18 Specifically in breast cancer, the recent results of AURORA (European multinational collaborative molecular screening initiative for advanced breast cancer patientsCitation19) show that 63% of patients have at least one targetable alteration while more than 40% have two or more. Not surprisingly, targeted treatment prioritization algorithm was not published.
Yet, certain limitations deserve to be mentioned. First, a significant effort is required to perform the EVOC system in well-standardized fashion. Second, the dosage of various drugs is partially arbitrary in the EVOC method and may not represent real-life therapeutic levels. Third, due to tumor heterogeneity and sampling issues; NGS may miss the actual driver mutations of the tumor.
In conclusion, this report provides a potential rationale for a strategy of utilizing EVOC system to predict tumor response to targeted therapy. Using EVOC on a given tumor with multiple genomic aberrations can provide additional information and prove a useful tool in the oncologist's armamentarium in the NGS era.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
2017CBT10803R-s02.docx
Download MS Word (14.2 KB)References
- Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016; 17;17(6):333–51. doi:10.1038/nrg.2016.49.
- Carr TH, McEwen R, Dougherty B, Johnson JH, Dry JR, Lai Z, Ghazoui Z, Laing NM, Hodgson DR, Cruzalegui F, et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer. 2016; 26;16(5):319–29. doi:10.1038/nrc.2016.35.
- American Society of Clinical Oncology. Targeted Agent and Profiling Utilization Registry (TAPUR) Study. [accessed April; 2017], https://www.tapur.org.
- National Cancer Institute. Molecular Analysis for Therapy Choice (MATCH) Trial. [accessed April; 2017], https://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/nci-match.
- Gingras I, Sonnenblick A, de Azambuja E, Paesmans M, Delaloge S, Aftimos P, Piccart MJ, Sotiriou C, Ignatiadis M, Azim HA, Jr. The current use and attitudes towards tumor genome sequencing in breast cancer. Sci Rep. 2016; 02;6:22517. doi:10.1038/srep22517.
- Le Tourneau C, Delord JP, Goncalves A, Gavoille C, Dubot C, Isambert N, Campone M, Tredan O, Massiani MA, Mauborgne C, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–34. doi:10.1016/S1470-2045(15)00188-6.
- Galsky MD, Stensland KD, McBride RB, Latif A, Moshier E, Oh WK, Wisnivesky J. Geographic accessibility to clinical trials for advanced cancer in the United States. JAMA Intern Med. 2015;175(2):293–5. doi:10.1001/jamainternmed.2014.6300.
- Meijer TG, Naipal KA, Jager A, van Gent DC. Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Sci OA. 2017;3(2):FSO190. doi:10.4155/fsoa-2017-0003.
- Brijwani N, Jain M, Dhandapani M, Zahed F, Mukhopadhyay P, Biswas M, Khatri D, Radhakrishna VD, Majumder B, Radhakrishnan P, et al. Rationally co-targeting divergent pathways in KRAS wild-type colorectal cancers by CANscript technology reveals tumor dependence on Notch and Erbb2. Sci Rep. 2017; 04;7(1):1502. doi:10.1038/s41598-017-01566-x.
- Donnadieu J, Lachaier E, Peria M, Saidak Z, Dakpe S, Ikoli JF, Chauffert B, Page C, Galmiche A. Short-term culture of tumour slices reveals the heterogeneous sensitivity of human head and neck squamous cell carcinoma to targeted therapies. BMC Cancer. 2016 ;16;16:273. doi:10.1186/s12885-016-2318-x.
- Gerlach MM, Merz F, Wichmann G, Kubick C, Wittekind C, Lordick F, Dietz A, Bechmann I. Slice cultures from head and neck squamous cell carcinoma: a novel test system for drug susceptibility and mechanisms of resistance. Br J Cancer. 2014; 21;110(2):479–88. doi:10.1038/bjc.2013.700.
- Majumder B, Baraneedharan U, Thiyagarajan S, Radhakrishnan P, Narasimhan H, Dhandapani M, Brijwani N, Pinto DD, Prasath A, Shanthappa BU, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun. 2015; 27;6:6169. doi:10.1038/ncomms7169.
- Merz F, Gaunitz F, Dehghani F, Renner C, Meixensberger J, Gutenberg A, Giese A, Schopow K, Hellwig C, Schafer M, et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro Oncol. 2013;15(6):670–81. doi:10.1093/neuonc/not003.
- Naipal KA, Verkaik NS, Sanchez H, van Deurzen CH, den Bakker MA, Hoeijmakers JH, Kanaar R, Vreeswijk MP, Jager A, van Gent DC. Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer. 2016; 09;16:78. doi:10.1186/s12885-016-2119-2.
- Roife D, Dai B, Kang Y, Perez MVR, Pratt M, Li X, Fleming JB. Ex Vivo Testing of Patient-Derived Xenografts Mirrors the Clinical Outcome of Patients with Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2016; 15;22(24):6021–30. doi:10.1158/1078-0432.CCR-15-2936.
- Goldman A, Majumder B, Dhawan A, Ravi S, Goldman D, Kohandel M, Majumder PK, Sengupta S. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun. 2015; 11;6:6139. doi:10.1038/ncomms7139.
- Radovich M, Kiel PJ, Nance SM, Niland EE, Parsley ME, Ferguson ME, Jiang G, Ammakkanavar NR, Einhorn LH, Cheng L, et al. Clinical benefit of a precision medicine based approach for guiding treatment of refractory cancers. Oncotarget. 2016; 30;7(35):56491–500. doi:10.18632/oncotarget.10606.
- Zick A, Peretz T, Lotem M, Hubert A, Katz D, Temper M, Rottenberg Y, Uziely B, Nechushtan H, Meirovitz A, et al. Treatment inferred from mutations identified using massive parallel sequencing leads to clinical benefit in some heavily pretreated cancer patients. Medicine (Baltimore). 2017;96(20):e6931. doi:10.1097/MD.0000000000006931.
- Maetens M, Brown D, Irrthum A, Aftimos P, Viale G, Loibl S, Laes JF, Campbell PJ, Thompson A, Cortes J, et al. The AURORA pilot study for molecular screening of patients with advanced breast cancer-a study of the breast international group. NPJ Breast Cancer. 2017;3:23. doi:10.1038/s41523-017-0026-6. PMID:28685159.