ABSTRACT
A 52 year-old man with Erdheim-Chester Disease (ECD) (a non-Langerhans polyostotic sclerosing histiocytosis) had next-generation sequencing (NGS) performed as part of his diagnostic workup. In addition to the tissue BRAF V600E mutation that is found in over 50% of ECD cases, he was also found to have a JAK2 V617F alteration in cell-free circulating tumor DNA (ctDNA) (liquid biopsy). The latter was thought to be an “incidental” finding, perhaps due to clonal hematopoiesis (though this usually occurs in older individuals), as his blood counts were normal and he had no splenomegaly. Approximately 13 months after the ctDNA test showing JAK2 V617F, he developed anemia, thrombocytopenia, and splenomegaly. Marrow biopsy then showed megakaryocytic atypia and markedly increased marrow fibrosis, consistent with WHO grade 2 of 3 myelofibrosis. Therefore, the patient was determined to have ECD with a typical BRAF V600E mutation, as well as primary myelofibrosis, with the latter diagnosis manifesting clinically over one year after the JAK2 V617F was first detected in ctDNA. He recently was started on the JAK2 inhibitor ruxolitinib. This case demonstrates that genomic alterations detected by liquid biopsy for evaluation of specific malignancies already present may serve as an early harbinger of hematological disease.
KEYWORDS:
Introduction
Next-generation sequencing (NGS) is being incorporated into the management of patients with cancer. In some rare tumors, such as Erdheim-Chester Disease (ECD) (a non-Langerhans histiocytosis), NGS can assist in confirming the diagnosis, as a majority cases have a mutation of BRAF V600E.Citation1,Citation2 However, in some cases, NGS testing will report mutations that may be unexpected or appear to be irrelevant. When NGS testing is performed on blood-derived circulating tumor DNA (ctDNA), mutations in TP53, IDH2, DNMT3A, TET2, ASXL1 and JAK2 may be found; these alterations are common in myelodysplastic syndrome or other myeloid disorders.Citation3 However, a subset of elderly individuals can harbor these alterations and be without any clinical signs of a hematologic disease, a condition now called clonal hematopoesis of indeterminate potential (CHIP).Citation4 CHIP is found in over 10% of patients that are 80 years old or more, but at much lesser frequencies in patients that are younger.Citation5 While CHIP is associated with a subsequent diagnosis of hematologic malignancy (or cardiovascular disease), the overall incidence is only about 0.5% to 1.0% per year for hematologic cancer.Citation6 Therefore, the clinical context can help in the assessment of NGS findings. Here we present a case in which a seemingly incidental mutation of JAK2 was found on NGS of ctDNA, with development of clinically overt myelofibrosis (a disease classically associated with the the JAK2 V617F alteration) about 13 months after the blood test. The implications for early diagnosis and monitoring of hematologic and other malignancies are discussed.
Case presentation
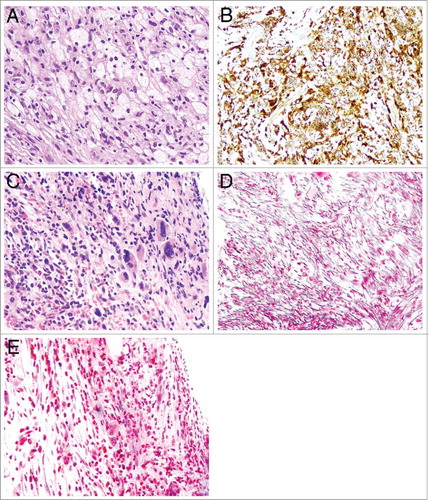
A 52 year-old Caucasian gentleman presented initially with eye swelling and a retro-orbital mass histologically consisting of CD68-positive foamy histiocytes ( and ) in the background of chronic inflammation. He was also found to have sclerotic lesions of long bones, including tibia, fibula, and femur, with epiphyseal sparing. Collectively these findings were most consistent with a diagnosis of Erdheim-Chester disease (ECD), as reviewed by our ophthalmic pathologist (JHL).Citation7
Figure 1. Microphotography of Erdheim Chester disease (ECD) and bone marrow fibrosis. A-B: There are numerous bland foamy histiocytes with abundant pale to clear cytoplasm (A) with immunoreactivity for CD68 (B) (original magnifications for A and B are 200x and 200x, respectively); C: The bone marrow core biopsy shows megakaryocytic hyperplasia, atypia and cellular streaming, indicative of fibrosis (original magnification 200x). D-E: Reticulin (D) and Trichrome (E) special stains show markedly increased reticulin fibrosis (D) and rare bundle of collagen (blue, E) (original magnifications of D and E are 200x and 200x, respectively).
To further evaluate the nature of the foamy histiocytes, the orbital mass tissue was assessed by targeted NGS, using the clinical-grade Foundation One panel (315 genes) (https://www.foundationmedicine.com). The NGS showed a BRAF V600E that is commonly associated with ECD, and further supported the diagnosis.Citation2 Sequencing of the orbital mass also demonstrated ASXL1 R693* and U2AF1 Q157P mutations, which were felt to be incidental and, at the time, not clinically relevant ().
Table 1. Summary of molecular alterations and clinical findings.
The patient was initially treated with vemurafenib and achieved disease stability. However, vemurafenib was poorly tolerated due to arthralgias, rash, and visual changes, despite dose reduction. He was then treated with trametinib, but similarly had difficulty tolerating it due to conjunctival swelling and uveitis.
At that time, plasma was sent for assessment of ctDNA both as a biomarker of response and to identify other potential actionable alterations. We used the Guardant360 test (Guardant Health, Inc., http://www.guardanthealth.com/) (digital sequencing (73 genes) in a Clinical Laboratory Improvement Amendments (CLIA)-licensed, College of American Pathologist (CAP)-accredited, New York State Department of Health-approved clinical laboratory).Citation8 The BRAF V600E mutation was no longer detected, consistent with the fact that he had stabilized on BRAF and MEK inhibitors, which slow cell growth and turnover, leading to decreased shedding of abnormal ctDNA in the blood.Citation9,Citation10 However, a mutation of JAK2 V617F was identified with a variant allele frequency (VAF) of 2.9%. In the absence of hematologic abnormalities, this anomaly was also interpreted as an incidental finding without clinical relevance.
He was then treated with pegylated interferon alfa-2b, with stability of ECD based on symptoms and radiographic findings. However, approximately 13 months after detection of the JAK2 alteration in ctDNA, he developed transfusion-dependent anemia (hemoglobin, 7 g/dL), thrombocytopenia (platelets 70–903/uL), and splenomegaly. A marrow core biopsy was performed and showed megakaryocytic atypia () and markedly increased marrow fibrosis by reticulin () and trichrome () stains, consistent with WHO grade 2 out of 3 myelofibrosis. There was no increase or aberrancy of myeloid blasts (not shown). Plasma was reassessed at this time by NGS of ctDNA and showed the following mutations: RIT1 M90V (VAF 4.0%), JAK2 V617F (3.2%), KRAS A59T (2.9%), and BRAF V600E (0.06%). () Although KRAS and RIT1 have been associated with other malignancies, such as lung cancer, workup including a PET/CT scan did not reveal other malignancy.
Collectively, therefore, the patient is assessed to have ECD with a typical BRAF V600E mutation, as well as primary myelofibrosis, with a typical JAK2 V617F. The myelofibrosis was not clinically apparent when first detected. He recently started ruxolitinib therapy. A repeat marrow biopsy has not yet been assessed; however, he has had a clinical response, with a decrease in splenomegaly, from 16.1 cm craniocaudal dimension (4.1 cm greater than upper range of normal) to 13.9 cm (2.9 cm). He still remains dependent on packed red blood cell transfusions approximately monthly.
Discussion
This case illustrates an increasingly common scenario in which genomic evaluation of malignancy reveals unexpected findings, and it may be difficult to determine their significance or make sense of seemingly discrepant results. In this case, with the appropriate clinical context, JAK2 V617F is very supportive of a diagnosis of a myeloproliferative neoplasm.Citation11 Similarly, mutations of U2AF1, ASXL1, and RIT1 have been shown to occur commonly in myeloid neoplasms ().Citation12–Citation15 The U2AF1 and ASXL1 genes found in tissue would not have been found in the ctDNA NGS targeted 73-gene panel because those genes are not covered. Why the RIT1 M90V mutation was found in plasma but not tissue may reflect a tissue false negative due to spatial heterogeneity (present in a different location than the biopsied location) or temporal heterogeneity (mutation acquired after the tissue biopsy).
Seemingly incidental gene mutations may therefore represent very early stages of malignancy. The presence of asymptomatic premalignant states in hematologic cancers is well established. Monoclonal gammopathy of undetermined significance and monoclonal B-cell lymphocytosis very commonly precede the diagnoses of multiple myeloma and chronic lymphocytic leukemia, respectively. Similarly, in our patient, blood counts were normal and there was no splenomegaly at the time of initial ctDNA positivity for JAK2 V617F. Indeed, it was recently reported that JAK2 V617F mutated clones may be present at very low levels sometimes years prior to diagnosis.Citation16
For those reasons, NGS may facilitate the detection of premalignant clones, and aid in determining prognosis. Indeed, the genes most commonly found to be mutated in healthy individuals include DNMT3A, TET2, and ASXL1, which have been shown to be early driver mutations that may result in a pre-leukemic state.Citation17,Citation18 Similarly, TP53 mutations have been identified at low levels several years before diagnosis of therapy-related acute myeloid leukemia (AML) and secondary myelodysplastic syndrome (MDS).Citation19 Monitoring for such mutations may be helpful in individuals at increased risk for developing secondary AML or MDS. Of relevance to our case is the recent finding that there is a high prevalence (approximately 10%) of myeloid neoplasms in adults with ECD.Citation20
On the other hand, clonal hematopoiesis may also be a benign condition, and this can confound the interpretation of NGS results. Blood cells of a proportion of healthy individuals without any clinically apparent hematologic abnormality can also be found to have such mutations, a condition called clonal hematopoiesis of indeterminate potential (CHIP).Citation21,Citation22 This appears to be an aspect of aging, and such mutations can be found in greater than 10% of individuals over the age of 80; however, at age 52 (our patient's age), such mutations are uncommon.Citation6 Interestingly, CHIP is associated with a nearly two-fold risk of coronary heart disease, possibly due to increased inflammation.Citation22 CHIP is also associated with an increased relative risk for subsequent diagnosis of hematologic cancer, approximately ten times higher than the general population, though the absolute risk of hematologic cancer remains low, approximately 0.5-1% per year in persons with an incidental mutation.Citation4,Citation6 Therefore, diagnosis of a hematologic malignancy is largely dependent on morphologic changes and clinical signs and symptoms.Citation23,Citation24 As in this case, knowledge of mutations and their association with specific conditions may allow for a more streamlined diagnostic workup should blood count abnormalities develop in the future.
This case also illustrates the potential utility of monitoring ctDNA for genomic aberrations and early diagnosis. ctDNA is released to the circulation from cells undergoing cell death, and has been useful in aiding with the diagnosis of ECD, as well as other cancers, as an alternative or an adjunct to tissue biopsy.Citation8,Citation25–Citation31 The amount of mutant DNA may also correlate with disease burden or prognosis.Citation25–Citation28,Citation32 In this case, it appears that the variant allele fraction (VAF) of BRAF V600E correlated well with response to BRAF inhibitor treatment, becoming undetectable when on therapy, and then expanding at the most recent assessment to a low but detectable level when off vemurafenib and on interferon.
The clinician should be aware that genomic testing for established tumors may reveal CHIP mutations of hematopoietic origin. There are several relatively simple means of distinguishing a CHIP mutation from a somatic mutation due to a solid tumor or a disorder such as ECD. First of all, some of the genes that drive many hematologic malignancies are relatively uncommon in solid tumor malignancies and in conditions such as ECD.Citation33 Although one can find case reports of JAK2 mutations associated with cancers such as lung cancer, these may be related to tissue biopsies heavily infiltrated with lymphocytes. Notably drivers of lung cancer such as EGFR and ROS1 are not reported as drivers of hematologic conditions (although KRAS mutations may play a role in myelodysplastic syndromes).Citation34 Secondly, the VAF of the CHIP mutation is unlikely to change when the established malignancy is treated, unless the treatment would also be expected to impact the hematological disorder. For example, in this case, the JAK2 V617F VAF in the liquid biopsy does not change over time (3.2% then 2.9%). A third potential differentiator is when the other somatic mutations cluster around one level of allele fraction, while the CHIP occurs at a much higher or lower VAF. This case was illustrated recently in a report by Zhang et al. where an IDH2 R140Q mutation was found at 19.6% VAF while the other somatic mutations clustered at VAFs two orders of magnitude lower, at 0.1%-0.2% VAF.Citation35
Most importantly, our results suggest potential clinical utility of ctDNA analysis in early detection of malignant states. Indeed, our patient showed the JAK2 V617F, a hallmark of myelofibrosis, in his ctDNA 13 months before any signs or symptoms suggestive of myelofibrosis. ctDNA may also be useful because dynamic changes in levels may indicate response to therapy, as seen in our patient and reported in the literature.Citation25 However, confounders to the interpretation of ctDNA for use as an early diagnostic tool exist, including the fact that benign lesions may harbor mutations often associated with malignancyCitation36 and that clonal hematopoiesis occurs in the elderly and does not always progress to malignancy.Citation5,Citation6 Even so, as illustrated by the individual presented herein, ctDNA alterations can be a very early harbinger of cancer, indicating that further exploration of this modality for early diagnosis or prevention of hematological malignancy is needed, particular in high-risk individuals.
Declarations
Richard Lanman is an employee of Guardant. Razelle Kurzrock receives consultant fees from X-biotech, Loxo Actuate Therapeutics, as well as speaker fees from Roche, and research funds from Incyte, Genentech, Pfizer, Sequenom, Guardant, Foundation Medicine and Merck Serono, and has an ownership interest in CureMatch Inc.
Additional information
Funding
References
- Blombery P, Wong SQ, Lade S, Prince HM. Erdheim-Chester disease harboring the BRAF V600E mutation. J Clin Oncol. 2012;30(32):e331–32. doi:10.1200/JCO.2012.43.2260. PMID:23008323.
- Haroche J, Charlotte F, Arnaud L, von Deimling A, Helias-Rodzewicz Z, Hervier B, Cohen-Aubart F, Launay D, Lesot A, Mokhtari K, et al. High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 2012;120(13):2700–703. doi:10.1182/blood-2012-05-430140. PMID:22879539.
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–478. doi:10.1038/nm.3733. PMID:25326804.
- Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi:10.1182/blood-2015-03-631747. PMID:25931582.
- Heuser M, Thol F, Ganser A. Clonal hematopoiesis of indeterminate potential. Dtsch Arztebl Int. 2016;113(18):317–22. PMID:27215596.
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–498. doi:10.1056/NEJMoa1408617. PMID:25426837.
- Haroun F, Millado K, Tabbara I. Erdheim-Chester disease: Comprehensive review of molecular profiling and therapeutic advances. Anticancer Res. 2017;37(6):2777–783. PMID:28551613.
- Janku F, Vibat CR, Kosco K, Holley VR, Cabrilo G, Meric-Bernstam F, Stepanek VM, Lin PP, Leppin L, Hassaine L, et al. BRAF V600E mutations in urine and plasma cell-free DNA from patients with Erdheim-Chester disease. Oncotarget. 2014;5(11):3607–610. doi:10.18632/oncotarget.1964. PMID:25003820.
- Scholer LV, Reinert T, Orntoft MW, Kassentoft CG, Arnadottir SS, Vang S, Nordentoft I, Knudsen M, Lamy P, Andreasen D, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23(18):5437–445. doi:10.1158/1078-0432.CCR-17-0510. PMID:28600478.
- Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra392. doi:10.1126/scitranslmed.aaf6219.
- Jamieson CH, Gotlib J, Durocher JA, Chao MP, Mariappan MR, Lay M, Jones C, Zehnder JL, Lilleberg SL, Weissman IL. The JAK2 V617F mutation occurs in hematopoietic stem cells in polycythemia vera and predisposes toward erythroid differentiation. Proc Natl Acad Sci U S A. 2006;103(16):6224–229. doi:10.1073/pnas.0601462103. PMID:16603627.
- Carbuccia N, Murati A, Trouplin V, Brecqueville M, Adelaide J, Rey J, Vainchenker W, Bernard OA, Chaffanet M, Vey N, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23(11):2183–186. doi:10.1038/leu.2009.141. PMID:19609284.
- Gomez-Segui I, Makishima H, Jerez A, Yoshida K, Przychodzen B, Miyano S, Shiraishi Y, Husseinzadeh HD, Guinta K, Clemente M, et al. Novel recurrent mutations in the RAS-like GTP-binding gene RIT1 in myeloid malignancies. Leukemia. 2013;27(9):1943–946. doi:10.1038/leu.2013.179. PMID:23765226.
- Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24(6):1128–138. doi:10.1038/leu.2010.69. PMID:20428194.
- Tefferi A, Finke CM, Lasho TL, Wassie EA, Knudson R, Ketterling RP, Hanson CA, Pardanani A. U2AF1 mutations in primary myelofibrosis are strongly associated with anemia and thrombocytopenia despite clustering with JAK2V617F and normal karyotype. Leukemia. 2014;28(2):431–33. doi:10.1038/leu.2013.286. PMID:24097336.
- McKerrell TPN, Chi J., Collord G., Moreno T., Ponstingl H., Dias J., Gerasimou P., Melanthiou K., Prokopiou C., Antoniades M., et al. JAK2 V617F hematopoietic clones are present several years prior to MPN diagnosis and follow different expansion kinetics. Blood Advances. 2017;1(14):968–71. doi:10.1182/bloodadvances.2017007047. PMID:29296738.
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118. doi:10.1126/scitranslmed.3004315. PMID:22932223.
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–33. doi:10.1038/nature13038. PMID:24522528.
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–55. doi:10.1038/nature13968. PMID:25487151.
- Papo M, Diamond EL, Cohen-Aubart F, Emile JF, Roos-Weil D, Gupta N, Durham BH, Ozkaya N, Dogan A, Ulaner GA, et al. High prevalence of myeloid neoplasms in adults with non-Langerhans cell histiocytosis. Blood. 2017;130(8):1007–013. doi:10.1182/blood-2017-01-761718. PMID:28679734.
- Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–487. doi:10.1056/NEJMoa1409405. PMID:25426838.
- Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111–21. doi:10.1056/NEJMoa1701719. PMID:28636844.
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the world health organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi:10.1182/blood-2016-03-643544. PMID:27069254.
- Tefferi A, Noel P, Hanson CA. Uses and abuses of JAK2 and MPL mutation tests in myeloproliferative neoplasms a paper from the 2010 William Beaumont hospital symposium on molecular pathology. J Mol Diagn. 2011;13(5):461–66. doi:10.1016/j.jmoldx.2011.05.007. PMID:21723416.
- Husain H, Melnikova VO, Kosco K, Woodward B, More S, Pingle SC, Weihe E, Park BH, Tewari M, Erlander MG, et al. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res. 2017;23(16):4716–723. doi:10.1158/1078-0432.CCR-17-0454.
- Husain H, Nykin D, Bui N, Quan D, Gomez G, Woodward B, Venkatapathy S, Duttagupta R, Fung E, Lippman SM, et al. Cell-Free DNA from ascites and pleural effusions: Molecular insights into genomic aberrations and disease biology. Mol Cancer Ther. 2017;16(5):948–55. doi:10.1158/1535-7163.MCT-16-0436. PMID:28468865.
- Janku F, Kurzrock R. Bringing blood-based molecular testing to the clinic. Clin Cancer Res. 2016;22(22):5400–402. doi:10.1158/1078-0432.CCR-16-1769. PMID:27663595.
- Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, Patel SP, Harismendy O, Ikeda M, Parker BA, et al. Use of liquid biopsies in clinical oncology: Pilot experience in 168 Patients. Clin Cancer Res. 2016;22(22):5497–505. doi:10.1158/1078-0432.CCR-16-0318. PMID:27185373.
- Schwaederle M, Patel SP, Husain H, Ikeda M, Lanman R, Banks KC, Talasaz A, Bazhenova L, Kurzrock R. Utility of genomic assessment of blood-derived circulating tumor DNA (ctDNA) in patients with advanced lung adenocarcinoma. Clin Cancer Res. 2017;23(17):5101–111. doi:10.1158/1078-0432.CCR-16-2497. PMID:28539465.
- Kato S, Krishnamurthy N, Banks KC, De P, Williams K, Williams C, Leyland-Jones B, Lippman SM, Lanman RB, Kurzrock R. Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary. Cancer Res. 2017;77(16):4238–246. doi:10.1158/0008-5472.CAN-17-0628. PMID:28642281.
- Schwaederle M, Chattopadhyay R, Kato S, Fanta PT, Banks KC, Choi IS, Piccioni DE, Ikeda S, Talasaz A, Lanman RB, et al. Genomic alterations in circulating tumor DNA from diverse cancer patients identified by next-generation sequencing. Cancer Res. 2017;77(19):5419–427. doi:10.1158/0008-5472.CAN-17-0885. PMID:28807936.
- Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O'Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P, Jackman DM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–705. doi:10.1158/1078-0432.CCR-13-2482. PMID:24429876.
- Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–134. doi:10.1016/j.cell.2012.08.024. PMID:22980976.
- Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, Przychodzen B, Nagata Y, Meggendorfer M, Sanada M, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204–12. doi:10.1038/ng.3742. PMID:27992414.
- Zhang BM, Aleshin A, Lin CY, Ford J, Zehnder JL, Suarez CJ. IDH2 mutation in a patient with metastatic colon cancer. N Engl J Med. 2017;376(20):1991–992. doi:10.1056/NEJMc1701072. PMID:28514606.
- Kato S, Lippman SM, Flaherty KT, Kurzrock R. The conundrum of genetic "drivers" in benign conditions. J Natl Cancer Inst. 2016;108(8). doi:10.1093/jnci/djw036.
