ABSTRACT
Introduction: Tissue sampling of gallbladder cancer (GBCa) is challenging because of the anatomy of the gallbladder. The aim of this study is to investigate the possibility of diagnosing GBCa patients by performing a liquid biopsy of bile in addition to current diagnostic methods.
Methods: Thirty patients with GBCa were enrolled in this study. Cytological examination was performed in all patients. Using next generation sequencing (NGS), DNA isolated from the bile and tumor tissue was analyzed for mutations in 49 oncogenes. We also compared these mutations with cytology results.
Results: 57.1% of DNA samples from tumor tissue were positive for a mutation. In these patients, 87.5% of the bile circulating tumor DNA (ctDNA) samples had the same mutation. The concordance rate between bile ctDNA and tissue DNA samples was 85.7%, and the mutation frequencies detected in ctDNA were approximately half of what was detected in tumor tissue DNA. On the other hand, the sensitivity of the cytological and bile ctDNA analyses was 45.8% and 58.3%, respectively. The concordance rate between cytology and bile ctDNA analyses was 87.5%.
Conclusions: Mutated tumor DNA could be detected in bile by NGS. Liquid biopsy of bile might help us to diagnose GBCa because of higher sensitivity and positive predict value compared to cytology with ERCP.
Introduction
Gallbladder cancer (GBCa) is a rare neoplasm with an incidence of 2.5 in 100,000 individuals.Citation1 Risk factors for its development include the presence of gallstones, chronic inflammation, anomalous pancreaticobiliary ductal junctionsCitation2,Citation3 and other environmental or metabolic factors such as diabetes.Citation4 GBCa cells are generally highly metastatic, but little is known about their pathogenesis. Surgical resection is the preferred therapy for GBCa. However, because patients with GBCa often do not show early symptoms, they often receive late diagnoses and unsatisfactory treatments leading to poor prognosis. In addition, tissue sampling of GBCa is challenging because of the anatomy of the gallbladder.
No clinical molecular marker for early diagnosis is known, and no effective targeted molecular therapies have been approved; therefore, the median survival time of individuals with GBCa is less than 1 y.Citation5 Because of this unmet medical need, the genomic and transcriptomic landscape, as well as the pathogenesis, of GBCa must be defined.
Knowledge of the spectrum of somatic mutations present in GBCa remains incomplete. A systematic review and meta-analysis of candidate gene studies in gallbladder cancer showed that the mutations reported to date were insufficient to confirm any association with GBCa.Citation6 However, a more comprehensive approach has been recently described. Jiao, Y et al. analyzed exome sequences from nine GBCa samples and identified TP53 as a significantly mutated gene.Citation7 Furthermore, Maolan Li et al. identified recurrent mutations in the ErbB pathway following whole genome sequencing of GBCa samples.Citation8
Computed tomography (CT), magnetic resonance imaging (MRI) and/or cytology are usually used to diagnose GBCa. To add support to the current methods of GBCa diagnosis, we investigated the possibility of performing a “liquid biopsy” of bile as a noninvasive diagnostic method to identify circulating tumor DNA (ctDNA). To achieve improved treatment effects in the future, it will be mandatory to utilize so-called “custom-made” approaches based on the character of each tumor.
Here, we characterized a GBCa cohort (30 cases, including tumor tissue and/or bile) of Japanese patients by analyzing a combination of 47 oncogenes, including potential therapeutic targets (Cancer panel; Haloplex, Agilent Technology, Santa Clara, CA, USA), and two other target genes (TERT and ERBB2). The aim of this study is to investigate the possibility of diagnosing GBCa patients by performing a liquid biopsy of bile in addition to current diagnostic methods.
Materials and methods
Patients and samples
Thirty patients with GBCa that were previously diagnosed histologically using material obtained by surgical (n = 20) or fine needle aspiration (FNA) methods (n = 10) between January 2012 and December 2016 were enrolled in this study. The median age of the patients was 77 y, and 9 patients (30%) were male. This cohort included stage I/II patients (12/30, 40%) (). Bile was obtained and cytological examination was performed in all patients. Using next generation sequencing (NGS), DNA isolated from the bile obtained from 24 of the 30 patients was analyzed for mutations in 49 oncogenes.
Table 1. Characteristics of patients with GBCa.
We also analyzed mutations in tumor tissue DNA obtained from 20 patients. As negative controls, 19 non-GBCa bile and 32 noncancerous gallbladder tissue samples were also analyzed for mutations in 49 oncogenes. All the samples were obtained prior to therapy.
All patients provided written informed consent for the examination of their samples and the use of their clinical data. The study protocol conformed to the ethical guidelines of the World Medical Association and the Declaration of Helsinki and was approved by the Okayama University Ethics Committee.
Cytology technique
Endoscopic retrograde cholangiopancreatography (ERCP) was performed by experienced endoscopists using a conventional therapeutic duodenoscope (TJF-260V, Olympus, Japan) and standard procedures. Endoscopic nasobiliary drainage (ENBD) tubes (pigtail nasal catheter, 5Fr, Cook Medical, USA) were placed, and bile (5 ml) was obtained 6 times over 2 d through continuous cytology. All aspirated materials were used for cytopathological evaluations. DNA extraction was performed using the first bile sample (3 ml out of 5 ml).
Extraction of DNA
Formalin fixed paraffin embedded (FFPE) samples were obtained by surgical resection. Histological examinations confirmed that these samples contained tumor cells at least 30%. We extracted DNA from 5-μm thick sections from FFPE samples using a QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions.
Bile (3 ml) for ctDNA analysis was collected in tubes (BD vacutainer blood collection tubes) and processed within 1 h after collection. The samples were stored at −80°C. ctDNA was extracted from aliquots (1 ml) of bile using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions.
Next-generation sequencing
We used NGS (Miseq, illumina, Hayward, CA, USA). We also performed deep targeted sequencing of 49 tumor-related genes in the target custom enrichment panel (Haloplex; Agilent Technologies, Santa Clara, CA, USA). These genes included the following: ABL1, AKT1, ALK, AR, ATM, BRAF, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB3, ERBB4, FANCA, FANCC, FANCF, FANCG, FGFR1, FGFR2, FGFR3, FLT3, HRAS, IDH1, IDH2, JAK2, JAK3, KIT, KRAS, MAP2K1, MAP2K2, MAP2K4, MET, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PIK3R1, PTEN, RET, RUNX1, SMAD4, SMO, SRC, STK11, TP53, VHL, WT1, hTERT, and ERBB2. These targets were selected in accordance with genes previously reported and identified as high priority in the COSMIC database. In the present study, we set the cut-off value to be a 0.05 frequency. In addition to these cut-off values, we set the coverage to be over 1000. We excluded the genes of which mutations were detected in noncancerous gallbladder tissue samples in further analysis.
Statistical analysis
Fisher's exact test was performed to analyze categorical data. Overall survival was measured from the d of diagnosis to the d of death or last follow-up examination. Survival curves were calculated using the Kaplan-Meier method. The log-rank test was used to compare survival curves. The Cox proportional hazards model was used to determine the significant variables contributing to survival. P-values <0.05 were considered statistically significant. All analyses were performed using JMP pro statistical software (ver. 13.0.0 SAS Institute, Japan).
Results
Comparison of ctDNA mutational analysis and cytological analysis of bile
Bile cytological analysis was performed in all 30 GBCa patients prior to treatments. Fourteen out of 30 patients with GBCa (46.7%) were diagnosed as having a definite class 5 cancer (). Bile ctDNA samples were obtained in 24 of 30 GBCa patients. Fourteen of these 24 (58.3%) bile ctDNA samples were positive for mutations. Mutations in TP53, ERBB2 and KRAS were detected in 11/24 (45.8%), 2/24 (8.3%), and 1/24 (4.2%) samples, respectively (). None of the 19 negative control samples had any mutations. Cytology was positive in 11 of 24 GBCa patients (45.8%) (). The sensitivity, specificity, positive predictive values (PPV) and negative predictive values (NPV) of the cytological analyses were 45.8%, 100%, 100% and 59.4%, respectively; these values were 58.3%, 100%, 100% and 65.5%, respectively, when the bile ctDNA analysis was performed. The concordance rate between cytology and bile ctDNA analyses was 87.5% (21/24; P < 0.01) (). Bile ctDNA could be analyzed in 6 of the stage I/II patients (12/30, 40%). Mutations in ctDNA were detected in 4 patients (66.7%). Conversely, cytology showed class 5 cancer in only 2 of these 6 patients (33.3%). In terms of sensitivity, ctDNA analysis of bile was superior to cytological analysis.
Table 2. GBCa profile.
Table 3. Comparison between mutation analysis and cytology with bile.
Concordance of mutations between tissue and bile
DNA analysis was performed in tumor tissues from 20 of the 30 GBCa patients. Eleven of 20 (55%) DNA samples were positive for a mutation. TP53, MET, SMAD4, CTNNB1 and AR were mutated in 7/20 (35.0%), 2/20 (10.0%), 1/20 (5.0%), 1/20 (5.0%) and 1/20 (5.0%) samples, respectively (). The frequency of the most common amino acid changes in each gene were as follows: TP53 (23.8%; E153K), TP53 (15.9%; R148K), TP53 (11.6%; R121K), TP53 (11.6%; P138L), TP53 (11.3%; A117D), TP53 (43.7%; R114C), TP53 (11.3%; A117D), TP53 (27.1%; S176R), MET (7.0%; P1246L), MET (42.9%; N375S), SMAD4 (18.5%; R361C), CTNNB1 (29.0%; T41I) and AR (8.0%; D223Y) (). None of the 32 negative control samples had mutations.
Both tumor tissue DNA and bile ctDNA samples were collected in 14 of the 30 GBCa patients. Eight of 14 (57.1%) DNA samples from tumor tissue were positive for a mutation. Seven (87.5%) of the bile ctDNA samples in these eight patients had the same mutation. The frequency of the most common amino acid changes in each gene were as follows: TP53 (15.0%; E153K), TP53 (5.0%; R148K), TP53 (5.7%; R121K), TP53 (5.7%; P138L), TP53 (5.7%; R114L), TP53 (9.1%; R114C), TP53 (5.7%; R114L), MET (49.8%; N375S) and ERBB2 (5.0%; 739Y) (). The concordance rate between bile ctDNA and tissue DNA samples was 85.7% (12/14; P < 0.01), and the mutation frequency detected in ctDNA was approximately half of what was detected in tumor tissue DNA. The bile samples of the remaining 6 patients without tumor tissue mutations also had no mutations.
Correlation with patients' survival
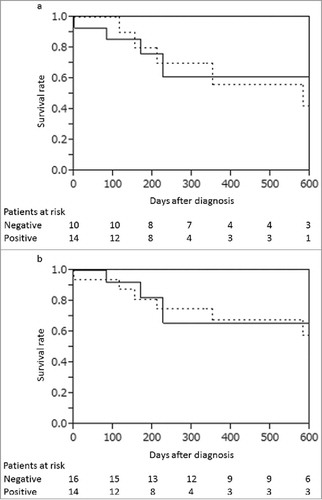
No significant difference of survival was observed between patients with and without the mutations in bile ctDNA analysis (P = 0.90; ), and between patients classified as class 5 positive and negative in bile cytology (P = 0.81; ). Univariate and multivariate analyses for potential prognostic factors such as age, tumor markers, diabetes status, the presence of gallstones, the presence of anomalous pancreaticobiliary ductal junctions, the presence of gene mutations in bile ctDNA or tissue DNA, and class 5 cytological status did not show significant differences. Tumor stage was the only significant prognostic factor (P = 0.01), while specific mutations, such as those found in TP53, were not correlated with prognosis. Association between allele frequency in bile ctDNA and tumor stage was observed (P = 0.03).
Figure 1. (a) The survival rates of the patients with bile ctDNA positive (solid line) and with bile ctDNA negative (dotted line). The survival rates were not significantly different (P = 0.90). (b) The survival rates of the patients with bile cytology class 5 positive (solid line) and negative (dotted line). The survival rates were not significantly different (P = 0.80).
Discussion
In this study, we first demonstrated that isolating ctDNA from bile was useful for the diagnosis of GBCa. The sensitivity of ctDNA (58.3%) was higher than that of cytology (45.8%). In addition, the mutation concordance rate between GBCa tissue DNA and bile ctDNA was high at 12/14 (85.7%) patients. The presence of ctDNA was not correlated with patient survival.
Although sensitivity is not high enough to be an alternative of tissue biopsy, liquid biopsy of bile is useful as a first step inspection since the same mutation pattern in tissue can be detected. Driver mutations were observed in approximately half of the cases by this noninvasive method.
TP53, ERBB2 and K-ras have been reported to be significant GBCa-related genes.Citation8 However, changes in these genes were not necessarily driver mutations, and the mutation frequencies were not high at 47.1%, 9.8%, and 7.8%, respectively.Citation8 The present study revealed that mutations were not only detectable in GBCa tissue DNA but were also detectable in bile ctDNA, with a high concordance rate (85.7%) between the two sample types. Our previous pancreatic cancer study using liquid biopsies of blood showed that K-ras mutation rates in EUS-FNA tissue DNA and blood ctDNA were 74.7% and 62.6%, respectively.Citation11 The K-ras mutation concordance rate between EUS-FNA tissue DNA and blood ctDNA was 77.3%. Surprisingly, in GBCa the mutation concordance rate between tumor tissue DNA and bile ctDNA was much higher than that of blood liquid biopsies in pancreatic cancer. This result means that performing a liquid biopsy of bile was useful for GBCa, although we were unable to detect any particular driver mutations as was achieved in pancreatic cancer, which displayed the characteristic driver mutation (K-ras) at rates ranging from 75% to 95%.Citation9,Citation10
The present study also revealed that in order to detect mutations in bile ctDNA, the mutation frequency in tumor tissue needed to be relatively high. In mutation positive cases in both tumor tissue DNA and bile ctDNA, the mutation frequencies in ctDNA were approximately half as much as those found in tumor tissue DNA. In one case with a detectable mutation in tissue DNA, mutations in bile ctDNA could not be detected. In this case, the mutation frequency detected in tumor tissue DNA (mutated DNA/not-mutated DNA) was low (7%). We have performed ctDNA analysis in blood in both pancreatic cancer and gastric cancer studies.Citation11,Citation12 These studies displayed results similar to the present study, as a tissue sample mutation frequency of greater than 10% was required to detect ctDNA mutations in blood. Interestingly, the present study showed that mutations in bile could be identified during stage I GBCa. It may not be necessary to wait until later stages to detect GBCa using a liquid bile biopsy, although for various cancers, the sensitivity of a liquid biopsy of blood has been reported to depend on the tumor stage.Citation13 This difference may be because there is a difference in the total amount of gene mutations present in bile and blood.
Cytological analysis often requires the repeated collection of 5 ml of bile (at least 6 times) until tumor tissue can be obtained because it has been difficult to obtain the requisite amount of sample at one time. All patients in this study needed continuous cytology. Conversely, liquid biopsy with bile required just one collection of 1 ml of bile. This method diagnosed GBCa quickly and easily.
Determining the origin of the ctDNA is a problem that has been reported when performing a liquid biopsy. It is especially difficult to estimate the ctDNA origin when performing a liquid biopsy of blood because several organs can produce ctDNA in blood. However, unlike previous reports using blood, the liquid biopsy in the present study was performed with bile so that ctDNA origin could be more easily identified. The hot spot positions and amino acid mutations in tumor tissue DNA were completely concordant with bile ctDNA. These findings convinced us that the bile ctDNA originated in GBCa tumor DNA.
Recently, circulating tumor cell (CTC) and ctDNA analysis of blood has been rapidly developed as a method of liquid biopsy. Our study is the first report of a liquid biopsy of bile. CTCs contain the whole genome; however, mutation detection rates remain low compared to ctDNA.Citation14,Citation15 On the other hand, ctDNA contains fragmented genomic information, but mutation detection rates are much higher than those in CTCs. In this study, we selected ctDNA for liquid biopsy, because ctDNA should exist in bile even if tumor tissue might not be detected in cytological analyses. We speculated that this characteristic might point to the superiority of bile ctDNA analyses over cytology.
Problems with obtaining sufficient tissue samples during biopsy lead to difficulties with GBCa diagnosis. Itoi et al reported that additional TERT analysis of the biopsy specimens obtained from GBCa patients improved the reliability of the diagnosis.Citation16 However, we have always faced problems with obtaining tumor tissue samples owing to their location. In view of this, performing a liquid biopsy of bile might overcome both problems at the same time and supersede conventional procedures.
This study was limited by the small sample size, the high cost, and the complex design. Mutation analysis was performed using NGS, which was not necessarily a cost effective method. Selecting appropriate target genes is very important, and digital PCR might be a candidate for the clinical application of bile biopsies, thus making analysis cheaper and easier. In addition to these limitations, a simple study design is needed to compare tissue DNA, bile ctDNA, and cytology in each GBCa patient.
In conclusion, ctDNA in bile could be detected by NGS. Liquid biopsies of bile might help us to diagnose GBCa because of higher sensitivity and PPV compared to cytology with ERCP. Prospective and large-scale studies are warranted for further verification of this technique in other patient populations.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authors' contributions
Dr. HK designed the study and drafted the manuscript. Dr. KN was responsible for the revision of the manuscript. Drs. KM and HK were responsible for the diagnosis and treatment of patients. Drs. SA, CD and HM assisted Dr. HK in analyzing the data. Dr. HO supervised the manuscript preparation. All authors approved the final manuscript.
Acknowledgments
This work was supported by KAKENHI (16K19345) and by a grant from the Japanese Society of Gastroenterology.
Additional information
Funding
References
- Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer. 2006;118:1591–602. doi:10.1002/ijc.21683. PMID:16397865.
- Wolpin BM, Mayer RJ. A step forward in the treatment of advanced biliary tract cancer. N Engl J Med. 2010;362:1335–7. doi:10.1056/NEJMe1001183. PMID:20375411.
- Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology. 2011;54:173–84. doi:10.1002/hep.24351. PMID:21488076.
- Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–29. doi:10.1053/j.gastro.2013.10.013. PMID:24140396.
- Boutros C, Gary M, Baldwin K, Somasundar P. Gallbladder cancer: past, present and an uncertain future. Surg Oncol. 2012;21:e183–191. doi:10.1016/j.suronc.2012.08.002. PMID:23025910.
- Srivastava K, Srivastava A, Sharma KL, Mittal B. Candidate gene studies in gallbladder cancer: a systematic review and meta-analysis. Mutat Res. 2011;728:67–79. doi:10.1016/j.mrrev.2011.06.002. PMID:21708280.
- Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–3. doi:10.1038/ng.2813. PMID:24185509.
- Li M, Zhang Z, Li X, Ye J, Wu X, Tan Z, Liu C, Shen B, Wang XA, Wu W et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet. 2014;46:872–6. doi:10.1038/ng.3030. PMID:24997986.
- Koorstra JB, Hustinx SR, Offerhaus GJ, Maitra A. Pancreatic carcinogenesis. Pancreatology. 2008;8:110–25. doi:10.1159/000123838. PMID:18382097.
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–54. doi:10.1016/0092-8674(88)90571-5. PMID:2453289.
- Kinugasa H, Nouso K, Miyahara K, Morimoto Y, Dohi C, Tsutsumi K, Kato H, Matsubara T, Okada H, Yamamoto K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271–80. doi:10.1002/cncr.29364. PMID:25823825.
- Kinugasa H, Nouso K, Tanaka T, Miyahara K, Morimoto Y, Dohi C, Matsubara T, Okada H, Yamamoto K. Droplet digital PCR measurement of HER2 in patients with gastric cancer. Br J Cancer. 2015;112:1652–5. doi:10.1038/bjc.2015.129. PMID:25897674.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi:10.1126/scitranslmed.3007094.
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi:10.1038/nature06385. PMID:18097410.
- Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–31. doi:10.1038/nrc3820. PMID:25154812.
- Itoi T, Shinohara Y, Takeda K, Takei K, Ohno H, Ohyashiki K, Yahata N, Ebihara Y, Saito T. Detection of telomerase activity in biopsy specimens for diagnosis of biliary tract cancers. Gastrointest Endosc. 2000;52:380–6. doi:10.1067/mge.2000.108303. PMID:10968854.
