ABSTRACT
Tobacco usage is a known risk factor associated with development of oral cancer. It is mainly consumed in two different forms (smoking and chewing) that vary in their composition and methods of intake. Despite being the leading cause of oral cancer, molecular alterations induced by tobacco are poorly understood. We therefore sought to investigate the adverse effects of cigarette smoke/chewing tobacco exposure in oral keratinocytes (OKF6/TERT1). OKF6/TERT1 cells acquired oncogenic phenotype after treating with cigarette smoke/chewing tobacco for a period of 8 months. We employed whole exome sequencing (WES) and quantitative proteomics to investigate the molecular alterations in oral keratinocytes chronically exposed to smoke/ chewing tobacco. Exome sequencing revealed distinct mutational spectrum and copy number alterations in smoke/ chewing tobacco treated cells. We also observed differences in proteomic alterations. Proteins downstream of MAPK1 and EGFR were dysregulated in smoke and chewing tobacco exposed cells, respectively. This study can serve as a reference for fundamental damages on oral cells as a consequence of exposure to different forms of tobacco.
Introduction
Tobacco is mainly consumed in two different forms viz. smoking and chewing, besides intake in the form of snuff. Cigarette smoking is prevalent in the West and epidemiological studies link cigarette smoking as a cause for development of oral cancer.Citation1,Citation2 The habit of chewing tobacco is mostly prevalent in the South East Asian countries such as India, Pakistan, China, Korea and Sri LankaCitation3 and in the African countries such as SudanCitation4 where smokeless tobacco-associated oral cancer cases are high. Case control studies undertaken on the Indian population have identified tobacco chewing as a major risk factor for multiple oral premalignant lesions compared to smoking.Citation5,Citation6 Significant differences exist in the etiology, prognosis and therapeutic response of oral cancer patients who are tobacco users compared to non-users.Citation7-9 The pathogenesis of cancer between chewers and smokers may vary significantly since the two forms of tobacco differ in their mode of intake and in their composition.
A number of in vitro and in vivo studies employ the use of research grade cigarettes to study quantitative and qualitative comparison of results across laboratories.Citation10 In addition, these cigarettes resemble cigarette brands available commercially. Upon burning of a cigarette, the smoke that is inhaled by a smoker is termed as mainstream smoke (MSS). Composition of MSS from a research grade cigarette smoked using standardized machine smoking protocols has been previously documented by Roemer et al.Citation10 Studies have identified a number of compounds including nicotine, aldehydes, aromatic amines, polycyclic aromatic hydrocarbons (such as Benzo[a]pyrene, Benz[a]anthracene), phenols, volatile N-nitrosamines and tobacco-specific N-nitrosamines. IARC categorizes at least 70 out of 5,300 identified components of cigarette smoke as carcinogenic.Citation11 Chewing tobacco is generally available as loose leaves, plugs or twists. The composition of tobacco leaves change as the leaves are cured, processed and stored. Smokeless tobacco is known to contain more than 3,000 chemical compounds including alkaloids (such as nicotine, nornicotine), aliphatic and aromatic hydrocarbons, aldehydes, ketones and amines.Citation12 Of these, 28 compounds have been classified as carcinogens by the IARC.Citation13
Molecular studies have implicated cigarette smoke and its components in sustained inflammation and suppression of immune response.Citation14 We have previously documented the adverse effects of chronic exposure to cigarette smoke condensate (CSC) on primary skin keratinocytes.Citation15 A number of in vitro studies have highlighted the carcinogenic effects of exposure to cigarette smoke or its constituents in different cell types including oral and esophageal cells.Citation16,Citation17 Acute exposure has been implicated in increased levels of oxidative stress markers and modulation of pathways involved in cell survival.Citation18,Citation19 Exposure to CSC or its constituents has been shown to activate markers of cell survival and metastasis and is known to result in irreversible genetic alterations in esophageal and oral cells.Citation20-22
Most studies on short term in vitro exposure to smokeless tobacco extract (STE) highlights the toxicity of STE in different cellular models. Decreased platelet adhesion,Citation23 increased ROS production and cellular membrane damage in cultured human oral epidermal carcinoma cellsCitation24 have been observed upon acute exposure to STE. Smokeless tobacco was also reported to significantly increase the proportion of aneuploid cells in HPV-positive oral keratinocytes thereby predisposing cells to malignant transformation.Citation25 A study on the effect of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) in Het1A cells highlights the role of nicotinic acetylcholine receptors (nAChRs) in tobacco-related carcinogenesis.Citation26 Another study identifies alterations in the expression of cell cycle regulators, such as cyclin D1, tumor suppressor retinoic acid receptor beta (RARb) and proteins involved in DNA methylation such as MGMT upon treatment with ‘Khaini’ on epithelial cells cultured from human oral leukoplakia.Citation27 In addition, components of cellular processes such as transcription, cell-cell adhesion, signaling, growth and transformation have been shown to be differentially expressed in oral pre-malignant lesions (OPLs) and oral squamous cell carcinomas (OSCCs) of smokeless tobacco consumers.Citation28
A number of high-throughput sequencing studies have investigated the dissimilarities exhibited at the genomic, transcriptomic or proteomic levels in different samples from smokers compared to non-smokers,Citation29-34 including those in lung cancer.Citation35,Citation36 In comparison, few studies discuss the impact of chewing or other forms of smokeless tobacco on gene expression using high-throughput techniques.Citation37-39 Our group has previously reported the proteome-wide alterations in oral keratinocytes upon chronic exposure to smokeless tobacco and identified stearoyl CoA desaturase as a potential therapeutic target in head and neck squamous cell carcinoma.Citation40
Metabolic differences have been reported between smokers and snuff users but the molecular/proteomic alterations between smokers and tobacco chewers are not well elucidated.Citation41 A recent study employed microarray analysis to elucidate gene expression differences in peripheral blood mononuclear cells (PBMCs) obtained from smokers and moist snuff users.Citation42 They reported differential expression of several genes including aryl-hydrocarbon receptor repressor (AHRR), G protein-coupled receptor 15 (GPR15), leucine rich repeat neuronal 3 (LRNN3), cochlin (COCH) and serine protease 23 (PRSS23) in smokers compared to moist snuff users. Though there are a few studies comparing the effects of different tobacco products in vitro, most investigate the adverse effects of acute exposure rather than chronic exposure to smoke or smokeless tobacco. A microarray-based profiling study on oral cell lines exposed to different tobacco product preparations for 24 h identified aldo-keto reductase gene, AKR1C1, as a potential candidate to differentiate between the biological effects of combustible and non-combustible tobacco products in a cellular model.Citation43 In another study, acute exposure to Swedish-type smokeless tobacco (Snus) extract or cigarette smoke extract resulted in altered gene expression profiles in human normal endothelial cells and fibroblasts.Citation44 However, acute treatment for up to 24 h with Snus extract increased cell proliferation while treatment with cigarette smoke extract for the same duration adversely affected cell proliferation.
Despite being one of the leading causes of oral cancer, the molecular mechanisms involved in tobacco-induced oncogenic transformation are yet to be fully elucidated. Simple, interference-free cellular models of chronic smoke or smokeless tobacco exposure could aid in expanding this understanding in oral cells. We performed whole exome sequencing of cigarette smoke and chewing tobacco exposed cells to elucidate the genetic alterations occurring in oral cells in response to the two forms of tobacco. We also employed a quantitative proteomic approach to investigate the proteome-wide changes in oral keratinocytes upon chronic exposure to cigarette smoke and smokeless (chewing) tobacco using a temporal approach.
Results
To study the effects of smokeless tobacco and cigarette smoke exposure on normal oral keratinocytes, we developed two in vitro models. OKF6/TERT1 cells were treated with cigarette smoke or smokeless tobacco for increasing durations of up to 8 months. Cells exposed to both insults were studied for cellular and molecular changes.
Chronic exposure to cigarette smoke and chewing tobacco induces phenotypic and molecular changes in oral keratinocytes
Chronic exposure to cigarette smoke or chewing tobacco resulted in changes in the cellular morphology of OKF6/TERT1 cells (). OKF6/TERT1-Smoke-8M cells were more elongated and granular compared to parental cells. In contrast, OKF6/TERT1-Tobacco-8M cells were found to be spindle-shaped and formed tightly clustered colonies compared to OKF6/TERT1-Parental cells.
Figure 1. Chronic exposure to cigarette smoke and chewing tobacco induces phenotypic changes in oral keratinocytes. A. Cellular morphology of OKF6/TERT1-Parental cells, OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells (magnification 20X) B. Growth curve depicting cellular proliferation rates of OKF6/TERT1-Parental, OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells. C. Western blot analysis of EMT markers –E-cadherin and Slug. β-actin was used as loading control. D. Invasive ability of OKF6/TERT-Parental and OKF6/TERT1-Smoke-8M cells and E. Invasive ability of OKF6/TERT1 and OKF6/TERT1-Tobacco-8M cells. Representative images were photographed for each time point at 10x magnification. Invaded cells were counted and relative changes in invasive ability of OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells at the indicated time points were calculated and represented graphically (***p < 0.0001).
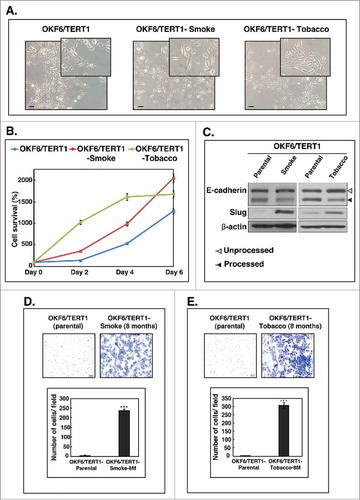
Increase in cellular proliferation is a well-known hallmark of cancer cells. We observed an increase in the proliferative capability of both OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells compared to OKF6/TERT1-Parental cells ().
Cancer progression and increased invasiveness in transformed cells is preceded in most cell types by the phenomenon of epithelial- mesenchymal transition.Citation45 We therefore studied the expression of some established epithelial–mesenchymal transition (EMT) markers in smoke and tobacco exposed cells. As shown in , we observed a decrease in processed E-Cadherin levels (lower band) in both OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells. In addition, there was a significant increase in Slug expression in both OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells (). Collectively, these results indicate that both smoke and tobacco exposure result in cellular transformation that may predispose oral cells to oncogenicity.
Chronic exposure to cigarette smoke and chewing tobacco increases the invasive ability of oral cells
Oncogenic transformation results in the acquisition of characteristic cellular features which includes increase in proliferation and invasion capabilities of cancer cells. As shown in and , OKF6/TERT1-Parental cells acquired an invasive phenotype upon chronic exposure to cigarette smoke and chewing tobacco. We observed a significant increase in the invasive ability of oral cells upon prolonged exposure to cigarette smoke and chewing tobacco.
Whole exome sequencing reveals distinct alterations in oral cells chronically exposed to cigarette smoke condensate and chewing tobacco extract
Cellular transformation to cancer phenotypes tends to occur through irreversible changes at the genomic level in transformed cells. We therefore sought to investigate the genetic alterations acquired by oral cells chronically exposed to cigarette smoke and chewing tobacco for 8 months. Whole exome sequencing revealed high levels of C>T transitions in both smoke and tobacco exposed cells. In conjunction with previous studies, we observed a higher rate of C>A transversions in OKF6/TERT1-Smoke-8M cells (). In contrast, we detected higher frequency of C>G transversions in OKF6/TERT1-Tobacco-8M cells compared to OKF6/TERT1-Smoke-8M cells ().
Figure 2. Exome sequencing of OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells. A. Transitions and transversions observed in OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M compared to OKF6/TERT1-Parental cells. B. Visual representation of log2 normalized read counts and annotated copy number profile in OKF6/TERT1-Smoke-8M cells relative to OKF6/TERT1-Parental cells. Each dot corresponds to an amplicon. Color code – green dots: outliers; dark grey surroundings: unchanged amplicons; plum color surroundings: 1-level gain; all purple dots above the red dotted line represent copy number amplifications >1-level gain in OKF6/TERT1-Smoke-8M cells. C. Visual representation of log2 normalized read counts and annotated copy number profile in OKF6/TERT1-Tobacco-8M cells relative to OKF6/TERT1-parental cells. Each dot corresponds to an amplicon. Color code – green dots: outliers; dark grey surroundings: unchanged amplicons; plum color surroundings: 1-level gain; all purple dots above the red dotted line represent copy number amplifications>1-level gain in OKF6/TERT1-Tobacco-8M cells.
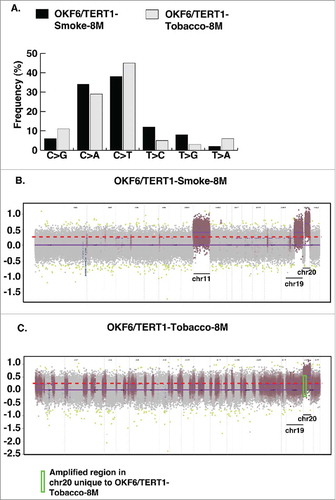
We acquired ∼11 million reads each upon whole exome sequencing of OKF6/TERT1-Parental and OKF6/TERT1-Tobacco-8M cells and ∼12 million reads for OKF6/TERT1-Smoke-8M cells. Using BWA, we achieved 99.9% alignment with exome target coverage of 99.8% with read coverage ≥1. We employed ONCOCNV for CNV analysis and identified somatic copy number gains corresponding to chromosomes 1, 5, 11, 19 and 20 affecting 2321 genes (Fold change ≥3, p value < 0.000001) in OKF6/TERT1-Smoke-8M cells (, Supplementary Table S1). In addition, 592 genes were affected (fold change ≥3, pvalue < 0.000001) on chromosomes 1, 3, 7, 13, 19 and 20 in OKF6/TERT1-Tobacco cells. Interestingly, copy number gains corresponding to 356 genes on chromosome 20 observed in OKF6/TERT1-Smoke-8M were also identified in OKF6/TERT1-Tobacco-8M cells, indicating an influence of tobacco exposure (, ). However, 184 genes in chromosome 20 were uniquely amplified only in OKF6/TERT1-Tobacco-8M cells (, Supplementary Table S2).
Exome sequencing of OKF6/TERT1-Smoke-8M cells revealed a total of 20 somatic non-synonymous SNVs and 2 stop gain variant in 21 genes (Supplementary Tables S3, S4). Variants in 4 genes including UbiA prenyltransferase domain containing 1 (UBIAD1), vav guanine nucleotide exchange factor 3 (VAV3), sodium voltage-gated channel alpha subunit 8 (SCN8A) and fibrillin 2 (FBN2) were predicted to be damaging/ deleterious by all prediction algorithms. Whereas, exome sequencing of OKF6/TERT1-Tobacco-8M cells revealed a total of 24 non-synonymous SNVs and 2 stop gain variants in 26 genes (Supplementary Tables S3, S5). Employing the same filtering criteria, we identified 3 non-synonymous SNVs of which variants in collagen type V alpha 1 chain (COL5A1), 3-oxoacid CoA-transferase 1 (OXCT1) and leucine rich repeat kinase 1 (LRRK1) that were predicted to be damaging/ deleterious by majority of the prediction algorithms employed. Overall, exome sequencing of OKF6/TERT1-Smoke-8M and OKF6/TERT1-Tobacco-8M cells reaffirms that chronic exposure to different forms of tobacco results in distinct molecular signatures in oral keratinocytes.
Cigarette smoke and chewing tobacco exposure result in proteome-wide changes in oral keratinocytes
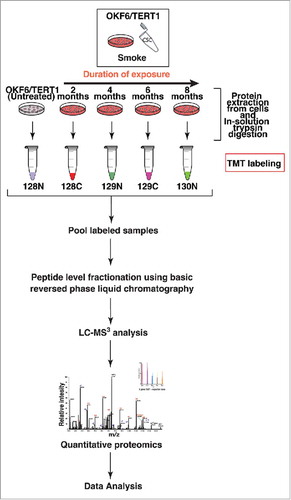
We employed Tandem Mass Tag (TMT)-based quantitative proteomic approach to investigate global proteome changes in OKF6/TERT1-Parental cells chronically exposed to cigarette smoke and chewing tobacco. The experimental strategy employed for proteomic analysis of smoke and tobacco treated cells are depicted in and Supplementary Figure S1, respectively. We quantified a total of 5,342 and 2,821 proteins in OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells, respectively. We observed overexpression of 68 proteins (≥2-fold) and downregulation of 70 proteins (2-fold) in OKF6/TERT1-Smoke-8M cells. In addition, 44 proteins were overexpressed and 26 proteins were downregulated (≥2-fold) in OKF6/TERT1-Tobacco-8M cells. Total quantified proteins and number of dysregulated proteins in each exposure condition are provided in Supplementary Tables S6 and S7. A complete list of quantified peptides for each exposure condition is provided in Supplementary Tables S8 and S9.
Figure 3. Workflow employed to identify proteomic alterations in normal oral keratinocytes chronically exposed to cigarette smoke for increasing durations of time (2-4-6-8 months). Proteins were extracted and quantified from OKF6/TERT1-Parental, OKF6/TERT1-Smoke-2M, OKF6/TERT1-Smoke-4M, OKF6/TERT1-Smoke-6M and OKF6/TERT1-Smoke-8M cells. In-solution trypsin digestion of equal amount of proteins from each condition was performed and peptides from OKF6/TERT1-Parental, OKF6/TERT1-Smoke-2M, OKF6/TERT1-Smoke-4M, OKF6/TERT1-Smoke-6M and OKF6/TERT1-Smoke-8M were labeled with TMT reagents 128N, 128C, 129N, 129C and 130N, respectively. Labeled samples were pooled, fractionated using basic reversed phase liquid chromatography and fractions were analyzed in triplicate by mass spectrometry.
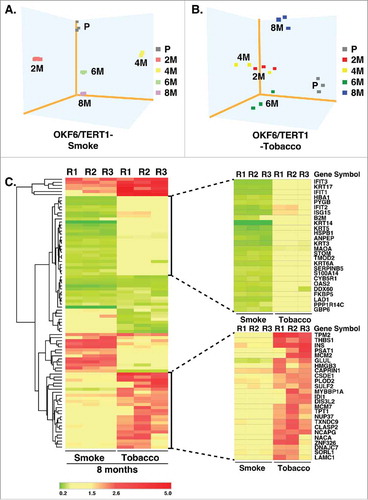
Principal component analysis of proteomic data from OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells shows that smoke and chewing tobacco exposed cells cluster independently of OKF6/TERT1-Parental cells (, ). Bioinformatics analysis of proteins dysregulated in OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells resulted in the identification of distinct patterns of dysregulation between the two exposure models (, Supplementary Figure S2).
Figure 4. Cluster analysis of smoke and tobacco exposed cells Principle component analysis of A. OKF6/TERT1-Parental and OKF6/TERT1-Smoke (2-4-6-8 months) and B. OKF6/TERT1-Parental and OKF6/TERT1-Tobacco (2-4-6-8 months) cells, C. Heat map depicting the expression pattern of proteins significantly dysregulated (p<0.05) in all three replicates of OKF6/TERT1-Smoke-8M and/or OKF6/TERT1-Tobacco-8M.
Chronic exposure to cigarette smoke and chewing tobacco enrich for unique upstream protein regulators
As mentioned earlier, we observed considerable differences in the dysregulation pattern of smoke and tobacco exposed cells. We therefore studied the signaling cascades involved in OKF6/TERT1-Smoke and OKF6/TERT1-Tobacco cells to elucidate these differences further.
Upstream regulator analysis revealed that proteins downstream of mitogen-activated protein kinase 1 (MAPK1) were downregulated in oral cells exposed to cigarette smoke (). A number of these downregulated proteins are involved in IFN signalling. Suppression of IFN signalling is associated with increased metastasis.Citation46 MAPK1 (ERK2) is a serine/threonine kinase and an integral member of MAPK/ERK signalling cascade. Western blot analysis confirmed overexpression and increased activation of MAPK1/2 (ERK1/2) in OKF6/TERT1-Smoke-8M cells compared to parental cells (). Besides its kinase activity, MAPK1 is also known to act as a transcriptional repressor for interferon gamma-induced genes.Citation47 Proteins involved in inflammatory responses such as all four members of interferon-induced proteins with tetratricopeptide repeats (IFIT) family – IFIT1, IFIT2, IFIT3 and IFIT5 were found to be downregulated in smoke exposed cells. Other proteins activated by interferon signalling such as ISG15 ubiquitin-like modifier protein, DExD/H-box helicase 58 (DDX58) and members of the 2′-5′-oligoadenylate synthetase family including OAS1, OAS2 and OAS3 were downregulated across different durations of exposure in OKF6/TERT1-Smoke cells. We also observed decreased expression levels of signal transducer and activator of transcription molecules, STAT1 and STAT2 in OKF6/TERT1-Smoke cells. Another protein downregulated in OKF6/TERT1-Smoke cells was phospholipid scramblase 1 (PLSCR1) which functions in various cellular processes, including apoptosis, innate immune response, epidermal differentiation, oxidative stress response and tumor suppression.Citation48 Interestingly, Ki67 (MKI67) which is an established marker of proliferation was observed to be overexpressed in OKF6/TERT1-Smoke cells. Ki67 is known to be overexpressed in oral epithelial dysplasia and oral squamous cell carcinoma.Citation49 Overall, proteins involved in cellular processes such as cellular proliferation, migration, apoptosis and cell survival were found to be dysregulated upon smoke exposure in oral cells.
Figure 5. Bioinformatics analysis of dysregulated proteins in smoke and tobacco exposed cells A. Ingenuity pathway analysis of proteins significantly dysregulated (p < 0.05) in cigarette smoke exposed cells B. Western blot analysis of phospho MAPK1 (T202/Y204) and total MAPK1 levels in smoke exposed (8 months) cells compared to parental cells. β-actin was used as loading control C. Ingenuity pathway analysis of proteins significantly dysregulated (p < 0.05) in chewing tobacco exposed cells D. Western blot analysis of phospho EGFR (Y1068) and total EGFR levels in chewing tobacco exposed (8 months) cells compared to parental cells. β-actin was used as loading control.
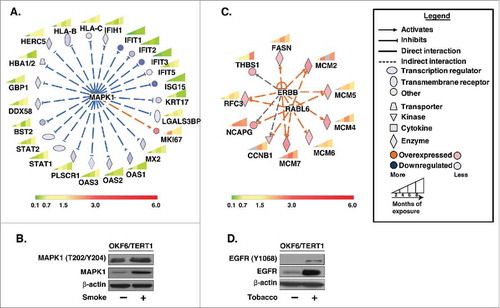
In contrast, analysis of the proteins significantly dysregulated in OKF6/TERT1-Tobacco cells predicted a signalling cascade downstream of the erb-b2 receptor tyrosine kinase family (ERBB) in OKF6/TERT1-Tobacco cells (). We observed significant increase in both expression as well as activation of epidermal growth factor receptor, EGFR (ERBB1) in OKF6/TERT1-Tobacco-8M cells compared to OKF6/TERT1-Parental cells () revealing possible activation of ERBB signalling in OKF6/TERT1-Tobacco cells. Proteins downstream of ERBB signalling such as fatty acid synthase (FASN) and Replication factor C subunit 3 (RFC3) were overexpressed in OKF6/TERT1-Tobacco cells. RFC3 is a subunit of the replication factor C required for the elongation of primed DNA templates by DNA polymerases.Citation50 RFC3 is also downstream of RAB which is a member of RAS oncogene family like 6 (RABL6). A number of proteins downstream of RABL6 that are involved in cell cycle regulation and DNA replication were also found to be overexpressed in OKF6/TERT1-Tobacco cells (). Proteins such as non-SMC condensin I complex subunit G (NCAPG) which is essential for chromosome stability and mitosis and CCNB1, a regulatory protein involved in cell cycle checkpoint control (G2 to M phase) were overexpressed in oral cells upon chronic exposure to chewing tobacco. In addition, most members of the minichromosome maintenance (MCM) complex or eukaryotic replicative helicase enzyme complex (MCM2, MCM4, MCM5, MCM6, MCM7) were overexpressed in OKF6/TERT1-Tobacco cells. Collectively, these observations indicate that chewing tobacco exposure may result in deregulation of key cellular processes including DNA replication and mitosis in OKF6/TERT1-Tobacco cells.
Discussion
Previous literature evidence links tobacco smoke exposure and dysregulation of signalling cascades involved in cellular proliferation, inflammatory response, cellular and tissue injury and apoptosis.Citation51 The involvement of signalling cascades mediated by proteins such as COX-2 and aryl hydrocarbon receptor have been reported in response to cigarette smoke exposure.Citation52,Citation53 Transcriptome studies on the effect of smokeless tobacco on oral epithelia reveal gene expression changes in cell cycle, cell adhesion, inflammation and apoptosis related cellular processes and activation of Akt signalling.Citation54 However, a comparative high throughput study to elucidate the differences between smoke and chewing tobacco exposure in an in vitro system is currently lacking. In our study, we aimed to develop cellular models of chronic exposure to different forms of tobacco and elucidate the different molecular alterations observed as a consequence of these exposures in normal oral cells. Chronic exposure to cigarette smoke and chewing tobacco resulted in an increased rate of proliferation and increased invasive ability in oral cells. Mesenchymal markers such as slug are seen to be overexpressed in aggressive tumors while expression of epithelial markers such as E-cadherin is decreased in cells acquiring a tumorigenic phenotype.Citation45 We found observable changes in expression of these established EMT markers in cells exposed to either form of tobacco.
Genomic studies have previously identified higher rates of G.C>T.A transversions in smokers.Citation55 Whole exome sequencing (WES) revealed an increased rate of C>A transversions in oral cells upon chronic smoke exposure. However, C>T and C>G transversions were higher in chewing tobacco exposed cells compared to smoke exposed cells. This could be interpreted as chewing tobacco exposure-specific responses which needs further validation in large cohort studies. WES analysis of oral cells chronically exposed to cigarette smoke for 8 months resulted in the identification of copy number gain on chromosomes 1, 5, 11, 19 and 20. Several proteins encoded by genes corresponding to chromosomes 11, 19 and 20 were also seen to be overexpressed at the proteomic level. These were classified under regulation of nucleic acid metabolism according to Gene Ontology (Supplementary Table S6). These include proteins involved in DNA replication such as topoisomerase (DNA) I (TOP1) and DNA ligase 1 (LIG1) and DNA damage repair mechanisms such as tripartite motif containing 28 (TRIM28), polynucleotide kinase 3′-phosphatase (PNKP) and ERCC excision repair 2, TFIIH core complex helicase subunit (ERCC2). Overexpression of proteins involved in DNA damage repair pathways have been previously implicated in cancer progression.Citation56,Citation57 Copy number gain and protein level overexpression of transcriptional factors such as zinc finger protein 217 (ZNF217) and CCAAT/enhancer binding protein beta (CEBPB) were also observed in smoke exposed cells. Overexpression of these proteins has been significantly associated with multiple cancers including breast and colon cancer.Citation58,Citation59
CNAs in regions of chromosome 11 including 11q13 are well documented in oral squamous cell carcinomas.Citation60,Citation61 Interestingly, we observed copy number gains for a majority of genes on q-arm of chromosome 20 in both smoke and chewing tobacco exposed cells. Amplification of q arm of chromosome 20 has been previously implicated in early tumorigenesis and linked to cancer initiation.Citation62 In addition, amongst the four genes with non-synonymous variants predicted to be deleterious in smoke exposed cells, UBIAD1 is a known tumor suppressor gene seen to be downregulated in a number of cancers including bladder and prostate cancer.Citation63,Citation64 In contrast, non-synonymous variants corresponding to three genes were predicted to be damaging in chewing tobacco exposed cells. Increased expression of one of these genes, namely, OXCT1 is known to correlate to high patient mortality in hepatocellular carcinoma.Citation65
Cigarette smoke and smokeless tobacco differ inherently in their constitution of pro-carcinogenic agents and other damaging constituents. This variation in composition could result in a difference in their effects on oral cells. Our analysis of proteomic data reveals that this difference begins as early as two months of smoke or tobacco exposure. This was observed as a distinct pattern of expression of dysregulated proteins between oral cells exposed to the two forms of tobacco.
Cellular signalling cascades are tightly regulated processes and cellular control of essential pathways is affected in oncogenic transformation.Citation66,Citation67 We identified distinct upstream regulators that control cellular processes in smoke exposed cells compared to chewing tobacco- exposed oral cells. Network analysis predicted transcriptional repression by MAPK1 in smoke exposed oral cells. This is in conjunction with previous studies that link MAPK1 (ERK) signalling to tobacco smoke induced cellular damage and pathogenesis.Citation68 Proteins downstream of MAPK1 belong to a diverse range of cellular processes including inflammatory response, cellular proliferation and cell survival. Some of these proteins belong to the group of interferon-stimulated genes (ISGs). ISGs can be induced by a variety of cellular stress responses.Citation69 IFN signalling has been implicated in suppression of oncogenic transformation.Citation70 Proteins involved in IFN signalling such as the OAS family of proteins were downregulated in our study. These proteins have been recently identified as negative regulators of cellular proliferation and as activators of apoptosis downstream of BRCA1 and STAT1 signaling.Citation71 Downregulation of proteins involved in IFN signalling in smoke exposed cells reinforces the possibility that prolonged smoke exposure may result in malignant transformation in oral keratinocytes.
In contrast, network analysis predicted signaling regulation by ERBB family and RABL6 in chewing tobacco exposed oral cells. We observed a marked overexpression of EGFR in chewing tobacco exposed cells. EGFR is a tyrosine kinase receptor from the ErbB family of receptors. Higher EGFR levels have been linked to tumour aggressiveness and poor survival in oral squamous cell carcinoma.Citation72 Proteins downstream of ERBB signalling such as FASN and RCF3 were overexpressed in tobacco exposed cells. Increased FASN expression has previously been reported in tongue squamous cell carcinoma.Citation73 Another upstream regulator, RABL6 is member of the Ras superfamily of small GTPases and is known to be involved in tumor cell growth, proliferation and linked to tumorigenesis in breast cancer.Citation74-76 Proteins involved in cell cycle regulation and DNA replication such as NCAPG, CCNB1 and components of mini-chromosome maintenance (MCM) complex were overexpressed in tobacco exposed cells. NCAPG is reported to be overexpressed in multiple cancers including paediatric high grade gliomas and melanoma.Citation77-79 CCNB1 overexpression has been previously associated with poor prognosis in esophageal squamous cell carcinoma (ESCC).Citation80 These observations point to a role of chewing tobacco in deregulating key cellular processes such DNA replication and mitosis during cellular transformation in oral cells.
We developed in vitro cellular models of normal oral keratinocytes chronically exposed to two different forms of tobacco – cigarette smoke and chewing tobacco. Chronic exposure to both cigarette smoke and chewing tobacco resulted in a significant increase in the proliferative and invasive abilities of OKF6/TERT1-Parental cells. Exome sequencing revealed discrete genetic variations such as increased C>A transversions in OKF6/TERT1-Smoke-8M cells and a prevalence of C>G and C>T transversions and OKF6/TERT1-Tobacco-8M cells. Copy number gains in the q-arm of chromosome 20 were observed in both smoke exposed and tobacco treated cells. Further, quantitative proteomic analysis revealed a distinct response signature based on type of tobacco insult. Molecules downstream of MAPK1 were predominantly downregulated in smoke exposed cells. These changes were associated with increased cellular proliferation and survival. In contrast, molecules downstream of ERBB2 and RABL6 which are involved in DNA replication and cell cycle progression were overexpressed in chewing tobacco exposed cells.
Our exome sequencing and mass spectrometric data recapitulate a number of known adverse molecular effects of cigarette smoke and chewing tobacco exposure in an in vitro setting. In addition, we identified unique regulators involved in cellular transformation of oral keratinocytes chronically exposed to either cigarette smoke or chewing tobacco. Further in-depth studies in a clinical setting are needed to validate and identify proteins which can serve as signature markers in oral cancer based on tobacco usage history.
This work provides a scaffold for future studies to systematically investigate cigarette smoke and chewing tobacco induced molecular alterations which may aid not only in diagnosis and prognosis of oral cancer but may also help in identification of therapeutic targets for oral patients based on their tobacco using habits. Further studies are essential to elucidate the role of key molecules in the molecular pathogenesis of smoke or chewing tobacco-induced oral cancer.
Materials and methods
Cell culture
Normal human oral keratinocytes, OKF6/TERT1, were a generous gift from Dr. James Rheinwald (Brigham and Women's Hospital, Boston, MA). OKF6/TERT1 were cultured and maintained in keratinocyte serum free medium (KSFM) supplemented with bovine pituitary extract (25 µg/ml), epidermal growth factor (EGF) (0.2 ng/ml) (ThermoFisher Scientific, MA), 1% penicillin/streptomycin and CaCl2 (0.4 mM). The cells were cultured at 37°C in a humidified air incubator with 5% CO2. OKF6/TERT1 was authenticated by short tandem repeat analysis at the Genetic Resources Core Facility of Johns Hopkins University School of Medicine.
Adaptation of oral keratinocytes to smokeless tobacco extract and cigarette smoke condensate
OKF6/TERT1 cells were chronically treated with 1% smokeless tobacco extract (STE) (preparation detailed in Supplementary Methods) for a period of 8 months as previously described.Citation40 Cell stocks of cells treated for two, four, six and eight months were periodically frozen and stored in liquid nitrogen for further experiments. STE treated cells were grown at 37°C in a humidified 5% CO2 incubator.
Cigarette smoke condensate (CSC) was purchased from Murty Pharmaceuticals, Inc., KY. OKF6/TERT1 cells were cultured in the presence of CSC in a dedicated smoke exposure incubator as described previously.Citation81 Briefly, cells were treated with 0.1% CSC chronically for a period of 8 months. Cell stocks of keratinocytes exposed to CSC for each month of exposure were periodically frozen and stored for further experiments. The dose of treatment reflects, as percentage, the volume of cigarette smoke condensate diluted by the volume of medium. Treatment concentration (0.1%) was selected based on dose response curve of OKF6/TERT1 cells to different concentrations of CSC (data not shown).
In the study, OKF6/TERT1 cells exposed to STE have been referred to as OKF6/TERT1-Tobacco cells and cells exposed to CSC have been referred to as OKF6/TERT1-Smoke cells. The suffix of 2M, 4M, 6M or 8M have been given to cells exposed to STE or CSC for the respective duration in months of exposure. In addition, OKF6/TERT1 cells were also cultured in a regular incubator without any exposure to either STE or CSC for the same duration. These cells served as control and have been referred to as OKF6/TERT1-Parental cells.
Cellular proliferation assays
OKF6/TERT1-Smoke-8M, OKF6/TERT1-Tobacco-8M and OKF6/TERT1-Parental cells were seeded at a density of 25 × 103 cells per well in 6-well plates. Cellular proliferation was monitored for 6 days where the cells were counted every 48h using trypan blue exclusion method. All assays were performed in replicates.
Western blot analysis
OKF6/TERT1-Parental, OKF6/TERT1-Tobacco-8M and OKF6/TERT1-Smoke-8M cells were grown to 75% confluence and washed thrice with ice-cold 1X phosphate buffer saline (PBS). Following this, cells were harvested in RIPA lysis buffer (10 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton-X-100, 0.1% SDS containing protease and phosphatase inhibitor cocktails) and protein concentration was determined using BCA assays.Citation82 Western blot analysis was carried out as described previously.Citation40 Antibodies for E-cadherin and Slug were purchased from Cell Signaling (Cell Signaling Technology, Danvers, MA). β-actin antibody was procured from Sigma (Sigma Aldrich, USA) and was used as loading control for all Western blots.
Cellular invasion assays
The invasive ability of OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells at 0,2,4,6 and 8 months was studied using a transwell system (BD Biosciences, San Jose, CA) with Matrigel (BD Biosciences, San Jose, CA) coated filters. Cellular invasion was evaluated after 48h. Briefly, invasiveness of the cells was assayed in the membrane invasion culture system using polyethylene terephthalate (PET) membrane (8-µm pore size) (BD Biosciences, San Jose, CA) coated with Matrigel. The cells were seeded at a density of 2.0 × 104 in 500 µl of serum free media on the Matrigel-coated PET membrane in the upper compartment and placed in a compartment filled with complete growth media. All plates were incubated at 37°C for 48h. Post incubation, upper surface of the membrane was wiped with a cotton-tip applicator to remove non-migratory cells. Cells that migrated to the lower side of the membrane were fixed and stained using 4% methylene blue. All experiments were performed in triplicate.
Whole exome sequencing and computational analysis
Details provided in Supplementary Methods.
Sample preparation for mass spectrometric analysis
OKF6/TERT1-Parental, OKF6/TERT1-Tobacco (2-4-6-8M) and OKF6/TERT1-Smoke (2-4-6-8M) cells were grown to 80% confluence and serum starved for 8h. Post-starvation, cells were washed with 1X PBS thrice and harvested in lysis buffer (2% SDS, 5 mM sodium fluoride, 1 mM β-glycerophosphate, 1mM sodium orthovanadate in 50 mM TEABC). The cell lysates were sonicated, centrifuged and protein concentration was determined by BCA. In-solution trypsin digestion of samples was carried out as described previously (details in Supplementary Methods).
TMT labeling
After trypsin digestion, peptides were lyophilized and labeled with TMT reagents as per manufacturers' instructions. Briefly, peptide samples were dissolved in 50mM TEABC (pH 8.0) and added to TMT reagents dissolved in anhydrous acetonitrile. Peptides from OKF6/TERT1-Parental, OKF6/TERT1-Smoke (2-4-6-8M) and OKF6/TERT1-Tobacco (2-4-6-8M) were labeled with TMT tags as detailed in Supplementary Tables S1 and S2, respectively. After incubation at room temperature for 1h, the reaction was quenched with 5% hydroxylamine. The labeled samples from all smoke exposed or chewing tobacco exposed cells were pooled in two separate experiments and subjected to fractionation. Fractionated samples were analyzed on Orbitrap Fusion Tribrid mass spectrometer (Thermo Electron, Bremen, Germany) as detailed in Supplementary Methods. Data from LC/MS analysis was searched using Proteome Discoverer (Version 2.1) software suite (Thermo Fisher Scientific, Bremen, Germany) (details of data analysis is provided in Supplementary Methods).
Data availability
Mass spectrometric data generated in this study was submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repositoryCitation83 with the dataset identifier PXD007127. Raw sequencing data is available in SRA (Sequence Read Archive) database with accession number SRP115679.
Bioinformatics analysis
Information on the subcellular localization of differentially expressed proteins and the biological processes was obtained from Human Protein Reference DatabaseCitation84 (HPRD; http://www.hprd.org). Principal Component Analysis of OKF6/TERT1-Parental, OKF6/TERT1-Tobacco and OKF6/TERT1-Smoke cells as well as p-value calculation for all quantified proteins was done using one sample t-test in Perseus (version 1.5.8.5).Citation85 Ingenuity Pathway Analysis (version 31813283) (Qiagen, USA) was employed for upstream regulator analysis of proteins dysregulated in smoke exposed or chewing tobacco treated cells compared to control cells. Analysis was performed with a list of proteins significantly dysregulated in response to CSC or STE with a fold change of ≥2.0 (p-value ≤0.05). Interaction networks were generated for Homo sapiens. Heatmaps were generated by supervised hierarchical clustering method based on Euclidean distance and average linkage using Morpheus (version 3.0.206) software (https://software.broadinstitute.org/morpheus/).
Authors' contributions
AC and HG participated in study conception and study design. PR, VN and TS were involved in cell culture and performed all assays and experiments. APJ, KKD and KKM carried out fractionation and mass spectrometric analysis of samples. CK and SM were involved in DNA extraction, quality control and next generation sequencing. PR, KP and APJ prepared the manuscript and manuscript figures. PR, APJ, KP, AK, MM, RaG and RoG were involved in data analyses and interpretation. AC, HG, DS, JC, AKG, PPM, TSKP and BN edited, critically read and revised the manuscript. All the authors have read and approved the final manuscript.
Abbreviations
| HPV | = | Human papillomavirus |
| MGMT | = | O-6-Methylguanine-DNA Methyltransferase |
| PBMC | = | Peripheral Blood Mononuclear Cell |
| EDTA | = | Ethylenediaminetetraacetic acid |
| SDS | = | Sodium Dodecyl Sulfate |
| NRF2 | = | Nuclear factor (erythroid-derived 2)-like 2 (NFE2L2) |
| TEABC | = | Triethyl ammonium bicarbonate |
| BCA | = | Bicinchoninic acid |
| SNV | = | Single Nucleotide Variant |
| CNV | = | Copy Number Variation |
| CNA | = | Copy Number Alteration |
supp_mat_1470724_KCBT.zip
Download Zip (34.8 MB)Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics (IOB), Bangalore. Pavithra Rajagopalan is a recipient of Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), New Delhi, India. Kiran K. Mangalaparthi is a recipient of Senior Research Fellowship from University Grants Commission (UGC), New Delhi, India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure statement
No potential conflicts of interest were disclosed.
References
- Sadri G, Mahjub H. Tobacco smoking and oral cancer: a meta-analysis. J Res Health Sci. 2007;7:18–23. PMID:23343867.
- Sasco AJ, Secretan MB, Straif K. Tobacco smoking and cancer: a brief review of recent epidemiological evidence. Lung Cancer. 2004;45 Suppl 2:S3–9. doi:10.1016/j.lungcan.2004.07.998. PMID:15552776.
- Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–16. doi:10.1016/j.oraloncology.2008.06.002. PMID:18804401.
- Idris AM, Ahmed HM, Mukhtar BI, Gadir AF, el-Beshir EI. Descriptive epidemiology of oral neoplasms in Sudan 1970–1985 and the role of toombak. Int J Cancer. 1995;61:155–8. doi:10.1002/ijc.2910610202. PMID:7705940.
- Kavarodi AM, Thomas M, Kannampilly J. Prevalence of oral pre-malignant lesions and its risk factors in an Indian subcontinent low income migrant group in Qatar. Asian Pac J Cancer Prev. 2014;15:4325–9. doi:10.7314/APJCP.2014.15.10.4325. PMID:24935392.
- Thomas G, Hashibe M, Jacob BJ, Ramadas K, Mathew B, Sankaranarayanan R, Zhang ZF. Risk factors for multiple oral premalignant lesions. Int J Cancer. 2003;107:285–91. doi:10.1002/ijc.11383. PMID:12949809.
- Bachar G, Hod R, Goldstein DP, Irish JC, Gullane PJ, Brown D, Gilbert RW, Hadar T, Feinmesser R, Shpitzer T. Outcome of oral tongue squamous cell carcinoma in patients with and without known risk factors. Oral Oncol. 2011;47:45–50. doi:10.1016/j.oraloncology.2010.11.003. PMID:21167767.
- Link JO, Kaugars GE, Burns JC. Comparison of oral carcinomas in smokeless tobacco users and nonusers. J Oral Maxillofac Surg. 1992;50:452–5. doi:10.1016/S0278-2391(10)80315-5. PMID:1573481.
- Pytynia KB, Grant JR, Etzel CJ, Roberts DB, Wei Q, Sturgis EM. Matched-pair analysis of survival of never smokers and ever smokers with squamous cell carcinoma of the head and neck. J Clin Oncol. 2004;22:3981–8. doi:10.1200/JCO.2004.02.133. PMID:15459221.
- Roemer E, Schramke H, Weiler H, Buettner A, Kausche S, Weber S, et al. Mainstream smoke Chemistry and in Vitro and in Vivo toxicity of the reference Cigarettes 3R4F and 2R4F. Contributions to Tobacco Research. 2012;25:316–35.
- Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004; 83:1–1438. PMID:15285078.
- Bhisey RA. Chemistry and toxicology of smokeless tobacco. Indian J Cancer. 2012;49:364–72. doi:10.4103/0019-509X.107735. PMID:23442400.
- Smokeless tobacco and some tobacco-specific N-nitrosamines. IARC Monogr Eval Carcinog Risks Hum. 2007;89:1–592. PMID:18335640.
- Lee J, Taneja V, Vassallo R. Cigarette smoking and inflammation: cellular and molecular mechanisms. J Dent Res. 2012;91:142–9. doi:10.1177/0022034511421200. PMID:21876032.
- Rajagopalan P, Nanjappa V, Raja R, Jain AP, Mangalaparthi KK, Sathe GJ, Babu N, Patel K, Cavusoglu N, Soeur J, et al. How Does Chronic Cigarette smoke exposure affect Human Skin? A Global Proteomics study in Primary Human Keratinocytes. OMICS. 2016;20:615–26. doi:10.1089/omi.2016.0123. PMID:27828771.
- Zhang L, Wu R, Dingle RW, Gairola CG, Valentino J, Swanson HI. Cigarette smoke condensate and dioxin suppress culture shock induced senescence in normal human oral keratinocytes. Oral Oncol. 2007;43:693–700. doi:10.1016/j.oraloncology.2006.08.008. PMID:17070097.
- Jin Z, Gao F, Flagg T, Deng X. Tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone promotes functional cooperation of Bcl2 and c-Myc through phosphorylation in regulating cell survival and proliferation. J Biol Chem. 2004;279:40209–19. doi:10.1074/jbc.M404056200. PMID:15210690.
- Rao P, Ande A, Sinha N, Kumar A, Kumar S. Effects of Cigarette smoke condensate on Oxidative Stress, Apoptotic Cell Death, and HIV Replication in Human Monocytic Cells. PLoS One. 2016;11:e0155791. doi:10.1371/journal.pone.0155791. PMID:27203850.
- Jiao S, Liu B, Gao A, Ye M, Jia X, Zhang F, Liu H, Shi X, Huang C. Benzo(a)pyrene-caused increased G1-S transition requires the activation of c-Jun through p53-dependent PI-3K/Akt/ERK pathway in human embryo lung fibroblasts. Toxicol Lett. 2008;178:167–75. doi:10.1016/j.toxlet.2008.03.012. PMID:18448277.
- Allam E, Zhang W, Al-Shibani N, Sun J, Labban N, Song F, Windsor LJ. Effects of cigarette smoke condensate on oral squamous cell carcinoma cells. Arch Oral Biol. 2011;56:1154–61. doi:10.1016/j.archoralbio.2011.03.008. PMID:21489402.
- Kim MS, Huang Y, Lee J, Zhong X, Jiang WW, Ratovitski EA, Sidransky D. Cellular transformation by cigarette smoke extract involves alteration of glycolysis and mitochondrial function in esophageal epithelial cells. Int J Cancer. 2010;127:269–81. PMID:19937795.
- Tan D, Goerlitz DS, Dumitrescu RG, Han D, Seillier-Moiseiwitsch F, Spernak SM, Orden RA, Chen J, Goldman R, Shields PG. Associations between cigarette smoking and mitochondrial DNA abnormalities in buccal cells. Carcinogenesis. 2008;29:1170–7. doi:10.1093/carcin/bgn034. PMID:18281252.
- Ljungberg LU, Persson K, Eriksson AC, Green H, Whiss PA. Effects of nicotine, its metabolites and tobacco extracts on human platelet function in vitro. Toxicol in Vitro. 2013;27:932–8. doi:10.1016/j.tiv.2013.01.004. PMID:23318728.
- Bagchi M, Bagchi D, Stohs SJ. In vitro effects of a smokeless tobacco extract on the production of reactive oxygen species by human oral epidermal cells and rat hepatic mitochondria and microsomes, and peritoneal macrophages. Arch Environ Contam Toxicol. 1996;30:418–22. doi:10.1007/BF00212303. PMID:8854974.
- Merne M, Rautava J, Ruutu M, Syrjanen S. Smokeless tobacco increases aneuploidy in oral HPV16 E6/E7-transformed keratinocytes in vitro. J Oral Pathol Med. 2014;43:685–90. doi:10.1111/jop.12185. PMID:24761760.
- Arredondo J, Chernyavsky AI, Grando SA. Nicotinic receptors mediate tumorigenic action of tobacco-derived nitrosamines on immortalized oral epithelial cells. Cancer Biol Ther. 2006;5:511–7. doi:10.4161/cbt.5.5.2601. PMID:16582591.
- Rohatgi N, Kaur J, Srivastava A, Ralhan R. Smokeless tobacco (khaini) extracts modulate gene expression in epithelial cell culture from an oral hyperplasia. Oral Oncol. 2005;41:806–20. doi:10.1016/j.oraloncology.2005.04.010. PMID:15979382.
- Rohatgi N, Matta A, Kaur J, Srivastava A, Ralhan R. Novel molecular targets of smokeless tobacco (khaini) in cell culture from oral hyperplasia. Toxicology. 2006;224:1–13. doi:10.1016/j.tox.2006.03.014. PMID:16730401.
- Titz B, Sewer A, Schneider T, Elamin A, Martin F, Dijon S, Luettich K, Guedj E, Vuillaume G, Ivanov NV, et al. Alterations in the sputum proteome and transcriptome in smokers and early-stage COPD subjects. J Proteomics. 2015;128:306–20. doi:10.1016/j.jprot.2015.08.009. PMID:26306861.
- Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8:e63812. doi:10.1371/journal.pone.0063812. PMID:23691101.
- Steiling K, Kadar AY, Bergerat A, Flanigon J, Sridhar S, Shah V, Ahmad QR, Brody JS, Lenburg ME, Steffen M, et al. Comparison of proteomic and transcriptomic profiles in the bronchial airway epithelium of current and never smokers. PLoS One. 2009;4:e5043. doi:10.1371/journal.pone.0005043. PMID:19357784.
- Airoldi L, Magagnotti C, Iannuzzi AR, Marelli C, Bagnati R, Pastorelli R, Colombi A, Santaguida S, Chiabrando C, Schiarea S, et al. Effects of cigarette smoking on the human urinary proteome. Biochem Biophys Res Commun. 2009;381:397–402. doi:10.1016/j.bbrc.2009.02.055. PMID:19222993.
- Jessie K, Pang WW, Haji Z, Rahim A, Hashim OH. Proteomic analysis of whole human saliva detects enhanced expression of interleukin-1 receptor antagonist, thioredoxin and lipocalin-1 in cigarette smokers compared to non-smokers. Int J Mol Sci. 2010;11:4488–505. doi:10.3390/ijms11114488. PMID:21151451.
- Boyle JO, Gumus ZH, Kacker A, Choksi VL, Bocker JM, Zhou XK, Yantiss RK, Hughes DB, Du B, Judson BL, et al. Effects of cigarette smoke on the human oral mucosal transcriptome. Cancer Prev Res (Phila). 2010;3:266–78. doi:10.1158/1940-6207.CAPR-09-0192. PMID:20179299.
- Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–34. doi:10.1016/j.cell.2012.08.024. PMID:22980976.
- Huang YT, Lin X, Liu Y, Chirieac LR, McGovern R, Wain J, Heist R, Skaug V, Zienolddiny S, Haugen A, et al. Cigarette smoking increases copy number alterations in nonsmall-cell lung cancer. Proc Natl Acad Sci U S A. 2011;108:16345–50. doi:10.1073/pnas.1102769108. PMID:21911369.
- Al-Hebshi NN, Li S, Nasher AT, El-Setouhy M, Alsanosi R, Blancato J, Loffredo C. Exome sequencing of oral squamous cell carcinoma in users of Arabian snuff reveals novel candidates for driver genes. Int J Cancer. 2016;139:363–72. doi:10.1002/ijc.30068. PMID:26934577.
- Chakrabarti S, Multani S, Dabholkar J, Saranath D. Whole genome expression profiling in chewing-tobacco-associated oral cancers: a pilot study. Med Oncol. 2015;32:60. doi:10.1007/s12032-015-0483-4. PMID:25663065.
- Bhatnagar R, Dabholkar J, Saranath D. Genome-wide disease association study in chewing tobacco associated oral cancers. Oral Oncol. 2012;48:831–5. doi:10.1016/j.oraloncology.2012.03.007. PMID:22503698.
- Nanjappa V, Renuse S, Sathe GJ, Raja R, Syed N, Radhakrishnan A, Subbannayya T, Patil A, Marimuthu A, Sahasrabuddhe NA, et al. Chronic exposure to chewing tobacco selects for overexpression of stearoyl-CoA desaturase in normal oral keratinocytes. Cancer Biol Ther. 2015;16:1593–603. doi:10.1080/15384047.2015.1078022. PMID:26391970.
- Prasad GL, Jones BA, Schmidt E, Chen P, Kennedy AD. Global metabolomic profiles reveal differences in oxidative stress and inflammation pathways in smokers and moist snuff consumers. Journal of Metabolomics. 2015;1:2. doi:10.7243/2059-0008-1-2.
- Arimilli S, Madahian B, Chen P, Marano K, Prasad GL. Gene expression profiles associated with cigarette smoking and moist snuff consumption. BMC Genomics. 2017;18:156. doi:10.1186/s12864-017-3565-1. PMID:28193179.
- Woo S, Gao H, Henderson D, Zacharias W, Liu G, Tran QT, Prasad GL. AKR1C1 as a Biomarker for Differentiating the Biological Effects of Combustible from Non-Combustible Tobacco Products. Genes (Basel). 2017;8. doi:10.3390/genes8050132. PMID:28467356.
- Laytragoon-Lewin N, Bahram F, Rutqvist LE, Turesson I, Lewin F. Direct effects of pure nicotine, cigarette smoke extract, Swedish-type smokeless tobacco (Snus) extract and ethanol on human normal endothelial cells and fibroblasts. Anticancer Res. 2011;31:1527–34. PMID:21617206.
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi:10.1038/nrm1835. PMID:16493418.
- Lu LM, Zavitz CC, Chen B, Kianpour S, Wan Y, Stampfli MR. Cigarette smoke impairs NK cell-dependent tumor immune surveillance. J Immunol. 2007;178:936–43. doi:10.4049/jimmunol.178.2.936. PMID:17202355.
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–22. doi:10.1016/j.cell.2009.08.037. PMID:19879846.
- Kodigepalli KM, Bowers K, Sharp A, Nanjundan M. Roles and regulation of phospholipid scramblases. FEBS Lett. 2015;589:3–14. doi:10.1016/j.febslet.2014.11.036. PMID:25479087.
- Birajdar SS, Radhika M, Paremala K, Sudhakara M, Soumya M, Gadivan M. Expression of Ki-67 in normal oral epithelium, leukoplakic oral epithelium and oral squamous cell carcinoma. J Oral Maxillofac Pathol. 2014;18:169–76. doi:10.4103/0973-029X.140729. PMID:25328294.
- Lee SH, Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci U S A. 1990;87:5672–6. doi:10.1073/pnas.87.15.5672. PMID:1974050.
- Gumus ZH, Du B, Kacker A, Boyle JO, Bocker JM, Mukherjee P, et al. Effects of tobacco smoke on gene expression and cellular pathways in a cellular model of oral leukoplakia. Cancer Prev Res (Phila). 2008;1:100–11. doi:10.1158/1940-6207.CAPR-08-0007. PMID:19138943.
- Sobus SL, Warren GW. The biologic effects of cigarette smoke on cancer cells. Cancer. 2014;120:3617–26. doi:10.1002/cncr.28904. PMID:25043526.
- Ono Y, Torii K, Fritsche E, Shintani Y, Nishida E, Nakamura M, Shirakata Y, Haarmann-Stemmann T, Abel J, Krutmann J, et al. Role of the aryl hydrocarbon receptor in tobacco smoke extract-induced matrix metalloproteinase-1 expression. Exp Dermatol. 2013;22:349–53. doi:10.1111/exd.12148. PMID:23614742.
- Rajapakse R, Basu A, Shahid S, P Timko M. Gene expression changes and Signal Transduction Pathway alterations in Primary Human Oral Epithelial Cells exposed to Smokeless Tobacco Extracts. Scientific Research. 2014;5:13.
- Alexandrov LB, Ju YS, Haase K, Van Loo P MI, Nik-Zainal S, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354:618–22. doi:10.1126/science.aag0299. PMID:27811275.
- Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, van den Oord JJ, Spatz A, Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–73. doi:10.1038/sj.onc.1210700. PMID:17891185.
- Broustas CG, Lieberman HB. DNA damage response genes and the development of cancer metastasis. Radiat Res. 2014;181:111–30. doi:10.1667/RR13515.1. PMID:24397478.
- Cohen PA, Donini CF, Nguyen NT, Lincet H, Vendrell JA. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget. 2015;6:41566–81. doi:10.18632/oncotarget.5893. PMID:26431164.
- Zahnow CA. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:e12. doi:10.1017/S1462399409001033. PMID:19351437.
- van Kempen PM, Noorlag R, Braunius WW, Moelans CB, Rifi W, Savola S, Koole R, Grolman W, van Es RJ, Willems SM. Clinical relevance of copy number profiling in oral and oropharyngeal squamous cell carcinoma. Cancer Med. 2015;4:1525–35. doi:10.1002/cam4.499. PMID:26194878.
- Ambatipudi S, Gerstung M, Gowda R, Pai P, Borges AM, Schaffer AA, Beerenwinkel N, Mahimkar MB. Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome. PLoS One. 2011;6:e17250. doi:10.1371/journal.pone.0017250. PMID:21386901.
- Tabach Y, Kogan-Sakin I, Buganim Y, Solomon H, Goldfinger N, Hovland R, Ke XS, Oyan AM, Kalland KH, Rotter V, et al. Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLoS One. 2011;6:e14632. doi:10.1371/journal.pone.0014632. PMID:21297939.
- Fredericks WJ, Sepulveda J, Lai P, Tomaszewski JE, Lin MF, McGarvey T, Rauscher 3rd FJ, Malkowicz SB. The tumor suppressor TERE1 (UBIAD1) prenyltransferase regulates the elevated cholesterol phenotype in castration resistant prostate cancer by controlling a program of ligand dependent SXR target genes. Oncotarget. 2013;4:1075–92. doi:10.18632/oncotarget.1103. PMID:23919967.
- Fredericks WJ, Yin H, Lal P, Puthiyaveettil R, Malkowicz SB, Fredericks NJ, Tomaszewski J, Rauscher FJ 3rd, Malkowicz SB. Ectopic expression of the TERE1 (UBIAD1) protein inhibits growth of renal clear cell carcinoma cells: altered metabolic phenotype associated with reactive oxygen species, nitric oxide and SXR target genes involved in cholesterol and lipid metabolism. Int J Oncol. 2013;43:638–52. doi:10.3892/ijo.2013.1985. PMID:23759948.
- Huang LT, Wang L, Zhang L, Yan R, Li K, et al. Hepatocellular carcinoma redirects to ketolysis for progression under nutrition deprivation stress. Cell Res. 2016;26:1112–30. doi:10.1038/cr.2016.109. PMID:27644987.
- Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015;5. doi:10.1101/cshperspect.a006098. PMID:25833940.
- Plati J, Bucur O, Khosravi-Far R. Dysregulation of apoptotic signaling in cancer: molecular mechanisms and therapeutic opportunities. J Cell Biochem. 2008;104:1124–49. doi:10.1002/jcb.21707. PMID:18459149.
- Zhong CY, Zhou YM, Douglas GC, Witschi H, Pinkerton KE. MAPK/AP-1 signal pathway in tobacco smoke-induced cell proliferation and squamous metaplasia in the lungs of rats. Carcinogenesis. 2005;26:2187–95. doi:10.1093/carcin/bgi189. PMID:16051644.
- Fensterl V, Sen GC. Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol. 2015;89:2462–8. doi:10.1128/JVI.02744-14. PMID:25428874.
- Chen HM, Tanaka N, Mitani Y, Oda E, Nozawa H, Chen JZ, Yanai H, Negishi H, Choi MK, Iwasaki T, et al. Critical role for constitutive type I interferon signaling in the prevention of cellular transformation. Cancer Sci. 2009;100:449–56. doi:10.1111/j.1349-7006.2008.01051.x. PMID:19076978.
- Mullan PB, Hosey AM, Buckley NE, Quinn JE, Kennedy RD, Johnston PG, Harkin DP. The 2,5 oligoadenylate synthetase/RNaseL pathway is a novel effector of BRCA1- and interferon-gamma-mediated apoptosis. Oncogene. 2005;24:5492–501. doi:10.1038/sj.onc.1208698. PMID:15940267.
- Chen IH, Chang JT, Liao CT, Wang HM, Hsieh LL, Cheng AJ. Prognostic significance of EGFR and Her-2 in oral cavity cancer in betel quid prevalent area cancer prognosis. Br J Cancer. 2003;89:681–6. doi:10.1038/sj.bjc.6601171. PMID:12915878.
- Krontiras H, Roye GD, Beenken SE, Myers RB, Mayo MS, Peters GE, Grizzle WE. Fatty acid synthase expression is increased in neoplastic lesions of the oral tongue. Head Neck. 1999;21:325–9. doi:10.1002/(SICI)1097-0347(199907)21:4%3c325::AID-HED6%3e3.0.CO;2-P. PMID:10376752.
- Tang H, Ji F, Sun J, Xie Y, Xu Y, Yue H. RBEL1 is required for osteosarcoma cell proliferation via inhibiting retinoblastoma 1. Mol Med Rep. 2016;13:1275–80. doi:10.3892/mmr.2015.4670. PMID:26676380.
- Hagen J, Muniz VP, Falls KC, Reed SM, Taghiyev AF, Quelle FW, et al. RABL6A promotes G1-S phase progression and pancreatic neuroendocrine tumor cell proliferation in an Rb1-dependent manner. Cancer Res. 2014;74:6661–70. doi:10.1158/0008-5472.CAN-13-3742. PMID:25273089.
- Montalbano J, Jin W, Sheikh MS, Huang Y. RBEL1 is a novel gene that encodes a nucleocytoplasmic Ras superfamily GTP-binding protein and is overexpressed in breast cancer. J Biol Chem. 2007;282:37640–9. doi:10.1074/jbc.M704760200. PMID:17962191.
- Doughty TW, Arsenault HE, Benanti JA. Levels of Ycg1 Limit Condensin Function during the Cell Cycle. PLoS Genet. 2016;12:e1006216. doi:10.1371/journal.pgen.1006216. PMID:27463097.
- Liang ML, Hsieh TH, Ng KH, Tsai YN, Tsai CF, Chao ME, Liu DJ, Chu SS, Chen W, Liu YR, et al. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget. 2016;7:19723–37. doi:10.18632/oncotarget.7736. PMID:26933822.
- Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One. 2007;2:e594. doi:10.1371/journal.pone.0000594. PMID:17611626.
- Nozoe T, Korenaga D, Kabashima A, Ohga T, Saeki H, Sugimachi K. Significance of cyclin B1 expression as an independent prognostic indicator of patients with squamous cell carcinoma of the esophagus. Clin Cancer Res. 2002;8:817–22. PMID:11895914.
- Chang SS, Jiang WW, Smith I, Glazer C, Sun WY, Mithani S, Califano JA. Chronic cigarette smoke extract treatment selects for apoptotic dysfunction and mitochondrial mutations in minimally transformed oral keratinocytes. Int J Cancer. 2010;126:19–27. doi:10.1002/ijc.24777. PMID:19634139.
- Walker JM The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol Biol. 1994;32:5–8. PMID:7951748.
- Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–9. doi:10.1093/nar/gks1262. PMID:23203882.
- Goel R, Muthusamy B, Pandey A, Prasad TS. Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol Biotechnol. 2011;48:87–95. doi:10.1007/s12033-010-9336-8. PMID:20927658.
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–40. doi:10.1038/nmeth.3901. PMID:27348712.
