ABSTRACT
Background: HER2+ metastatic breast cancer (MBC) is a poor prognosis disease, unusually curable. To date, no predictive factors have been clearly correlated with long-term response to anti-HER2 agents.
Methods: 54 HER2+ MBC patients treated with HER2 targeted therapy as first line treatment were analysed: 40 with a time to progression longer than 3 years in Long Responders (LR) group and 14 with a progression disease within one year of anti-HER2 therapy in a control group named Early Progressors (EP). The expression of 770 genes and 13 molecular pathways were evaluated using Nanostring PanCancer pathway panel performed on FFPE BC tissues.
Results: Considering baseline patients and tumor characteristics, EP women had more CNS spread and more metastatic burden of disease compared to LR (p > 0.05). Gene expression analysis identified 30 genes with significantly different expression in the two cohorts; five were driver genes (BRCA1, PDGFRA, AR, PHF6 and MSH2). The majority of these genes were over-expressed, mainly in LR patients, and encoded growth factors, pro- or anti-inflammatory interleukins and DNA repair factors. Only four genes were down regulated, all in EP group (TNFSF10, CACNG1, IL20RB and BRCA1). Most of these genes were involved in MAPK and PI3K pathways. MAPK pathway was differently expressed between LR and EP (p = 0.05). PI3K was the only pathway overexpressed in EP patients.
Conclusions: Whole genome expression analysis comparing LR vs. EP identified a group of genes that may predict more favourable long-term outcomes. Up-regulation of MAPK and down-regulation of PI3K pathway could be a positive predictive factors. Further clinical implications are warranted.
Abbreviations: BC: breast cancer; MBC: metastatic breast cancer; LR: long responder; EP: early progressor; FFPE: formalin-fixed paraffin-embedded; CNS: central nervous system; PFS: progression free survival; OS: overall survival.
Background
Epidermal growth factor receptor 2 (HER2) positive metastatic breast cancer (MBC) is considered an incurable disease, with poor prognosis.Citation1 In the last decade, the introduction of HER2 targeted therapies (such as trastuzumab) markedly improved the clinical outcomes of MBC patients. Recently, the approval of HER2 dual-block has further improved the median overall survival (32–42 months).Citation2 Unusually, long-term survival and durable complete responses have been reported in literature suggesting that cure is possible in a small subset of HER2 positive metastatic patients. Unfortunately, predicting disease outcome is particularly difficult because of heterogeneity of the disease. To date, no clinical and/or molecular factors have been univocally correlated with long-term response to anti-HER2 agents.Citation3–Citation6
In the recent years, several multigene tests have changed the understanding of breast cancer (BC), providing an opportunity to perform more detailed and individualized treatments. Gene expression analysis with its ability to simultaneously analyse hundreds of genes, has become a powerful tool in understanding all the aspects of cancer biology.Citation7,Citation8 The current challenge is the discovery of new prognostic and predictive markers in order to assist clinicians in treatment decision-making process, allowing a personalized cancer care.
The present study has been designed to investigate clinical and molecular factors that could differentiate long-term responders from metastatic anti-HER2-treated patients, with early progression. To investigate this issue, we analysed data from a retrospective database of HER2 positive MBC patients treated with anti-HER2 targeted therapies as first line treatment in our institution. Molecular analysis were performed comparing patients with a progression free for at least 3 years after starting an anti-HER2 therapy versus a control group.
Material and methods
Population
Patients diagnosed with HER2 positive MBC treated with a first line anti-HER2 therapy at the University Hospital of Modena were considered. HER2 status was determined on the primary BC tissue using immunohistochemistry and/or fluorescence in situ hybridization (FISH) according to institutional guidelines. Patients were included in the study in accordance to their survival outcomes and availability of formalin-fixed paraffin-embedded (FFPE) BC tissue. Patients were divided in two cohorts: Long Responders (LR) group that included patients with first line anti-HER2 therapy time to progression (TTP) longer than 3 years and Early Progressors (EP) group that included patients with TTP shorter than one year. Regarding molecular analysis we extracted RNA from FFPE diagnostic core biopsy or surgical resection BC tissues. All tumor and patients’ characteristics and treatment information were collected, including: date of birth, dates of primary BC and MBC diagnosis, date of first progression, date of last follow up or death, systemic treatments received in the neo/adjuvant setting, first line treatment in the metastatic setting and sites of metastatic disease at relapse. Radiological and clinical responses were assessed by local investigators using the Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1. This mono-istitutional retrospective study was approved by our ethical committee, alive patients signed a written consent form.
RNA isolation and gene expression analysis
RNA was extracted from FFPE of BC. The hematoxylin and eosin stayned sections were reviewed by pathologist and areas containing not less than 20 mm2 of invasive breast carcinoma were outlined on the slides. After removal of the not neoplastic breast tissue by manual microdissection, the RNA was extracted from two to four 10 μm sections. RNA extracted was purified using High Pure FFPE RNA Isolation Kit according to manufacturer’s instructions. RNA concentration was measured by spectrophotometry using a Xpose Instrument (Trinean).
Gene expression on the nCounter platform (prep Station and Digital Analyzer – Nanostring technology, Seattle, USA) was assessed with the nCounter PanCancer Pathway panel. The panel analyses the expression of 770 genes (606 Pathway genes, 124 Driver genes and 40 Housekeeping genes) involved into 13 molecular pathways (Notch, Wnt, Hedgehog, TGFB, MAPK, STAT, P13K, RAS, Chromatin Modification, Transcriptional Regulation, DNA Damage Control, Cell Cycle and Apoptosis). The platform estimates the quantity of each mRNA transcript using a multiplexed hybridization system and digital readouts of fluorescent barcoded probes that are hybridized to each transcription. Row counts resulting from the analysis were normalized against reference genes, genes selected to have the least variance with the geNorm algorithm. Normalized data were analysed using NanoString’s nSolver version 3.0 software with the Advanced Analysis application tool based on Pathifier algorithm.Citation9 Using this algorithm, genes that were significantly up- or down-regulated between patients group were identified (p value ≤ 0.05). The same algorithm was used to calculate the molecular pathways deregulation score for each tumor sample on the basis of gene expression data. This score was graphically represented by a heat-map. Using pathways score, we compared pathways expression in LR versus EP group, the boxplots were designed, one for each pathways.
Statistical analysis
Statistical analysis was performed using STATA 13 (StataCorp. 2013. Stata Statistical Software: Release 13. College Station, TX: StataCorp LP). Baseline differences in prognostic factors were assessed by chi-square test or Fisher exact test for categorical variables and t test for continuous variables. Survival outcomes of interest were OS defined as the time from the diagnosis of BC to the death/last follow up and progression Free Survival (PFS) defined as the time from the date of the diagnosis of metastatic disease to the date of the first documented progression disease/death. OS and PFS were estimated using the Kaplan Meier method. A p-value < 0.05 was considered statistically significant; hazard ratio was estimated with 95% of confidence limits.
Results
A total of 54 HER2 positive MBC patients were included in the analysis: 40 patients in LR group and 14 patients in EP group. Median PFS and OS were 4.5 years (min 2.9 – max 12) and 17.4 (min 3.2 – max 20) years in LR versus 4.1 months (min 30 – max 103) and 5.05 years (min 0.8 – max 13) in EP patients, respectively. Main baseline patients and tumor characteristics, were balanced between the two groups, although EP patients were more likely to have central nervous system spread and more metastatic burden of disease compared to LR (29% vs. 5%, p = 0.04 and 57% vs. 17%, p = 0.009 respectively) (). A greater proportion of LR patients (22%) had de novo MBC compared with EP (14%). Regarding early stage treatment, chemotherapy was administered in 65% of LR patients compared to 71% of EP. Trastuzumab, as adjuvant treatment, was administered in 25% of patients in LR group and 43% of EP (). Considering systemic therapies for metastatic disease, most of the patients in both group received trastuzumab and chemotherapy or hormonal therapy as first line treatment. Capecitabine and lapatinib was the first line choice for six LR patients and for one EP patient. Two patients in EP group received the association of trastuzumab, pertuzumab and taxane. Two patients in both cohorts were treated with T-DM1 as first line treatment ().
Table 1. Baseline patients and tumor characteristics.
Table 2. Summary of treatments.
Table 3. Summary of first line treatments.
Whole genome expression analysis comparing LR versus EP showed 30 genes with significantly different expression between the two cohorts (p value < 0.05, ). Raw data files and normalized gene counts are available in . Five of these genes were driver genes: PDGFRA driver gene of MAPK and PI3K pathway, AR driver gene of MAPK pathway, BRCA1 driver gene of PI3K and DNA damage control pathway, PHF6 driver gene of NOTCH pathway and MSH2 driver gene of DNA damage control pathways. The majority of these genes were over-expressed, mainly in LR patients, and encoded growth factors, pro- or anti-inflammatory interleukins and DNA repair factors. Only four genes were down regulated, all in EP group (TNFSF10, CACNG1, IL20RB and BRCA1). Most of these genes were involved in MAPK and PI3K pathways (9 and 8, respectively) (). Considering differential pathway expression between LR and EP, eleven out of thirteen pathways were over-expressed in LR. Only PI3K and APC/WNT were up regulated in EP group (). MAPK pathway was significantly increased in LR compared to EP (p = 0.05). Even if not statistically significant, the different expression of PI3K pathway between the two groups was clinically relevant (median expression LR: 1441 ± 485 vs 1759 ± 762 in EP group; p = 0.1). APC/WNT, DNA Damage Control and TGF-β had the least different expression between the two groups ().
Table 4. Differential gene expression between LR patients and EP patients. FC = fold change.
Table 5. Pathway deregulation score in LR patients and EP patients.
Figure 1. Hierarchical clustering based on RNA expression levels of 30 genes out of 770 genes analysed by PanCancer pathways panel. Rows, genes; columns: samples. Expression level of each gene in a single sample was related to its median level across all samples and is depicted according to a colour scale show above. Red and green represented the expression levels above and below the median, respectively.
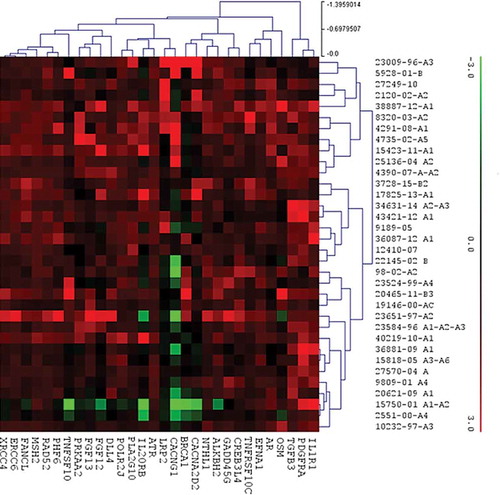
Figure 2. Distribution of the 30 genes differently expressed between the LR and EP groups within the molecular pathways.
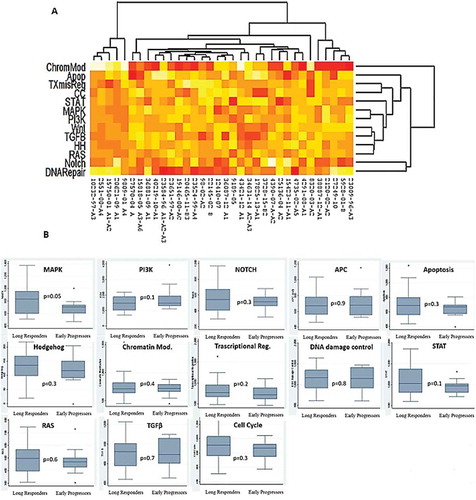
Figure 3. (A) Pathways deregulation score for each tumor sample on the basis of gene expression data. Each row corresponds to a pathway and each column to a sample. Pathways and samples are clustered according to pathways deregulation score. Red colour represents low score, yellow colour high score. (B) Boxplots of pathways deregulation scores. The distribution of Pathifier deregulation scores of each pathways is plotted for LR and EP. The top and the bottom of the box delineate the upper and lower quartile, while the thick line within each box represents the median. Whiskers extend capture all data within two standard deviations of the means.
Discussion
Long-term survival without evidence of disease progression is possible in a small subgroup of HER2-positive MBC patients.Citation10 To date, in this setting, there are no univocal clinical or biological factors able to predict disease outcome. In our study-population, main clinical factors such as grade of differentiation, stage at initial diagnosis, histological type, hormonal receptors (HR) status and Ki67 fail to correlate with long-term response (). Only patients with more metastatic sites, central nervous system involvement seemed to have worse outcome (). In the literature, only few studies analysed the clinical characteristics of HER2 positive long-term responder patients, with controversial results. The Regist-HER study identified 3 factors associated with prolonged response: i. hormone positive disease; ii. metastasis to node/local sites, iii. first line trastuzumab plus taxane.Citation3 On the other hand, trastuzumab, as part of adjuvant therapy was the only negative predictor identified in the LongHER patients.Citation5 As positive prognostic factors, other studies identified the ductal histotype,Citation4 younger age, stage I-II, HR positive status, one organ involvement at the diagnosis,Citation10 as well as the surgical management with resection of both the primary tumor and the metastasis.Citation11 Regarding molecular analysis, only the LongHer study demonstrated that mutations in PI3K pathway were associated with poor response to trastuzumab-based therapy. In particular, tumours in the LR group had at most two genes mutation while tumours in the control group usually had four or five alterations.Citation5
On these premises, we focused our analysed on the molecular profile of HER2 positive BC patients according with their prognosis. In our analysis we identified 30 genes with significantly different expression in the two cohorts of patients, correlating with the survival and sensitivity to the treatment. Five of these genes are known to be driver genes of main molecular pathways (AR, BRCA1, PDGFRA, PHF6 and MSH2) whereas other are widely known to be involved in BC metastatization and progression.
In particular, considering Androgen Receptor (AR), it is known to be associated with well-differentiated HR positive BC and more favourable survival outcome in triple negative BC subtype respectively.Citation12 Its expression has a significant variability depending on HR and HER2 status. In HR positive disease, the co-expression of estrogen receptors and ARs was found to be 75% whereas AR expression was range from 20% to 40% in HER2 positive BCs.Citation13–Citation15 Considering HER2 positive disease, recent evidence suggest a cross regulation genes between AR and HER2. AR seems to contribute to the growth of HER2 positive tumors by promoting ERK and AKT activation via up-regulation of HER2 gene transcription. Preclinical evidence suggested that inhibition of AR impairs the growth of HER2 positive BC cells.Citation16 The prognostic significance of AR in HER2 disease seems to either show no association with survival, however, many of the studies reported in literature are limited by smaller sample size.Citation16 In our study population, AR was overexpressed in LR cohort, suggesting a possible positive prognostic value.
BRCA1 is a known gene involved in familiar BC. It encodes for a protein that plays a critical role in maintaining the stability of cells’ genetic information and in repairing DNA damage.Citation17 Preclinical data suggested that low levels of BRCA1 were related with increased tumour grade and poor prognosis.Citation18,Citation19 In our analysis, down-regulation of BRCA1 was found in EP group supporting the idea of more aggressive behaviour of these cancers. Moreover, BRCA1 crosstalk with the Fanconi Anemia (FA) family genes (such as FANCL). FA pathway is essential for tumor cells to resist killing by DNA cross-linking agents. The BRCA/FA pathway’s defect lead to DNA damage and repair barriers making cancer cells sensitive to chemotherapy.Citation20 In our study population in LR subgroup FANCL was overexpressed suggesting that the disruption of this pathway may reverses drug resistance. As well as FANCL, MSH2 (MutS Homolog 2) is a gene involved in DNA repair. Recent evidence suggests that MSH2 is able to modulate BRCA1 activity too.Citation21 It’s likely that the ongoing cross-talk between these genes involved in the mechanism of DNA repair may sensitize cancer cells to improve or decrease the efficacy of different anticancer drugs.
The role of PDGFRA (platelet derived growth factor receptor alpha) in BC biology is less clear. It can be considered a modulator of tumor microenvironment and essential in tumor angiogenesis by interaction with VEGFR family and FGF family. Clinical data suggested that PDGFR is highly expressed in stromal cells of invasive carcinoma and it is associated with lymph node metastasis.Citation22 Repressing the expression of PDGFRA or inhibiting its kinase activity blocked rapamycin induced phosphorylation of AKT and decrease tumor cells viability.Citation22 In our series, PDGFRA was down regulated in EP group probably as a consequence of its interaction with the other genes involved PI3K and MAPK pathways. As well as PDGFR, FGF is involved in tumor microenvironment. Deregulation of FGF/FGFR signalling occurs through various mechanisms, including amplification, aberrant ligand expression, receptor mutation and translocation resulting in down or up regulation of the mRNA expression. Preclinical evidence suggests that resistance to HER2-targeted therapy may results from a switch in dependency on the ER/HER2 signalling pathway to the FGFR signalling pathway.Citation23 Lapatinib-resistant BC cells had an overexpression of FGFR2 and a reduction in HER2 overexpression.Citation24
Regarding molecular pathways, most of the identified genes were involved in MAPK and PI3K pathways (9 and 8 genes respectively), two molecular pathways known to be involved in tumorigenesis process. Mainly, genetic alterations in PI3K pathway were commonly observed in BC patients and correlated with drug-resistance.Citation25,Citation26 Several studies have shown how the expression and the activation of PI3K are frequently observed in trastuzumab-refractory cancers.Citation27 Even in our analysis PI3K pathway was overexpressed in EP group underling its involvement in drug resistance mechanisms. On the other hand, the clinical meaning of MAPK up-regulation in BC is less clear. Higher MAPK activity seems to be more evident in patients with more aggressive early triple negative BC, not clear evidence regarding its meaning in the metastatic setting and in HER2 positive tumours are known.Citation28 In our series, MAPK pathways was up regulated in LR patients.
In our opinion, considering the genes expression profile instead of the single genes mutations we offer a clearer overview of the complicated molecular architecture of BC. In fact, the genes expression prolife highlights not only the effect of a single gene mutation but also the consequence of several genes interactions and different epigenetic phenomenon. These events occurs during cancer progression and are able to influence treatment response. Nowadays, it is well known how the extreme complexity of cancer biology cannot be explain only with the detection of the single gene mutation but needs more amplified analysis involving genes and pathways expression and interaction and epigenetic phenomenon, such as abnormalities in DNA methylation. Our findings suggest that gene expression profile may be instrumental for individualized pathway-directed therapies and may be a major step in defining optimal treatments for BC patients.
The major limitation of our study is the small sample size due both to a rare subgroup of patients (HER2+ MBC with very good prognosis) and tissues sample availability for RNA extraction.
Conclusion
In summary, long-term survival is possible in a selected population of patients with advanced HER2 positive MBC. To the best of our knowledge, this is the first study in literature that performed a molecular analysis, comparing the expression of 770 genes and 13 pathways, in Long responders vs Early Progressor HER2 positive MBC patients. Gene expression analysis identified 30 genes with significantly different expression in the two cohorts of patients. More than half of these genes were involved in MAPK and PI3K pathways. Our findings suggest that gene expression profile may be instrumental for individualized pathway-directed therapies. In our opinion gene expression analysis could be an useful tool for assisting clinicians in decision-making process, in order to modulate the treatment intensity based on the intrinsic BC biological profile. Owing to the small sample size and the retrospective nature of the study, these correlations are purely exploratory and need to be validate in large studies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Ethics apEPoval and consent to participate
This study was apEPoved by the Ethical Committee of Azienda Ospedaliero Universitaria Policlinico di Modena (EPotocol number: CE 267/15). All patients signed a written, informed consent.
Additional information
Funding
References
- Slamon DJ. Proto-oncogenes and human cancers. N Engl J Med. 1987;317(15):955–957. doi:10.1056/NEJM198710083171509.
- Swain SM, Baselga J, Kim S-B, Ro J, Semiglazov V, Campone M, Ciruelos E, Ferrero J-M, Schneeweiss A, Heeson S, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372(8):724–734. doi:10.1056/NEJMoa1413513.
- Yardley DA, Tripathy D, Brufsky AM, Rugo HS, Kaufman PA, Mayer M, Magidson J, Yoo B, Quah C, Ulcickas Yood M. Long-term survivor characteristics in HER2-positive metastatic breast cancer from registHER. Br J Cancer. 2014;110(11):2756–2764. doi:10.1038/bjc.2014.174.
- Spano J.P, Beuzeboc P, Coeffic D, Arnould L, Lortholary A, Andre F, Ferrero J-M. Long term HER2+ metastatic breast cancer survivors treated by trastuzumab: results from the French cohort study LHORA. Breast. 2015;24(4):376–383. doi:10.1016/j.breast.2015.02.035.
- Gámez-Pozo A, Pérez Carrión RM, Manso L, Crespo C, Mendiola C, López-Vacas R, Berges-Soria J, López IÁ, Margeli M, Calero JLB, et al. The Long-HER study: clinical and molecular analysis of patients with HER2+ advanced breast cancer who become long-term survivors with trastuzumab-based therapy. PLoS One. 2014;9(10):e109611. doi:10.1371/journal.pone.0109611.
- Extra J.M, Antoine EC, Vincent-Salomon A, Delozier T, Kerbrat P, Bethune-Volters A, Guastalla J-P, Spielmann M, Mauriac L, Misset J-L, et al. Efficacy of trastuzumab in routine clinical practice and after progression for metastatic breast cancer patients: the observational Hermine study. Oncologist. 2010;15(8):799–809. doi:10.1634/theoncologist.2009-0029.
- van ‘T Veer LJ, Dai H, Van De Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, Van Der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi:10.1038/415530a.
- Li L.F, Xu X.J, Zhao Y, Liu Z.B, Shen Z.Z, Jin W.R, Shao Z.M. Integrated gene expression profile predicts prognosis of breast cancer patients. Breast Cancer Res Treat. 2009;113(2):231–237. doi:10.1007/s10549-008-9925-4.
- Drier Y, Sheffer M, Domany E. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci U S A. 2013;110(16):6388–6393. doi:10.1073/pnas.1219651110.
- Murthy P, Kidwell KM, Schott AF, Merajver SD, Griggs JJ, Smerage JD, Van Poznak CH, Wicha MS, Hayes DF, Henry NL. Clinical predictors of long-term survival in HER2-positive metastatic breast cancer. Breast Cancer Res Treat. 2016;155(3):589–595. doi:10.1007/s10549-016-3705-3.
- Harano K, Lei X, Gonzalez-Angulo AM, Murthy RK, Valero V, Mittendorf EA, Ueno NT, Hortobagyi GN, Chavez-MacGregor M. Clinicopathological and surgical factors associated with long-term survival in patients with HER2-positive metastatic breast cancer. Breast Cancer Res Treat. 2016;159(2):367–374. doi:10.1007/s10549-016-3933-6.
- Park SL, Fesinmeyer MD, Timofeeva M, Caberto CP, Kocarnik JM, Han Y, Love S-A, Young A, Dumitrescu L, Lin Y, et al. Androgen receptor expression and outcomes in early breast cancer: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106(1):djt319. doi:10.1093/jnci/dju061.
- Niemeier LA, Dabbs DJ, Beriwal S, Striebel JM, Bhargava R. Androgen receptor in breast cancer: expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol. 2010;23(2):205–212. doi:10.1038/modpathol.2009.159.
- Micello D, Marando A, Sahnane N, Riva C, Capella C, Sessa F. Androgen receptor is frequently expressed in HER2-positive, ER/PR-negative breast cancers. Virchows Arch. 2010;457(4):467–476. doi:10.1007/s00428-010-0964-y.
- Aleskandarany MA, Abduljabbar R, Ashankyty I, Elmouna A, Jerjees D, Ali S, Buluwela L, Diez-Rodriguez M, Caldas C, Green AR, et al. Prognostic significance of androgen receptor expression in invasive breast cancer: transcriptomic and protein expression analysis. Breast Cancer Res Treat. 2016;159(2):215–227. doi:10.1007/s10549-016-3934-5.
- He, L., Du Z, Xiong X, Ma H, Zhu Z, Gao H, Cao J, Li T, Li H, Yang K, et al. Targeting androgen receptor in treating HER2 positive breast cancer. Sci Rep. 2017;7(1):14584. doi:10.1038/s41598-017-14623-2.
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62(3):676–689.
- Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9(4):444–450. doi:10.1038/ng0495-444.
- Pistelli M, Page DB, Li BT, Connell LC, Schindler K, Lacouture ME, Postow MA, Wolchok JD. BRCA mutations and IGF-R1 expression in modulating sensitivity to trastuzumab in HER2-positive breast cancer. Ann Oncol. 2016;27(suppl 6). doi:10.1093/annonc/mdw141.
- Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26(13):1393–1408. doi:10.1101/gad.195248.112.
- Maresca L, Spugnesi L, Lodovichi S, Cozzani C, Naccarato AG, Tancredi M, Collavoli A, Falaschi E, Rossetti E, Aretini P, et al. MSH2 role in BRCA1-driven tumorigenesis: A preliminary study in yeast and in human tumors from BRCA1-VUS carriers. Eur J Med Genet. 2015;58(10):531–539. doi:10.1016/j.ejmg.2015.09.005.
- Carvalho I, Milanezi F, Martins A, Reis RM, Schmitt F. Overexpression of platelet-derived growth factor receptor alpha in breast cancer is associated with tumour progression. Breast Cancer Res. 2005;7(5):R788–R795. doi:10.1186/bcr1304.
- Andre F, Cortes J. Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res Treat. 2015;150(1):1–8. doi:10.1007/s10549-015-3301-y.
- Azuma K, Tsurutani J, Sakai K, Kaneda H, Fujisaka Y, Takeda M, Watatani M, Arao T, Satoh T, Okamoto I, et al. Switching addictions between HER2 and FGFR2 in HER2-positive breast tumor cells: FGFR2 as a potential target for salvage after lapatinib failure. Biochem Biophys Res Commun. 2011;407(1):219–224. doi:10.1016/j.bbrc.2011.03.002.
- Paplomata E, O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6(4):154–166. doi:10.1177/1758834014530023.
- Dieci MV, Prat A, Tagliafico E, Paré L, Ficarra G, Bisagni G, Piacentini F, Generali DG, Conte P, Guarneri V. Integrated evaluation of PAM50 subtypes and immune modulation of pCR in HER2-positive breast cancer patients treated with chemotherapy and HER2-targeted agents in the CherLOB trial. Ann Oncol. 2016;27(10):1867–1873. doi:10.1093/annonc/mdw262.
- Gagliato DM, Fontes Jardim DL, Pereira Marchesi MS, Hortobagyi GN. Mechanisms of resistance and sensitivity to anti-HER2 therapies in HER2+ breast cancer. Oncotarget. 2016. doi:10.18632/oncotarget.7043
- Eralp Y, Derin D, Ozluk Y, Yavuz E, Guney N, Saip P, Muslumanoglu M, Igci A, Kücücük S, Dincer M, et al. MAPK overexpression is associated with anthracycline resistance and increased risk for recurrence in patients with triple-negative breast cancer. Ann Oncol. 2008;19(4):669–674. doi:10.1093/annonc/mdm522.
