ABSTRACT
Colorectal cancer (CRC) is ranked third as the most common malignancy, and it develops into metastasis at a high rate. Importantly, distant metastasis is considered to be a key factor for colorectal therapy. In the present study, we identified FOXD4, a transcription factor belonging to the forkhead/winged helix-box (FOX) family, as a novel biomarker for diagnosis and treatment of patients with CRC. We revealed that FOXD4 was up-regulated in CRC tissues and increased the metastatic ability of CRC cells. Additionally, FOXD4 affected the metastasis of CRC by inducing the epithelial-mesenchymal transition (EMT) process. Furthermore, FOXD4 could directly bind the SNAI3 promoter during EMT in CRC and then facilitate CRC metastasis. In summary, the present research strongly suggests that FOXD4 is a valuable marker for CRC, and that targeting FOXD4 may be a novel strategy for enhancing the treatment outcomes of CRC therapy
Introduction
Colorectal cancer (CRC) is ranked third as the most common malignancy, for which the consequent mortality is also the third highest in the worl.Citation1 Moreover, cancer metastasis is the major cause of poor prognosis of CRC patient.Citation2–Citation4 Approximately 30% of CRC patients show the final development of metastatic disease. Despite the development of new therapeutic strategies for clinical treatment, many patients still die from CR.Citation5,Citation6 Thus, it is important to find novel biomarkers to predict the risk of cancer progression and metastasis, which will be useful for improving the prognosis for CRC patient.Citation7,Citation8
FOXD4 is a transcription factor (TF) that belongs to the forkhead/winged helix-box (FOX) famil.Citation9 FOX transcription factors participate in several key processes, such as metabolism, cell proliferation, and metastasi.Citation10,Citation11 It is reported that mutations in FOXD4 transcription are related to multiple phenotypes including dilated cardiomyopathy, obsessive-compulsive disorders, and suicidalit.Citation9
Previous studies have revealed that epithelial-mesenchymal transition (EMT) is an important process preceding tumor metastasi.Citation12,Citation13 EMT is an evolutionarily conserved developmental process characterized by the conversion of polarized immotile epithelial cells to motile mesenchymal cell.Citation14 During EMT, cell characteristics show obvious changes, which contribute to cell motility and invasivenes.Citation15
Recent studies have considered FOXD4 as a TF; however, the expression and mechanisms of FOXD4 in cancer are unclea.Citation16 Moreover, further research is required to clarify the function of FOXD4 in CRC. In the present study, we revealed that a high level of FOXD4 expression in CRC was significantly associated with poor prognosis. Both functional assays in vivo and in vitro indicated that FOXD4 upregulation is essential for the promotion of CRC progression.
Materials and methods
Patient tissue specimens
We obtained the CRC tissues and matched adjacent non-tumor tissues from CRC patients from Qilu Hospital between November 2012 and November 2014. All of the patients who participated in the research provided written informed consent. The patients included had not undergone chemotherapy or radiotherapy before surgery. The ethics committee of Qilu Hospital had approved the use of tissue in the present study. There were 94 paired CRC tissues and adjacent tissues selected for use in our research. Additionally, clinical information obtained from the patients included age, gender, and tumor stage.
Cells and antibodies
In the present study, the three CRC cells used – LoVo, SW480, and SW620 – were obtained from the Cell Bank of the Chinese Academy of Sciences (China). DMEM was used to culture the cells in a 5% CO2 atmosphere, which was supplement with 10% fetal bovine serum (FBS).
The antibodies used in this study included GAPDH (Santa Cruz, USA), FOXD4 (Santa Cruz, USA), Snail1, Snail2, Snail3 (Santa Cruz, USA), E-cadherin (Cell Signaling, USA), and vimentin (Cell Signaling, USA).
Bioinformatic analysis
RNA-Seq of Level 3 HiSeq data was obtained from The Cancer Genome Atlas (TCGA) website for 380 cancer and 50 normal samples. Moreover, the microarray data for CRC patients were downloaded from the Gene Expression Omnibus (GEO) database, which holds a larger amount of tissue sample data and clinical information. The expression of FOXD4 in different tissues was compared using Student’s t-test. Kaplan-Meier survival analysis was carried out to determine the association of FOXD4 with CRC prognosis.
Immunohistochemical staining
IHC staining and images analysis were carried out following the instructions described in previous studie.Citation17 Anti-FOXD4 antibody (1:100, Santa Cruz, USA) was used in the present study. The antibody dilution medium was used as a negative control.
Plasmids, RNA interference, and transfection
The FOXD4-overexpressing plasmid (pcDNA3.1-FOXD4) was built according to themanufacturer’s instructions. At first, full-length FOXD4 cDNA was cloned into the pcDNA3.1 plasmid. Moreover, Lipofectamine 2000 reagent (Life Technologies, Carlsbad, CA, USA) was used to transiently transfect the plasmid into cells following the manufacturer’s instructions. After 72 h, western blotting or real-time-PCR was performed to detect the protein or mRNA levels of specific genes, respectively. An RNA interference assay was used to knockdown the expression of target genes. FOXD4-specific siRNA, SNAI3-specific siRNA, and negative control siRNAs were obtained from RiboBio (Guangzhou, China).
Quantitative real-time PCR
Total RNA was extracted from cultured cells though TRIzol (Invitrogen, USA), and then reverse transcribed to cDNA following the manufacturer’s protocol. SYBR Green (Toyobo) was used to amplify the cDNA. Primers were designed according to the NCBI database. Additionally, the primers were synthesized by Invitrogen (Shanghai, China). GAPDH was used to normalize the RNA expression. The results were calculated using the Ct (2−ΔΔCt) method, which was applied to calculate the respective expression levels.
Western blot analysis
Nonidet P 40 (Beyotime, Shanghai, China) was used to extract the total protein, which was supplemented with a protease inhibitor cocktail according to the manufacturer’s instructions. We used a bicinchoninic acid (BCA) assay to measure the concentration of protein. After sodium dodecyl sulfate-PAGE and incubating antibodies, an enhanced chemiluminescence kit (Millipore, Billerica, MA, USA) was applied to examine the protein levels using Molecular Imager® (Bio-Rad).
Tumor cell invasion and migration assay
Boyden chambers were used to perform the invasion and migration assays, which contain filter insets. In addition, a Matrigel coat was applied to the chambers when the invasion assay was carried out. We suspended the cells into DMEM without FBS, and then these cells, with and without FOXD4, were seeded into the upper chamber. Moreover, the DMEM was supplemented with 20% FBS. After a 24-h incubation, cells in the upper chamber were fixed in 4% formaldehyde and stained with 0.05% crystal violet. We considered the cells in the lower chamber as invasion cells.
Chromatin immunoprecipitation (chip)
ChIP assay was performed following the instructions given in the ChIP Assay Kit (Millipore, Billerica, MA, USA). Anti-FOXD4 antibodies were used to capture the DNA fragments, and real-time PCR was then carried out to detect the expression level of SNAI3.
Luciferase assay
A dual luciferase assay kit was used to perform the luciferase assay. All steps taken were according to the manufacturer’s instructions (Promega, Madison, WI, USA). At first, 4 × 104 cells were cultured in 24-well plates and transfected with SNAI3 promoter-luciferase plasmids and pcDNA3.1-FOXD4 or siRNA-FOXD4. TK plasmids (Beyotime, Shanghai, China) were used to normalize the efficiency. After 24 h, the dual luciferase assay kit (Promega, Madison, WI, USA) was applied to detect the luciferase activity.
Statistical analysis
R software, SPSS 17.0 statistical software, and GraphPad Prism 5.0 software were used to carry out the statistical analysis. Student’s t-test and one-way analysis of variance (ANOVA) were used to describe the differences between two or multiple groups. Kaplan-Meier survival analysis was used to present the survival differences between groups. We considered P < 0.05 to be significant.
Results
FOXD4 was overexpressed in CRC and was associated with poor survival
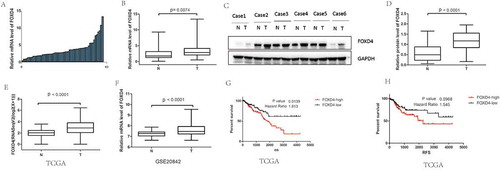
To investigate the expression status of FOXD4 in CRC, 12 pairs of frozen CRC tissue samples were used for analysis by qPCR and western blot. The result of qPCR showed that FOXD4 was significantly overexpressed in CRC tissues compared with that in matched normal colorectal samples (P < 0.01; -B). Furthermore, the protein level of FOXD4 in CRC samples was also higher than that in their adjacent normal controls (P < 0.05; -D). Moreover, we next calculated mRNA expression using the TCGA colorectal cancer database. The TCGA colorectal cancer database contains 381 CRC samples and 50 normal colorectal samples. Interestingly, FOXD4 expression was overexpressed in CRC tissues compared to that in normal tissues, which is consistent with the above results (P < 0.0001; ). Similar results can be found in another cohort from the GEO database. The results showed that higher FOXD4 mRNA expression levels were observed in human CRC tissues than in normal tissues (P < 0.0001; ). Moreover, Kaplan-Meier survival analysis was performed to compare the prognosis of different groups, classified by FOXD4 expression The results revealed that patients with high FOXD4 expression had significantly worse prognosis, as shown by the TCGA database (P < 0.0001; ). Additionally, the relapse-free survival showed similar results. The prognosis of CRC patients with FOXD4 overexpression was poorer than that in patients with low FOXD4 expression (P < 0.001; ). To further confirm the function of FOXD4 in CRC, IHC staining was used to examine the expression of FOXD4 in CRC specimens. Next, the relationship between the clinicopathological characteristics of the CRC patients and the expression of FOXD4 was analyzed (). The results showed that FOXD4 expression was related to advanced TNM stages and invasion depth. Notably, these data indicate that FOXD4 expression could represent a novel prognostic indicator for patients with CRC.
Table 1. Relative expression levels of FOXD4 according to clinicopathological parameters.
Figure 1. FOXD4 was over-expressed in CRC and was associated with poor survival. (A-B) The expression of FOXD4 was examined by quantitative real-time PCR of colorectal cancer tissues and adjacent noncancerous tissues. (C-D) The expression of FOXD4 was examined by western blots derived from colorectal cancer tissues and adjacent noncancerous tissues. (E-F) The expression of FOXD4 as documented for CRC cohorts on TCGA and GEO databases. (F-G) The Kaplan-Meier method was used to analyze the overall survival and recurrence-free survival of patients based on their FOXD4 levels. (*p < 0.05, **p < 0.01, ***p < 0.001).
FOXD4 promoted invasion and migration ability in vitro
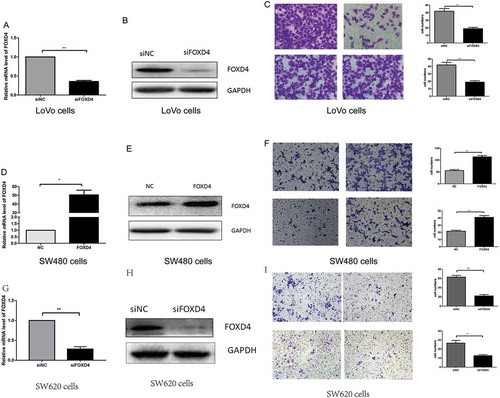
Our results demonstrated that FOXD4 has an effect on CRC carcinogenesis and prognosis; however, the molecular mechanisms are still undefined. In our previous research (Fig S1), we examined the expression of FOXD4 in five CRC cells. The results showed that the expression of FOXD4 was the highest in LoVo cells and lowest in SW480 cells. According to the above data, we selected LoVo cells for downregulation and SW480 cells for overexpression. To further test the effects of FOXD4 in CRC, CRC cells were transfected with siRNAs specific for FOXD4 and the pcDNA3.1-FOXD4 plasmid. qPCR and western blot were used to confirm the expression of FOXD4 in presence of transfected siRNA and plasmid (, A-B, D-E). Next, the biological function of FOXD4 in CRC cell migration and invasion was explored using Transwell assays. The results showed that down-regulation of FOXD4 significantly suppressed the invasion and migration capacity of LoVo cells (), while overexpression of FOXD4 greatly promoted SW480 cell migration and invasion (). In addition, SW620 cells were used to confirm the function of FOXD4 in cell migration and invasion. These cells were derived from the same patient as the SW480 cells. The results showed that down-regulation of FOXD4 significantly suppressed the invasion and migration capacity of SW620 cells (-I) Therefore, the above data strongly suggest that FOXD4 promotes the metastatic ability of CRC cells in vitro.
Figure 2. FOXD4 promoted cell invasion and migration ability in vitro. (A-B) FOXD4 knockdown by siRNA in LoVo cells was measured via quantitative real-time PCR and western blot. (C) Transwell assay revealed that knockdown of FOXD4 inhibited migration and invasion of LoVo cells. (D-E) FOXD4 up-regulation by pcDNA3.1- FOXD4 in SW480 cells were examined via quantitative real-time PCR and western blot. (F) Transwell assay was used to reveal that upregulation of FOXD4 promoted migration and invasion of SW480 cells. (G-H) FOXD4 knockdown by siRNA in SW620 cells was examined via quantitative real-time PCR and western blot. (I) Transwell assay was used to reveal that knockdown of FOXD4 inhibited migration and invasion of SW620 cells. (*p < 0.05, **p < 0.01, ***p < 0.001)
FOXD4 regulated EMT through SNAI3/CDH1
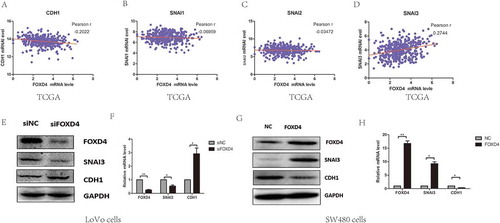
It has been confirmed that EMT progression is a key factor in tumor metastasi.Citation18 Molecular markers associated with EMT include E-cadherin, ZO-1, vimentin, and β-catenin. Analysis of these markers was performed to clarify EMT in various cell lines. In addition, we investigated Snail expression to identify the role of FOXD4 in CRC EMT progression. In order to understand the mechanism by which FOXD4 promotes CRC EMT progression, we next identified which Snail proteins are associated with FOXD4 in CRC cells by bioinformatic analysis. According to the TCGA colorectal data, we found that the expression of FOXD4 was correlated with SNAI3, but not SNAI1 or SNAI2 (. A-D). In order to further confirm this result, the protein expression levels of CDH1, and SNAI3 were determined using western blot analysis in FOXD4-knockdown or control CRC cells (. E-F). Western blot showed FOXD4 knockdown decreased the level of SNAI3, while the CDH1 level was increased. Meanwhile, SW480 cells were transfected with pcDNA3.1-FOXD4. The results demonstrated that FOXD4 could promote the expression of SNAI3 and inhibit the expression of CDH1 (. G-H). Additionally, these findings indicated that FOXD4 significantly increased SNAI3 expression, thus contributing to CRC metastasis.
Figure 3. FOXD4 regulated EMT through SNAI3/CDH1. (A-D) The co-expression of FOXD4 and EMT markers was checked on TCGA database for their colorectal cancer cohort. (E-F) The expression of SNAI3 was examined via quantitative real-time PCR in LoVo cells transfected with siFOXD4 or control siRNA. (G-H) The expression of SNAI3 was examined via quantitative real-time -PCR in SW480 cells transfected with pcDNA3.1- FOXD4 or the control vector. (*p < 0.05, **p < 0.01, ***p < 0.001).
FOXD4 bounded to the promoter of SNAI3 and regulated SNAI3/CDH1 expression
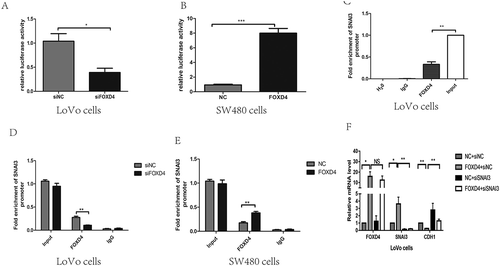
In order to analyze the interaction of FOXD4 with SNAI3, a promoter vector, pGL-SNAI3, was constructed and co-transfected into LoVo or SW480 cells with either siFOXD4 or pcDNA3.1-FOXD4. The results showed that when FOXD4 was silenced, SNAI3 was transcriptionally repressed. In contrast, up-regulation of FOXD4 increased the transcription ( and B). To verify binding between FOXD4 and the SNAI3 promoter, ChIP analysis was performed to detect enrichment at the SNAI3 promoter in LoVo cells (). FOXD4 knockdown decreased its occupancy at the SNAI3 promoter, as revealed by anti-FOXD4 antibody; while up-regulation of FOXD4 caused the reverse phenomenon in SW480 cells (-E). To further validate the role of FOXD4 through its regulation of CDH1, we co-transfected SW480 cells with pcDNA3.1-FOXD4 and siSNAI3. The data again showed that the CDH1 mRNA level was restored ().
Figure 4. FOXD4 bounded to the promoter of SNAI3 and regulated SNAI3/Ecad expression. (A-B) Luciferase activity was measured after co-transfecting siFOXD4 or pcDNA3.1- FOXD4, and luciferase reporters containing the promoter region of SNAI3, into LoVo and SW480 cells. (C-E) The enrichment of FOXD4 on the promoter of SNAI3 was detected by ChIP assay of LoVo and SW480 cells transfected with siFOXD4 or pcDNA3.1- FOXD4. (F)The expression of CDH1 was measured by quantitative real-time PCR of LoVo cells co-transfected with pcDNA3.1- FOXD4 and siSNAI3. (*p < 0.05, **p < 0.01, ***p < 0.001).
Knockdown of FOXD4 suppressed cancer progression in vivo
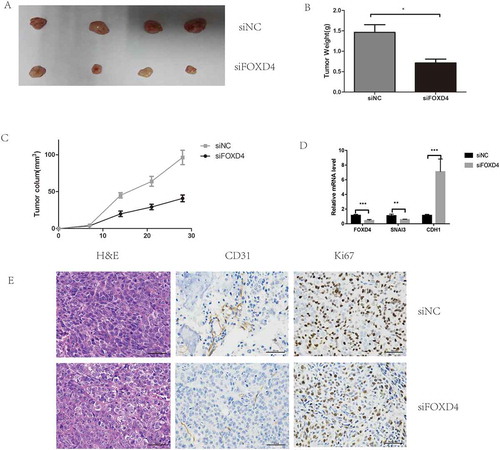
Finally, xenograft assays were used to test the effects of FOXD4 on tumor progression in vivo. The results showed that tumor growth in the short-hairpin-FOXD4 (sh-FOXD4) group was slower than that in the control group (), and the weight and size of the tumors in the sh-FOXD4 group were obviously smaller than those derived from the control group (-C). In addition, the expression of FOXD4 was lower in the shRNA group than in the control group ().
Figure 5. Silencing of FOXD4 inhibited colorectal cancer progression in vivo. (A) Photographs of tumors were obtained from nude mice. (B-C) Tumor weight and volume were examined after injection of colorectal cancer cells stably transfected with sh- FOXD4 or control. (D) Quantitative real-time PCR was used to measure the expression of FOXD4,SNAI3 and CDH1 in the tumors. (E) Immunohistochemical was used to measure the expression of CD31 andKi67.
(*p < 0.05, **p < 0.01, ***p < 0.001)
Discussion
FOXD4 is a TF of the FOX family; it has been reported that this family of Winged helix factors has been highly conserved during evolutio.Citation19 From their crystal structure, it has been shown that there is a DNA binding domain consisting of 101 amino acids common to the Winged helix factor.Citation20 Increasing evidence has demonstrated that the Winged helix factors not only participate in embryonic development and gene expression but also contribute to oncogenesi.Citation11 It is reported that FOXD4 is expressed in leukemia cell lines, and may be involved in leukemia progressio.Citation19 However, the expression of FOXD4 in CRC tumors has not been clarified. In the present study, we examined the expression of FOXD4 in CRC tissues and analyzed the relationship between clinical features and FOXD4 expression in CRC. In this study, we demonstrated that both FOXD4 mRNA and protein levels were significantly higher in CRC than in the normal tissues. We validated this conclusion through a TCGA colorectal cancer cohort and a GEO cohort. Furthermore, IHC results showed that most CRC tissue samples exhibit a positive FOXD4 expression. The above findings clarified that the up-regulation of FOXD4 protein contributed to the progression of CRC.
The FOX family has been reported to participate in several biological processes, including oncogenesis, cell proliferation, and gene expression.Citation21–Citation24 It seems that FOXD4 could potentially be used to predict the prognosis of CRC patients. Interestingly, we noted that patients with high levels of FOXD4 had a poor prognosis. These findings revealed that FOXD4 may play an important role in the oncogenesis of CRC, and could predict poor prognosis of CRC patients. Further experiments in vitro and in vivo are required to explore the biological role of FOXD4 during the cell migration and invasion processes in CRC. Additionally, we investigated FOXD4 function by up-regulation and silencing of FOXD4 in CRC cells. The results showed that overexpression of FOXD4 can promote cell migration and invasion in SW480 cells. In contrast, silencing of FOXD4 suppressed those abilities in LoVo cells.
To further clarify the target genes that FOXD4 regulates during CRC invasion and metastasis, co-expression of other genes was checked in a TCGA colorectal cancer cohort. It has been reported that EMT is correlated with tumor metastasi.Citation25 Therefore, we focused on the EMT-related genes, such as E-cadherin, vimentin, Snail1, Snail2, and Snail.Citation26 The results showed that the expression of FOXD4 was correlated with expression of E-cadherin and SNAI3, but not SNAI1 or SNAI2. In addition, western blot assay and qPCR assay showed that FOXD4 knockdown decreased the level of SNAI3 while increasing the E-cadherin expression. Meanwhile, the cell migration and invasion abilities were restored on co-transfection with siFOXD4 and SNAI3-overexpression vector, compared with cells only transfected with siFOXD4. Additionally, these findings indicated that FOXD4 significantly increased CRC metastasis via SNAI3. Furthermore, ChIP and luciferase assays demonstrated that FOXD4 could directly bind the promoter of SNAI3, up-regulating the transcription of SNAI3. Together, our data suggest that the up-regulation of FOXD4 is associated with a mechanism driving CRC progression. Finally, xenograft assays showed that silencing of FOXD4 suppressed cancer progression in vivo.
In conclusion, our research identified that FOXD4 was overexpressed in CRC, and that FOXD4 overexpression was related to CRC progression and poor outcomes for CRC patients. The present study contributes to the identification of novel biomarkers of CRC and the understanding of the mechanism of CRC metastasis.
Conflicts of interest
The authors declare that they have no competing interests.
Supplemental Material
Download MS Word (120.9 KB)Supplemental data
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Ca Cancer J Clin. 2016;66(1):7. doi:10.3322/caac.21332.
- Mellas N, Benbrahim Z, Mesbahi OE. Colorectal cancer: new developments after the 2013 ECCO/ESMO congress. Chin J Cancer. 2014;33(4):218.
- Kopetz S, Chang GJ, Overman MJ, Eng C, Sargent DJ, Larson DW, Grothey A, Vauthey JN, Nagorney DM, Mcwilliams RR. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. 2009;27(22):3677. doi:10.1200/JCO.2008.20.5278.
- Manisha V, Mona S, Durado B, Wender RC. Population-based programs for increasing colorectal cancer screening in the United States. Ca Cancer J Clin. 2015;65(6):496–510. doi:10.3322/caac.21295.
- Chen W, Zheng R, Zeng H, Zhang S. The incidence and mortality of major cancers in China, 2012. Chin J Cancer. 2016;35(8):73. doi:10.1186/s40880-016-0137-8.
- Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. Ca Cancer J Clin. 2016;66(4):271. doi:10.3322/caac.21349.
- Mellas N, Benbrahim Z, El MO. Colorectal cancer: new developments after the 2013 ECCO/ESMO congress. Chin J Cancer. 2014;33(4):218–221. doi:10.5732/cjc.013.10203.
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502.
- Minoretti P, Arra M, Emanuele E, Olivieri V, Aldeghi A, Politi P, Martinelli V, Pesenti S, Falcone C. A W148R mutation in the human FOXD4 gene segregating with dilated cardiomyopathy, obsessive-compulsive disorder, and suicidality. Int J Mol Med. 2007;19(3):369–372(364).
- Myatt SS, Lam EWF. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859. doi:10.1038/nrc2223.
- Freyaldenhoven BS, Freyaldenhoven MP, Iacovoni JS, Vogt PK. Aberrant cell growth induced by avian winged helix proteins. Cancer Res. 1997;57(1):123–129.
- Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796(2):75.
- Huber MA, Azoitei N, Baumann B, Grünert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Investig. 2004;114(4):569. doi:10.1172/JCI200421358.
- Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Investig. 2003;112(12):1776. doi:10.1172/JCI200320530.
- Bates RC, Mercurio A. The epithelial-mesenchymal tansition (EMT) and colorectal cancer progression. Cancer Biol Ther. 2005;4(4):365–370. doi:10.4161/cbt.4.4.1655.
- Neilson KM, Klein SL, Mhaske P, Mood K, Daar IO, Moody SA. Specific domains of FoxD4/5 activate and repress neural transcription factor genes to control the progression of immature neural ectoderm to differentiating neural plate. Dev Biol. 2012;365(2):363–375. doi:10.1016/j.ydbio.2012.03.004.
- Du PJ, Newman M, Williams D. GATA3 immunohistochemical stain is a highly sensitive tool for distinguishing between upper urothelial tract urothelial carcinoma and lung squamous cell carcinoma and adenocarcinoma. Pathology. 2016;48:S67–S67. doi:10.1016/j.pathol.2015.12.168.
- Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15(3):195. doi:10.1016/j.ccr.2009.01.023.
- Freyaldenhoven BS, Fried C, Wielckens K. FOXD4a and FOXD4b, two new winged helix transcription factors, are expressed in human leukemia cell lines. Gene. 2002;294(1–2):131. doi:10.1016/S0378-1119(02)00702-3.
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364(6436):412–420. doi:10.1038/364412a0.
- Lin L, Miller CT, Contreras JI, Prescott MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM. The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 2002;62(18):5273.
- Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62(16):4773–4780.
- Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 2001;61(24):8820–8829.
- Perreault N, Dutton-Sackett S, Lee CS, Mcnally S, Kaestner KH. The winged helix transcription factor foxa2 is required for the control of gastrointestinal epithelial cell proliferation. Gastroenterology. 2003;124(4):A277–A277. doi:10.1016/S0016-5085(03)81388-4.
- Philip R, Heiler S, Mu W, Büchler MW, Zöller M, Thuma F. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget. 2015;6(4):2046–2063. doi:10.18632/oncotarget.2858.
- Wang SC, Lin XL, Wang HY, Qin YJ, Chen L, Li J, Jia JS, Shen HF, Yang S, Ry X. Hes1 triggers epithelial-mesenchymal transition (EMT)-like cellular marker alterations and promotes invasion and metastasis of nasopharyngeal carcinoma by activating the PTEN/AKT pathway. Oncotarget. 2015;6(34):36713–36730. doi:10.18632/oncotarget.5457.
