ABSTRACT
Targeting FGFRs is one of the most promising therapeutic strategies in squamous non-small cell lung cancer (SQCC). However, different FGFR genomic aberrations can be associated with distinct biological characteristics that result in different clinical outcomes or therapeutic consequences. Currently, the full spectrum of FGFR gene aberrations and their clinical significance in SQCC have not been comprehensively studied. Here, we used Next-generation sequencing to investigate the presence of FGFR gene mutations in 143 tumors from patients with stage I, II or III SQCC and who had not been treated with chemotherapy or radiotherapy prior to surgery. FGFR gene mutations were identified in 24 cases, resulting in an overall frequency of 16.9%. Among the mutations, 7% (10/143) were somatic mutations, and 9.8% (14/143) germline mutations. FGFR mutations were significantly associated with an increased risk of lymph node metastasis. SQCC patients with a FGFR somatic mutation had shorter OS (overall survival, log rank P = 0.005) and DFS (disease-free survival,log rank P = 0.004) compared with those without an FGFR mutation. The multivariate analysis confirmed that a somatic mutation was an independent poor prognostic factor for OS (HR: 4.26, 95% CI: 1.49–12.16, P = 0.007) and DFS (HR: 3.16, 95% CI: 1.20–8.35, P = 0.020). Our data indicate that FGFR genes mutation is an independent prognostic factor and associated with lymph node metastasis in stage I to III Chinese SQCC patients.
Introduction
Lung cancer is the leading cause of cancer-related death worldwide.Citation1 Although squamous-cell carcinoma (SQCC) and adenocarcinoma constitute the vast majority of diagnosed cases of lung cancer,Citation2 they exhibit different somatic genomic profiles.Citation3,Citation4 Activated oncogenes, such as mutant alleles of EGFR, KRAS, and BRAF, or translocations involving ALK, RET, and ROS1 are commonly observed in lung adenocarcinoma cancer (Ad-NSCLC), whereas these mutations are rarely detected in SQCC.Citation4,Citation5 Therapies targeted against specific molecular and genomic aberrations have resulted in improved survival of some patients.Citation6–Citation8 However, these therapies are unlikely to be of benefit in patients with SQCC, since the most frequent aberrations among SQCC case known to date are TP53, PTEN, NFE2L2, and KEAP1 mutations and changes in SOX2 and CDKN2A copy number, all of which are not clinically actionable. Among the genes associated with SQCC genetic abnormalities, fibroblast growth factor receptor (FGFR) appears to be one of the most promising therapeutic targets.Citation5,Citation9–Citation11
The FGFR tyrosine kinases are encoded by 4 genes (FGFR1, FGFR2, FGFR3, and FGFR4) and composed of an extracellular component with 3 immunoglobulin-like domains, a transmembrane domain and an intracellular tyrosine kinase domain. FGFR activation triggers a cascade that leads to the activation of multiple signal transduction pathways, including the four primary downstream pathways Ras/Raf/MapK, PI3K/Akt, Stats and PLCγ, which can promote cell survival, motility and invasiveness, cell proliferation, epithelial-to-mesenchymal transition (EMT) and angiogenesis.Citation12 FGFR genes amplifications, somatic missense mutations and chromosomal translocations can promote aberrant receptor tyrosine kinase activation.Citation13–Citation17 Germline mutations in the FGFR genes family have been described in craniofacial and skeletal syndromes,Citation18 and subsequent studies have reported that some known FGFR SNPs are also observed in tumor samples as somatic mutation which might contribute to tumorigenesis.Citation16,Citation19,Citation20
Previous studies have demonstrated that FGFR genes aberrations are associated with transformative potential and mediate inhibitor sensitivity in SQCC.Citation15,Citation16,Citation21,Citation22 Preliminary data from early-phase trials evaluating FGFR inhibitors have provided proof-of-concept support for the effective inhibition of FGFR signaling by TKIs in patients with lung SQCC.Citation23 However, the underlying FGFR mutations in SQCC patients are not well known. In order to help identify patients who may benefit from therapies targeted against FGFR activation, the full spectrum of somatic FGFR mutations and their clinical significances remains to be characterized. In this study, we characterized FGFR genes mutations completely in SQCC from Chinese patients and investigated the correlation between various types of FGFR aberrations and clinical outcomes.
Materials and methods
Patients and samples
Patients who had been diagnosed with SQCC and treated in Peking University Cancer Hospital (Beijing, China) between 2003 and 2013 were recruited for this study. All of the recruited patients were of Chinese ethnicity, and the clinic stage of the tumors diagnosed were stage I to III, with no patients with stage IV disease included in this study. All of the patients received anatomical resection, including lobectomy, sleeve lobectomy and pneumonectomy,except for one who received segmentectomy. All patients received SMLD (systematic mediastinal lymph node dissection). Surgeons systematically resected the mediastinal lymph nodes. Stations resected included no. 2, 4, 7, and 9 on the right side, and no. 4, 5, 6, 7, and 9 on the left side. Tumor samples were obtained by surgical resection, and patients treated with chemotherapy or radiotherapy prior to surgery were excluded from the study. Among the recruited patients, 51 of them received adjuvant treatment. 46 patients received adjuvant chemotherapy only, 1 patient received adjuvant radiotherapy, and 4 patients received both chemotherapy and radiotherapy. All of the patients with stage IIA-IIIB were suggested to accept adjuvant chemotherapy, while patients with N2 lesion were advised to receive adjuvant radiotherapy. Some patients with stage IB were also suggested to receive chemotherapy due to the big size of resected tumor or other risk factors of recurrence. However, a portion of patients could not afford the side effect, or they refused adjuvant treatment from their own will. Written informed consent from each patient was obtained prior to sample collection. This study was approved by the medical ethics committee of Peking University Cancer Hospital (#2013KT31).
Genomic DNA was isolated using a DNeasy Blood and Tissue Kit (QIAGEN). DNA quantification and purity estimation were performed by NanoDrop ND 1000 spectrophotometer (Thermofisher).Baseline demographic and clinical characteristics, including smoking, age, gender, family history, pathological subtype, tumor node metastasis (TNM) classification, tumor stage, lymph node stage, differentiation status, adjuvant therapy after surgery and duration of survival after surgery, were obtained from medical records. Tumors were staged according to the UICC/AJCC (7th edition) for the lung.Citation24
Mutation analysis
An oncogene panel targeting 108 SCC-related genes as determined by a custom SeqCap EZ system (Roche NimbleGen, Madison, WI, USA) was designed in our previous study (data not published). The full FGFR (1–4) coding sequences (CDS) were included in this panel (supplementary Table S1) and the FGFR sequence data were used in this study. Approximately 1microgram of quantified genomic DNA from each SCC sample was used to prepare the DNA library, and the indexed samples were pooled for sequencing on a Hiseq2500 machine (Illumina, San Diego, CA, USA) with 2 × 150 bp pair-end reads. BWA, SAMtools and GATK methods were conducted to detect the SNVs. Based on data derived from NCBI dbSNP build 134, major germline SNVs were excluded. In addition, rare germline SNVs were excluded using the VN filtering method,Citation25 which is available as a tool from the Galaxy platform (http://galaxy-demo.ctmm-trait.nl/u/saskia-hiltemann/p/virtual-normal-analysis).
SQCC samples with FGFR genes mutations and the corresponding adjacent tissues samples were selected for further validation. PCR primers (primers are listed in Table S2) providing coverage of the mutant loci were designed using primer3 online software.Citation26 PCR fragments were amplified using standard procedures with the Q5® High-Fidelity DNA Polymerase (NEB, Ipswich, England), and the PCR products were sequenced using the ABI 3730 Genetic Analyzer (Life Technologies, Carlsbad, CA).
Each FGFR missense mutation was computationally analyzed for a predicted effect on protein function. Sorting Intolerant From Tolerant was used to calculate a SIFT probability score for the likelihood of the mutation to affect protein function.Citation27,Citation28 Scores of 0.05 or less were predicted to affect protein function. Polymorphism phenotyping was also used, predicting either unknown (insufficient data for a prediction), benign, possibly damaging, or probably damaging mutations based upon characterization of the substitution site.Citation29
Statistical analysis
Spearman correlation coefficients and the corresponding P values were used to evaluate the association between two continuous variables. Associations between qualitative variables were tested using the chi-square test or Fisher’s exact test, contingent on distributional assumptions. Continuous variables were analyzed using the non-parametric Wilcoxon’s rank sum test. Univariate and multivariate regress analysis were employed to evaluate the risk ratio of FGFR mutation to clinicopathological features. Age (categorical, >60 years), sex, tumor differentiation, family history of malignant tumor and smoking history were used in the multivariate model. When evaluate the FGFR germline mutation, FGFR somatic mutation cases were censored in the control group; the same way was used to evaluate the FGFR somatic mutation. Overall survival (OS; date of operation until the date of death from any cause or last date of follow-up) and disease-free survival (DFS; date of operation until date of any sign of tumor relapse) were the primary endpoints. Patients without the event were censored at the date of the last follow-up. Survival curves were plotted using the Kaplan-Meier method, and significance was determined using the log-rank test. Multivariate analysis was performed using the Cox proportional hazard regression model with stepwise model selection. The hazard ratio (HR) and 95% confidence interval (CI) were calculated for each factor evaluated. All P values were 2-sided with a significance level of 0.05 Statistical calculations were performed using Stata 12.1 (StataCorp LP).
Results
Spectrum of FGFR mutations identified in 143 SCC patients without preoperative therapy
Of the 143 tested samples, we observed 22 different FGFR mutations among 24 tumors, amounting to an overall FGFR mutation rate of 16.8% (24/143). Twenty-one of the identified aberrations were missense mutations, and only one of them was a cds-indel (FGFR3- E135del). FGFR3-A500T, FGFR2-W290C and FGFR3-S249C were observed in the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/), whereas the other mutations appeared to be novel. FGFR2-W290C and FGFR3-S249C were also found in TCGA SQCC samples.Citation3
Eight distinct somatic mutations were identified by confirming the absence of the mutation in the paired adjacent lung cancer tissues. The remaining were also present in the apparently normal adjacent tissue so we defined them as potential germline mutations (hereafter referred to as germline mutation). The frequency of somatic mutations was 7% (10/143), and the frequency of FGFR germline mutation was 9.8% (14/143). Among the different FGFR genes, FGFR1 was associated with 3 somatic mutations and 3 germline mutations, FGFR2 was associated with 2 somatic mutations and 2 germline mutations, FGFR3 was associated with 2 somatic mutations and 6 germline mutations, and FGFR4 was associated with 1 somatic mutation and 3 germline mutations. Most mutations were unique, with the exception of FGFR3-S249C and FGFR3-A500T, which were observed in two cases each (). Although FGFR3-S249C is predicted to be damaging by Polyphen and SIFT,Citation27,Citation29 it was also present in Exac and in dbSNP, as was FGFR3 which encodes A500T predicted to be tolerated by Polyphen and SIFT. Therefore we cannot exclude that they may represent bystander mutations.
Table 1. FGFR mutations identified in 143 patients with squamous lung cell cancer.
According to the protein topology characterization, the observed mutations resided in both the extracellular and cytoplasmic domains. Six germline mutations and one somatic mutation were located in the kinase domain (). The somatic FGFR mutations tended to occur in an alternative exon or in an Ig-like domain (), but no significant difference was observed in the distribution between somatic mutations and germline mutations (P = 0.193).
Figure 1. Alignment of altered amino acids encoded by the mutations observed in FGFRs.
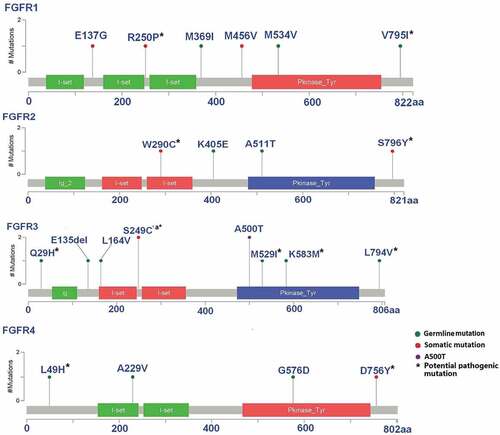
All of the listed FGFR aberrations were associated with a mutant allele frequency (MAF) greater than 0.22 (), which indicating that the Sanger sequencing method is sufficiently sensitive to detect FGFR mutant alleles for validation.
FGFRs mutation is associated with lymph node metastasis
To further evaluate the impact of the various types of FGFR mutations on the clinicopathological features of patients with SQCC, we analyzed FGFR somatic mutations, germline mutations and the overall FGFR mutation status separately.
Non-smokers were more likely to harbor a somatic FGFR mutation (P = 0.037) compared with smokers, whereas the frequency of FGFR germline mutation in smokers was not significantly greater compared with non-smokers (P = 0.425). TNM stage was correlated with the presence of FGFR overall mutations (P = 0.037, Fisher’s exact test). Patients at more advanced TNM stages exhibited a higher frequency of somatic or germline FGFR mutations. When FGFR somatic and germline mutation were considered separately, the correlation with TNM stage was no longer significant ().
Table 2. Clinicopathological features of lung squamous cell carcinoma patients harboring FGFR mutations.
Univariate logistic regression analysis revealed that the presence of any FGFR mutation in SQCC was associated with an increased risk of lymph node metastasis [odds ratio (OR) = 5.00, 95% confidence interval (CI) = 1.98–12.64, P = 0.001). When analyzing the data separately, a similar association was observed between FGFR germline mutation and lymph node metastasis (OR = 4.85, 95% CI = 1.52–15.55, P = 0.008). Association between FGFR somatic mutations and lymph node metastasis was also significant (OR = 4.05, 95% CI = 1.07–15.25, P = 0.039) (). After adjusting for age (categorical, >60 years), sex, tumor differentiation, family history of malignant tumor and smoking history, FGFR overall mutation (OR = 4.75, 95% CI = 1.78–12.7, P = 0.002) and germline mutation (OR = 5.56, 95% CI = 1.63–18.95, P = 0.006) remained positively associated with lymph node metastasis. TNM stage was not included in the multivariate unconditional logistic analysis because it was strongly correlated with lymph node metastasis (Spearman’s rho = 0.812, P < 0.001).
Survival outcome according to FGFR somatic mutation status
OS and DFS data were available for 140 patients (three patients were lost to follow-up). The log-rank test of the Kaplan-Meier survival analysis demonstrated that SQCC patients without an FGFR somatic mutation experienced longer OS (log rank P = 0.005) and DFS (log rank P = 0.004) than those with the FGFR somatic mutation (). Patients with any type of FGFR mutation also experienced significantly shorter DFS (log rank P = 0.036) and OS (log rank P = 0.070). In contrast, no association was observed between FGFR germline mutation and patient survival.
Figure 2. FGFR genes mutation status and overall (OS) and disease-free survival (DFS) in SCC patients.
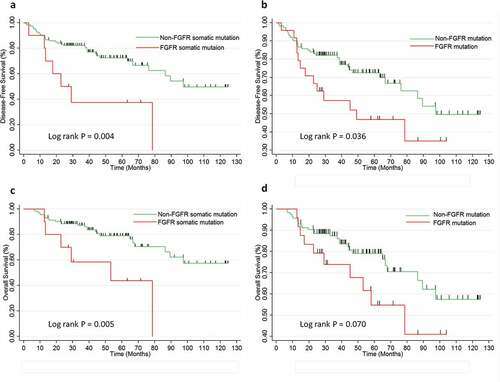
To eliminate potential confounding effects, a multivariate Cox regression model was applied to adjust for confounding factors, such as sex, age, family history of malignant tumor, smoking history, tumor differentiation, TNM stage and postoperative chemotherapy/radiotherapy. The multivariate analysis confirmed that the presence of an FGFR somatic mutation was an independent poor prognostic factor for OS (HR: 4.26, 95% CI: 1.49–12.16, P = 0.007). Patients with FGFR somatic mutations also tended to have shorter DFS (HR: 3.16, 95% CI: 1.20–8.35, P = 0.020). In addition, higher TNM stages were also associated with poorer survival in DFS and OS, as expected (Table S3).
Discussion
Accumulating in vivo evidence has shown that tumors with FGFR aberrations frequently respond to FGFR inhibitors,Citation15,Citation21,Citation30–Citation34 and these findings provide a rationale for conducting clinical trials with these agents. Many FGFR inhibitors have been developed, such as JNJ-42756493 (Johnson & Johnson), AZD4547 (AstraZeneca) and BGJ398 (Novartis). In addition, some have been approved by the U.S. Food and Drug Administration (FDA), for example lenvatinib, which is the most recently FDA-approved FGFR-inhibiting drug for iodine-refractory, well-differentiated thyroid carcinoma. However, genomic FGFR aberrations are diverse, and patients with different FGFR aberrations may respond differently to different therapies. Therefore, it is important to both identify the spectrum and determine the clinical implications of different FGFR aberrations.
The functional effects of some known FGFR somatic mutations have been identified in vivo, the most frequent mutations are FGFR2-W290C and FGRF3-S249C,Citation35,Citation36 which are implicated in driving cell transformation and mediating sensitivity to FGFR inhibitors in lung cancer cell line and mouse model.Citation16 Two FGFR2 extracellular domain insertion mutations, FGFR2-A266_S267ins and FGFR2-290_291WI>C were proved to be both oncogenic and sensitive to inhibition by FGFR kinase inhibitors in NSCLS recently.Citation37 In other cancer such as endometrial carcinoma and human rhabdomyosarcomas, multiple FGFR2 mutations exhibit oncogenic potential and suggested to be novel therapeutic target.Citation14,Citation34 FGFR2-S290C is in the immunoglobulin-like domain in the extracellular domain (ECD) that mediates the activation of the FGFR2 kinase by forming covalently bound receptor dimers.Citation16,Citation37 The FGFR3-S249C mutation resides in the receptor’s extracellular linker region between the two Ig-like domains, a key site for ligand binding. In our dataset, among the 8 identified FGFR somatic mutations, 4 represented ECD mutations () and we hypothesise that these mutations might exert effects similar to FGFR2-W290C or FGFR3-S249C in SCC. The remaining somatic mutations identified were located either in the TK domain or in an alternative exon of FGFR ().
It is intriguing that FGFR2-W290C and FGRF3-S249CCitation31 have also been detected in the germline causing the congenital developmental disorder Apert Syndrome and the related Pfeiffer and Crouzon syndromes.Citation18,Citation38 Furthermore, functional germline mutation such as FGFR4-G388R has been described in lung cancer and predicted poor survival in lung adenocarcinoma patientsCitation39,Citation40 and recent genome-wide association studies have suggested that FGFR genes polymorphisms are associated with an increased risk of developing breast cancer.Citation41,Citation42 The germline mutations that we identified were mostly localized to the tyrosine kinase and extracellular ligand binding domains, indicating likely pathogenicity as has been observed for other germline mutation of proto-oncogenes, for example RET mutations causing multiple endocrine neoplasia type 2.Citation43
To date, few studies have comprehensively evaluated the relationships of FGFR mutations with patient features and clinical outcomes in SQCC. Since different FGFR members have homogeneous protein structures and exerting similar biological functions in cellular signal transduction, we considered it reasonable to define them together as a whole. For example, both FGFR2-W290C and FGRF3-S249C mutants are sensitivity to FGFR TKIs through the same mechanism. In the meanwhile, the incidence of each mutation of FGFR genes is so low that it may not be helpful or meaningful to analyze them separately. We found that non-smokers were more likely to have somatic mutations than smokers. To our knowledge, this is the first study investigating a possible correlation between FGFR mutation status and smoking in lung cancer although such a correlation has been previously demonstrated for other tyrosine kinase receptors, such as EGFR, in lung adenocarcinoma.Citation44 However, the mechanisms by which smoking status contribute to tyrosine kinase receptors mutation is still largely unknown. Tumors with more advanced TNM stages or with lymph node metastasis were found to exhibit a higher frequency of either FGFR somatic or germline mutation. TNM stage is determined by tumor size and the presence of lymph node and distal metastases, and the strong correlation between TNM and lymph node metastasis was proven in our dataset (Spearman’s rho = 0.812, P < 0.001). Therefore, we presumed that an actual correlation exists between lymph node metastasis and the presence of an FGFR mutation, and the association with TNM stage might simply be a secondary consequence of the association with lymph node metastasis. FGFR activating mutations promote rhabdomyosarcomas metastasis in xenotransplant models,Citation14 and our data suggests that both germline and somatic FGFR mutation may contribute to lymph node metastasis in SQCC. The association between somatic mutation and lymph node metastasis was not significant statistically in multivariate analysis, which may be due to inadequate statistical power resulting from the small number of FGFR somatic mutation cases. Therefore, we hypothesize that there are functional somatic and germline mutations among these FGFR mutations we found in this study, and the impact of each FGFR mutation on tumor metastasis merits further investigation.
The prognostic role of FGFR mutations in lung cancer was recently investigated. Wang et al. evaluated FGFR fusions and mutations, and no correlation between FGFR aberrations and survival in SQCC was observed.Citation36 However, this previous study had two significant limitations. First, the full coding regions of FGFR2 and FGFR3 were not evaluated and secondly, FGFR mutations and the FGFR fusions were combined for the survival analysis rather than evaluated individually. Evaluating every FGFR aberration independently is not feasible because the number of cases associated with each is limited, however; stratifying FGFR abnormalities by type is reasonable for association studies. We evaluated the prognostic roles of FGFR somatic mutations, germline mutation and the overall set of FGFR mutations in SQCC separately and found that patients harboring an FGFR somatic mutation experienced significantly worse DFS and OS. We also observed that more advanced TNM stage was a predictor of worse survival, as expected. Poorer rates of OS and DFS in SQCC patients with FGFR somatic mutations is consistent with our finding that tumors with somatic FGFR mutations had a greater frequency of lymph node metastasis, which is regarded to be associated with a poor prognosis. However, no association between FGFR germline mutation and survival was observed, suggesting that somatic mutations may have a greater impact on survival compared with germline mutations.
FGFR inhibitors have been developed as potential new therapeutic options in SCC with preclinical and early clinical data suggest that targeting certain FGFR aberrations with cognate inhibitors exerts beneficial therapeutic effects.Citation45 Therefore characterizing the landscape of FGFR genes aberrations and determining which mutations are clinically relevant is important to identify patients most likely to benefit from FGFR-targeted therapies and optimizing treatment outcomes.
The main limitation of our study resides in the relatively small sample size and patient selection. The patients in this study were from a Chinese population and the study did not include those patients with stage IV SQCC. Therefore, more studies will be warranted to validate our findings in an independent cohort, particularly in a Western population. Furthermore, the presumed germline mutations identified in this study were based on the finding of the FGFR mutations detected to be also present in the surrounding normal lung tissue (no paired blood samples were available for analysis) and so we were unable to exclude the possibility that they may represent somatic mutation. Although we verified the FGFR mutations identified against publically available databases we cannot exclude that some of the variants identified may represent previously unrecognized polymorphisms.
Our observed association between FGFR mutation and clinical outcome in SQCC, along with preclinical and early clnical data already reported suggest that targeting FGFR mutation with cognate inhibitors has therapeutic potential. There is also evidence that there are FGFR alterations that confer resistance to other types of cancer treatment and that some specific FGFR aberrations may demonstrate differential sensitivity/resistance to distinct FGFR inhibitors. FGFR mutation also have prognostic value. Because many FGFR changes appear to activate signaling, it is also important to characterize the clinically relevant effects of the many potent FGFR inhibitors that are currently in clinical trials. Molecular interrogation of patients for FGFR muations in the clinical research and practice setting may be warranted.
In this comprehensive analysis of FGFR genes mutations in SQCC, we demonstrated that FGFR mutation may increase the risk of lymph node metastasis in SQCC, and the presence of an FGFR somatic mutation specifically is a predictive independent factor for poor prognosis. Thus the findings from this study will help identify patients with SQCC most likely to benefit from FGFR-targeted therapies and so facilitate future clinical studies.
Abbreviations
Supplemental Material
Download ()Acknowledgments
The authors acknowledge the technical contributions of Jia Jian (Shandong Mental Hospital) in DNA preparation. It thanked very specically to the Professors Hong Cai and Yang Ke who contributed to the study conecept and design.
Disclosure Statement
The authors have no conflict of interest.
Supplementary data
Supplemental data for this article can be accessed on the publisher's website.
Additional information
Funding
References
- Global Burden of Disease Cancer C, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et al. The Global Burden of Cancer 2013. JAMA oncology. 2015;1:505–527. doi:10.1001/jamaoncol.2015.0735.
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. New Eng J Med. 2008;359:1367–1380. doi:10.1056/NEJMra0802714.
- Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi:10.1038/nature11404.
- Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi:10.1038/nature13385.
- Moreira AL, Eng J. Personalized therapy for lung cancer. Chest. 2014;146:1649–1657. doi:10.1378/chest.14-0713.
- Cataldo VD, Gibbons DL, Pérez-Soler R, Quintás-Cardama A. Treatment of non–small-cell lung cancer with Erlotinib or Gefitinib. New Eng J Med. 2011;364:947–955. doi:10.1056/NEJMct0807960.
- Sequist LV, Soria JC, Goldman JW, Wakelee HA, Gadgeel SM, Varga A, Papadimitrakopoulou V, Solomon BJ, Oxnard GR, Dziadziuszko R, et al. Rociletinib in EGFR-mutated non–small-cell lung cancer. N Engl J Med. 2015;372:1700–1709. doi:10.1056/NEJMoa1413654.
- Solomon BJ, Mok T, Kim D-W, Wu Y-L, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. New Eng J Med. 2014;371:2167–2177. doi:10.1056/NEJMoa1408440.
- Drilon A, Rekhtman N, Ladanyi M, Paik P. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol. 2012;13:e418–26. doi:10.1016/S1470-2045(12)70291-7.
- Greulich H, Pollock PM. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol Med. 2011;17:283–292. doi:10.1016/j.molmed.2011.01.012.
- Filipits M. New developments in the treatment of squamous cell lung cancer. Curr Opin Oncol. 2014;26:152–158. doi:10.1097/CCO.0000000000000049.
- Semrad TJ, Mack PC. Fibroblast growth factor signaling in non–small-cell lung cancer. Clin Lung Cancer. 2012;13:90–95. doi:10.1016/j.cllc.2011.08.001.
- Gartside MG, Chen H, Ibrahimi OA, Byron SA, Curtis AV, Wellens CL, Bengston A, Yudt LM, Eliseenkova AV, Ma J, et al. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol Cancer Res. 2009;7:41–54. doi:10.1158/1541-7786.MCR-08-0021.
- Taylor J, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, Yu Y, Chen Q-R, Shah K, Youngblood V, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–3407. doi:10.1172/JCI39703.
- Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Science translational medicine. 2010;2:62ra93. doi:10.1126/scitranslmed.3001451.
- Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, Liu Q, Pugh TJ, Pedamallu CS, Hayes DN, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–5205. doi:10.1158/0008-5472.CAN-12-3950.
- Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P, Marie Y, Bruno A, Boisselier B, Giry M, et al. Detection, characterization and inhibition of FGFR-TACC fusions in IDH wild type glioma. Clin Cancer Res. 2015;21:3307–3317. doi:10.1158/1078-0432.CCR-14-2199.
- Shotelersuk V, Ittiwut C, Srivuthana S, Mahatumarat C, Lerdlum S, Wacharasindhu S. Distinct craniofacial-skeletal-dermatological dysplasia in a patient with W290C mutation in FGFR2. Am J Med Genet. 2002;113:4–8. doi:10.1002/ajmg.10449.
- Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi:10.1158/0008-5472.CAN-05-1855.
- Dodurga Y, Tataroglu C, Kesen Z, Satiroglu-Tufan NL. Incidence of fibroblast growth factor receptor 3 gene (FGFR3) A248C, S249C, G372C, and T375C mutations in bladder cancer. Genet Mol Res. 2011;10:86–95. doi:10.4238/vol10-1gmr923.
- Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi:10.1371/journal.pone.0020351.
- Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, Ware KE, Edwards MG, Böhm D, Perner S, Helfrich BA, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20:3299–3309. doi:10.1158/1078-0432.CCR-13-3060.
- Tiseo M, Gelsomino F, Alfieri R, Cavazzoni A, Bozzetti C, De Giorgi AM, Petronini PG, Ardizzoni A. FGFR as potential target in the treatment of squamous non small cell lung cancer. Cancer Treat Rev. 2015;41:527–539. doi:10.1016/j.ctrv.2015.04.011.
- Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi:10.1097/JTO.0b013e31812f3c1a.
- Hiltemann S, Jenster G, Trapman J, Van Der Spek P, Stubbs A. Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res. 2015;25:1382–1390. doi:10.1101/gr.183053.114.
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi:10.1093/nar/gks596.
- Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. doi:10.1038/nprot.2015.123.
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–7. doi:10.1093/nar/gks539.
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet/Edit Board, Jonathan L Haines [et al]. 2013;Chapter 7:Unit720.
- Gudernova I, Vesela I, Balek L, Buchtova M, Dosedelova H, Kunova M, Pivnicka J, Jelinkova I, Roubalova L, Kozubik A, et al. Multikinase activity of fibroblast growth factor receptor (FGFR) inhibitors SU5402, PD173074, AZD1480, AZD4547 and BGJ398 compromises the use of small chemicals targeting FGFR catalytic activity for therapy of short stature syndromes. Hum Mol Genet. 2016; 25:9–23.
- Xie L, Su X, Zhang L, Yin X, Tang L, Zhang X, Xu Y, Gao Z, Liu K, Zhou M, et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res. 2013;19:2572–2583. doi:10.1158/1078-0432.CCR-12-3898.
- Ren M, Hong M, Liu G, Wang H, Patel V, Biddinger P, Silva J, Cowell J, Hao Z. Novel FGFR inhibitor ponatinib suppresses the growth of non-small cell lung cancer cells overexpressing FGFR1. Oncol Rep. 2013;29:2181–2190. doi:10.3892/or.2013.2386.
- Andre F, Bachelot T, Campone M, Dalenc F, Perez-Garcia JM, Hurvitz SA, Turner N, Rugo H, Smith JW, Deudon S, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19:3693–3702. doi:10.1158/1078-0432.CCR-13-0190.
- Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, Nicoletti R, Winckler W, Grewal R, Hanna M, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci USA. 2008;105:8713–8717. doi:10.1073/pnas.0803379105.
- Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by Next-Generation sequencing. Clinical Cancer Research. 2016; 22:259–267.
- Wang R, Zhang Y, Pan Y, Li Y, Hu H, Cai D, Li H, Ye T, Luo X, Zhang Y, et al. Comprehensive investigation of oncogenic driver mutations in Chinese non-small cell lung cancer patients. Oncotarget. 2015; 6:34300–34308.
- Tanizaki J, Ercan D, Capelletti M, Dodge M, Xu C, Bahcall M, Tricker EM, Butaney M, Calles A, Sholl LM, et al. Identification of oncogenic and drug-sensitizing mutations in the extracellular domain of FGFR2. Cancer Res. 2015;75:3139–3146. doi:10.1158/0008-5472.CAN-14-3771.
- Lajeunie E, Heuertz S, El Ghouzzi V, Martinovic J, Renier D, Le Merrer M, Bonaventure J. Mutation screening in patients with syndromic craniosynostoses indicates that a limited number of recurrent FGFR2 mutations accounts for severe forms of Pfeiffer syndrome. Eur J Hum Genet. 2006;14:289–298. doi:10.1038/sj.ejhg.5201558.
- Falvella FS, Frullanti E, Galvan A, Spinola M, Noci S, De Cecco L, Nosotti M, Santambrogio L, Incarbone M, Alloisio M, et al. FGFR4 Gly388Arg polymorphism may affect the clinical stage of patients with lung cancer by modulating the transcriptional profile of normal lung. Int J Cancer. 2009;124:2880–2885. doi:10.1002/ijc.24302.
- Spinola M, Leoni V, Pignatiello C, Conti B, Ravagnani F, Pastorino U, Dragani TA. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23:7307–7311. doi:10.1200/JCO.2005.17.350.
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder, S, Wang, Z, Welch, R, Hutchinson, A, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi:10.1038/ng2075.
- Jiang Y, Sun S, Wei W, Ren Y, Liu J, Pang D. Association of FGFR3 and FGFR4 gene polymorphisms with breast cancer in Chinese women of Heilongjiang province. Oncotarget. 2015;6:34023–34029. doi:10.18632/oncotarget.5850.
- Mulligan LM. RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer. 2014;14:173–186. doi:10.1038/nrc3680.
- Zhou J, Song XB, He H, Zhou Y, Lu XJ, Ying BW. Prevalence and clinical profile of EGFR mutation in non- small-cell lung carcinoma patients in Southwest China. Asian Pac J Cancer Prev. 17;2016:965–971.
- Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC. Targeting FGFR signaling in cancer. Clin Cancer Res. 2015;21:2684–2694. doi:10.1158/1078-0432.CCR-14-2329.
