ABSTRACT
Objective: The study aimed to investigate candidate circular RNAs (circRNAs) in regulating the pathogenic process of esophageal carcinoma.
Methods: Specimens were collected from the patients with esophageal carcinoma. Total RNA was purified and treated with RNase R followed by RNA-seq in the purpose of screening the circRNAs in significant differentially expression. The expression level of the screened circRNAs were further validated using RT-PCR. The circular structure of the circRNA was validated with divergent and convergent primers. Overexpression vector was prepared in the purpose of raising the expression level of circ0043898 in the ECA-109 and Kyse-520 cells. The cell colony assay and MTS assay were conducted to determine the capacity of cell proliferation. Chamber assays were applied to determine the capacity of cell migration and invasion while flowcytometry was applied to determine the cell cycle and cell apoptosis. In vivo animal assay was conducted by injecting the cells to the chest of the mice. RNA-seq was performed followed by GO and KEGG study to further verify the regulation mechanism of circ0043898.
Results: circ0043898 was validated that down-regulated expressed in the specimens from the patients with esophageal carcinoma. The cell assays proved that overexpression of circ0043898 can obviously inhibit the cell proliferation, cell migration and invasion and induce cell apoptosis and death in the cancerous cells. The in vivo animal study also suggested that the circ0043898 performed inhibitory functions on oncogenesis. The RNA-seq presented the potential regulation mechanism of circ0043898. Histone H3 and BMI1 were presented significantly differential expression in both ECA-109 and Kyse-520 cells, indicating they might be the targets of circ0043898.
Conclusion: circ0043898 is presented as tumor inhibitor and could be a candidate biomarker in the therapeutic target and diagnosis of esophageal carcinoma.
Introduction
Esophageal carcinoma is one of the most common malignancies worldwide, accounting for > 400,000 cases annually with the 5-year survival rate of 10–15%.Citation1,Citation2 Esophageal squamous cell carcinoma is the most predominant type of cancer in China.Citation3 Epidemiological evidences proved that the prognosis of the patients with early-stage of esophageal carcinoma is better than those in the late stage.Citation4 Therefore, early-diagnosis of esophageal is important. However, the traditional diagnosis is lack of sensitivity and the underlying mechanism of esophageal carcinoma is still poor that limits the diagnosis and prognosis of esophageal carcinoma development and progression.Citation5,Citation6 Therefore, it is critical to identify new biomarkers and therapeutic targets to improve the diagnosis and treatment of esophageal carcinoma.
Circular RNA (circRNA) was found more than 20 years but these molecules were considered as byproducts of splicing errors.Citation7 Depending on the development of RNA sequencing, more circRNAs were detected in eukaryotic cells. The circRNAs were formed by back-splicing covalently joined 3ʹ- and 5ʹ-ends. These specific RNAs normally cannot encode protein. However, these RNAs can occur in any regions of the genome and 85% of the circRNAs are aligned in the sense orientation with known protein-coding genes and can span 1–5 exons.Citation8–Citation10 Recent data also suggested that the circRNAs can sponge miRNAs and are enriched with functional miRNA binding sites.Citation11,Citation12 Therefore, it helps to perform important regulatory roles in cell growth, proliferation, differentiation and death.Citation13 However, the circRNA involved in esophageal carcinoma is rarely reported so far. Whether circRNA performs regulatory roles in esophageal carcinoma is still unknown.
In this study, we performed RNA-seq for circRNAs in the adjacent normal tissues and esophageal cancerous tissues in order to screen the significant differentially expressed circRNAs involved in esophageal carcinoma. The functional studies based on the in vitro cell assays and in vivo animal assays were performed in the purpose of investigating the regulatory role of the circRNAs. RNA-seq based on the mRNA in the circRNA overexpressed cells and normal cells was applied to further verify the potential regulation pathways of the circRNA in esophageal carcinoma.
Materials and methods
Specimens collection
Specimens were obtained from the archives of the department of the Second Affiliated Hospital of Guangzhou Medical University. These specimens were tissue sections from 25 patients with esophageal carcinoma who did not have a history of chemotherapy or radiotherapy that collected between 2015 and 2016. Additionally, normal esophagus tissue specimens that were obtained 2cm from the periphery of cancer site were used as controls. The patients were diagnosed according to the TNM system of the American Joint Committee on Cancer (AJCC).
Cell culture
The HEEC, Kyse-140, Kyse-510, Kyse-520, Kyse-30, TE-1 and ECA-109 cell lines were obtained from ATCC, USA. The cell lines were maintained in a humidified 5% CO2 in low-glucose DMEM culture medium containing 10% FBS, 100U/ml penicillin, and 100 μg/ml streptomycin. For subculturing purposes, cells were detached by 0.05% trypsin-EDTA treatment at 37°C.
RNA-seq
RNeasy Mini Kit (QIAGEN, Germany) was used to extract total RNA from the specimens and cells. The RNA integrity was evaluated by Agilent bioanalyzer 2100 (Agilent, CA, USA). RNA Clean XP Kit (Beckman Coulter, CA, UA) and RNase-free DNase Set (QIAGEN, Germany) were used for RNA cleanup. The quality and concentration of the RNA were determined by NanoDrop 2000 (Thermo Fisher, USA). For circRNA sequencing, total RNA was digested with RNase R (Epicenter, USA) in order to remove the linear RNAs. 1μg of the RNA was used for library preparation by VAHTSTM mRNA-seq v2 library Prep Kit for illumina (Vazyme, Nanjing, China). For mRNA sequencing, the polyA mRNA was purified via the hybridization to Dynaloligo beads. The RNA was fragmented and then the double strand cDNA was synthesized. End repair and A-addition were thereby performed in order to ligate the cDNA fragments to adapters. The ligated cDNA was then subjected to PCR amplification. The quality of the library was determined using Agilent bioanalyzer 2100. Illumina Hiseq 4000 was used for the RNA-sequencing. R software was applied for quantile normalization and subsequent data processing. The differentially expressed circRNAs or mRNAs were screened according to a fold-change and q value (q value < 0.05 and fold change > 2.0).
GO and pathway analysis
The potential functions of the parental genes of differential circRNAs were analyzed by DAVID (Database for Annotation, Visualization and Integrated Discovery). The parental gene function was then predicted by GO functional annotation. The results of the GO analysis are presented in a scatter plot. The related pathways of the parental genes of differential circRNAs were analyzed by KEGG.
RT-PCR
circRNAs and genes were verified by RT-PCR. cDNA was synthesized from the total RNA using M-MLV Reverse Transcriptase (Promega, USA) according to the manufacturer’s instructions. PCR reaction was performed with GoTaq qPCR Master Mix (Promega, USA). The PCR amplification was performed on the ABI 7500 system (Applied Biosystem, USA). GAPDH was used as an internal control.
MTS assay
The cells were at the density of 1 × 106/ml and 100μl/well were seeded into 96-well plates. The cells were incubated at 37°C with 5% CO2. After culture for 1day, 2days and 3days, the MTS/PMS mixture (Promega, USA) was added. After 3h incubation, the optical density at 490nm of the cells was detected.
Colony formation assay
Cells (ranging from 100, 200 and 400 cells) were seeded into 6-well plates, and incubated at 37°C and 5% CO2 for 14 days. Cells were then fixed in methanol for 15min and stained with 0.05% crystal violet and were counted.
Migration and invasion assays
A 24-well plate containing 8mm-pore size chamber inserts (Corning, USA) was used to evaluate the migration and invasion of tumor cells. For the migration assay, 1 × 105 cells were seeded in the upper chamber. For the invasion assay, the membrane was coated with Matrigel (BD Biosciences, USA) to form a matrix barrier, and then 2 × 105 cells were placed in the upper chamber. In each lower chamber, 600ml of DMEM medium with 10% FBS was added. Cells were incubated at 37°C and allowed to migrate for 36h or invade for 48h. After incubation, the cells that had migrated through the pore were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The, the cells were counted and photographed under an IX71 inverted microscope (Olympus, Tokyo, Japan).
Cell cycle apoptosis assay
5 × 105 cells were seeded per well in 6-well plate and allowed to adhere overnight at 37°C. The cells were trypsinized, washed with PBS, fixed with 70% ethanol on ice followed by resuspension in 500μl of propidium iodide solution to stain the cells. The cell suspensions were assayed using a FACSCalibur flow cytometer (BD Biosciences, USA) at the excitation wavelength 488nm and emission wavelength 620nm. The 10,000 gated events were collected by CellQuest Pro software (BD Sciences), and flow cytometry data were analyzed by WinList and ModFit from Verity Software House (Topsham, USA). For cell apoptosis assay, the cells were resuspended in 1× binding buffer and added 5μl of fluorochrome-conjugated Annexin V and 5μl of propidium iodide staining solution. The samples were tested using FACSCalibur flow cytometer and the percentage of the apoptotic cells was measured.
Tumor formation study
Male BALB/c nude mice (5 weeks old) were used for animal studies. 3 × 106 cells were injected subcutaneously into the chest of mice. Tumors were allowed to grow for 28 days. The mice were then sacrificed, and the cancerous tissues were removed by sterile surgery and were placed in RNase-free saline. The weight and size of the cancerous tissues were then examined. The collected cancerous tissues were fixated by formalin and embedded by paraffin.
Statistical analysis
When data is corresponded to normal distribution, comparisons were performed using independent t-tests, one-way ANOVA and two-way ANOVA. Non-parametric Mann-Whitney U-test, K-S test, Kruskal-Wallis test and Wilcoxon test were performed if data is not corresponded to normal distribution. The significance was established as P < 0.05. SPSS and GraphPad Prism software were used for the statistical analysis.
Results
Screen the candidate circrnas involved in esophageal carcinoma
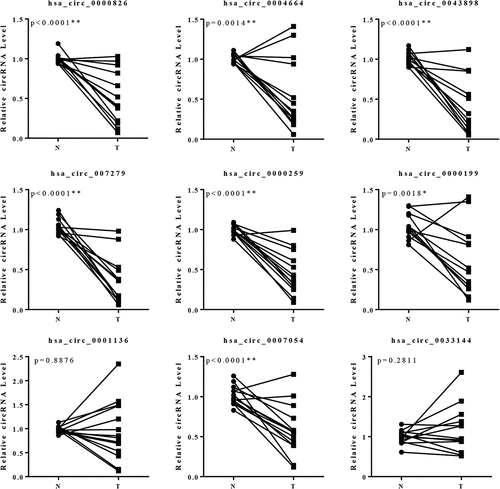
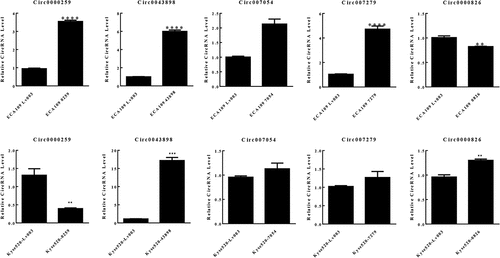
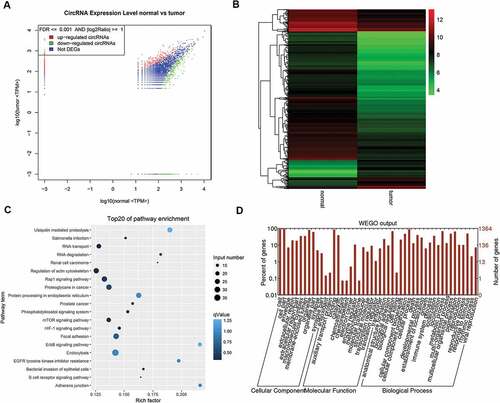
The specimens from patients with esophageal carcinoma were collected followed by RNA-seq. The cancerous tissue and the adjacent normal tissue from one patient were firstly analyzed so that to preliminarily screen candidate circRNAs. Depending on the bioinformatics analysis, large amounts of circRNAs were detected with significant differential expression signal, and presented up-regulated or down-regulated expression in the cancerous tissue, compared with the control group (). The cluster graph was presented in . The GO and KEGG pathway analysis were conducted in order to help to filter the candidate circRNAs. However, the results did not give very significant evidence to interpret the functions of the circRNAs. Based on the results, the circRNAs with differential expression were concentrated at the functions including Rap1 signaling pathway, proteoglycans in cancer, and mTOR signaling (). The GO annotation indicated that most of the circRNAs are cellular components and attend in various biological processes (). Since the functional analysis did not provide enough evidences to filter the candidate circRNAs for further study, we therefore chose the top 9 circRNAs presented significant differential expression signal in the results of RNA-seq for further investigation. 14 pairs of esophageal cancerous tissues and adjacent normal tissues were collected and the RNA level of the candidate circRNAs were determined by qPCR. Within the 14 pairs of the samples, 13 of them presented consistent results in the quantification of the candidate circRNAs, but only one pair presented different results against the others. Therefore, by focusing on the 13 pair of samples, 5 circRNAs presented significant differentially expression signal were screened, including circ0000259, circ0043898, circ007054, circ007279 and circ0000826 (). The corresponding over-expression vectors of these 5 circRNAs were thereby synthesized and transfected into both KYSE520 and ECA-109 cells for validation. The over-expression vectors were designed to raise the expression level of the corresponding circRNAs. From the results, the vectors worked fine in the KYSE520 cell line that most of the circRNAs were successfully raised in expression level, except circ0000826. However, in the ECA-109 cells, the expression level of circ0000259 was decreased in expression level after transfected with the vector, but for the rest of the circRNAs, the expression level was accordingly increased (). It might be due to the difference of the cell lines that the intra-cellular regulation mechanisms are different, so that the transfected vector inside cannot be expressed successfully or induced unexpected regulation and therefore the level of circRNAs did not present obvious change. Among these circRNAs, circ0043898 presented consistent results in both cell lines and the expression level was significant higher than the control group after transfected with the vector. Therefore, we determined to apply circ0043898 for further verification.
Figure 1. RNA-seq focused on circRNAs for the specimens of esophageal cancerous tissues and adjacent tissues. A. Dot graph of the differentially expressed circRNAs. B. Cluster graph of the differentially expressed circRNAs in up-regulated and down-regulated expression. C. Top 20 of the enriched pathways. D. GO analysis of the differentially expressed circRNAs.
Verification the functions of circ0043898 in esophageal carcinoma
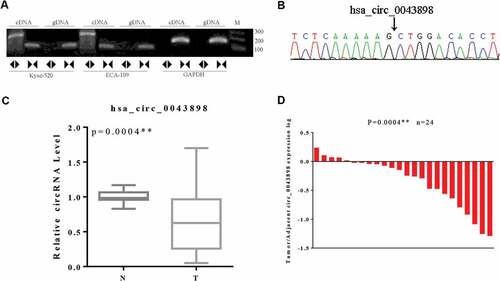
In total of 24 pairs of the collected clinical samples were thereby determined for the expression level of circ0043898 via qPCR. On the other hand the convergent and divergent primers were designed and synthesized in order to validate the circular structure of circ0043898. For the circular structure validation, the circRNA was reverse-transcribed to cDNA and validated using PCR. As it presented in the gel image, the convergent primer can amplify fragments in both cDNA and gDNA, and the length of the PCR products were consistent (). However, for the divergent primer, it can only amplify PCR product in the cDNA, but for the gDNA, the PCR products cannot be detected with obvious band in the agarose gel. We thereby conducted Sanger sequencing with the divergent primer amplified PCR product, As it showed in the sequencing peaks graph, the sequences of the spliced junction was presented clearly there (). Upon these evidences, the circular structure of the circ0043898 was proven valid. Its expression level in esophageal cancerous tissues were determined and compared with that in the normal tissues. The results presented that the expression level of circ0043898 was stable among the esophageal cancerous tissues, but generally it was lower than that in the normal tissues. The difference was statistically significant (P < 0.05; &).
Figure 4. Validation of the circular structure of circ0043898. A. The cDNA of circ0043898 can be amplified by both divergent and convergent primers, but gDNA can be amplified only by convergent primers. B. Spliced joint position was validated using Sanger sequencing. C. RT-PCR for in total of 24 pairs of specimens proved that circ0043898 was down-regulated in the esophageal cancerous tissues. D. Plot graph for the expression level of circ0043898 in 24 pairs of specimens.
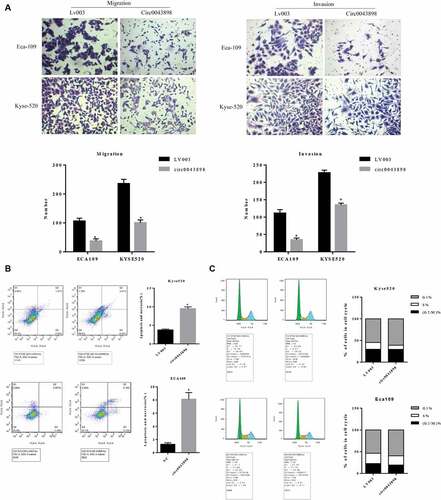
For investigation of the functions of circ0043898, the over-expression vector of circ0043898 was applied. Before the transfection of the vector, different cell lines were utilized in order to determine the expression level of circ0043898. The mRNA level of the circ0043898 was various across different cell lines. TE-1 cells presented the highest level of circ0043898 while Kyse-140 presented the least level within the cell lines (). We purposely looked for the cell lines which contains comparatively equal than the control cell line, HEEC, in order to avoid the bias which due to the initial expression level of circ0043898. As a result, Kyse-520 and ECA-109 cells were chosen for the following study. Both of the cell lines were transfected with circ0043898 over-expression vector in order to raise the level of circ0043898. The effect of the vector was obvious that the level of circ0043898 was raised 6-fold in the ECA-109 cells, while was raised ~ 18-fold in the Kyse-520 cells (). The capacity of cell proliferation for the cell lines with/without circ0043898 overexpression treatment was determined by MTS assay. The OD values were therefore compared. As it presented, the OD values were nearly the same at the first day among different cell lines with/without circ0043898 overexpression treatment. However, on 2nd and 3rd day of the experiment, the difference became significant that overexpression of circ0043898 inhibited the cell proliferation and yielded lower OD value that the control group in both Kyse-520 and ECA109 cells (). Cell colony formation assay also provided similar results as the MTS assay. The cells transfected with the overexpression vector generated fewer colonies that the control group in both cell lines. For the ECA109 and Kyse-520 cells, the colony formation ratio was reduced to 50% compared with the control group (). On the other hand, the capacity of cell migration and invasion was determined. As we can observe in the results, for both cell migration and invasion were inhibited in both cell lines when circ0043898 was overexpressed. The migrated cells and invasive cells were decreased ~ 60% when circ0043898 was raised in expression, compared with the control group (). For the flowcytometry studies, the cell cycle and cell apoptosis were determined for both cell lines. As the assay determined, the cell apoptosis percentage was significantly increased 2-fold in the Kyse-520 cells, and 4-fold in the ECA109 cells when circ0043898 was overexpressed (). In addition, the cell cycle assay proved that the Kyse-520 and ECA109 cells in S phase were decreased while those in G1 phase were increased after transfected with circ0043898 overexpression vector (). It indicated that the most of the cells did not actively proliferate but decreased in number because of the occurrence of cell apoptosis. It is consistent with the results of MTS assay. By summarizing the results above, the circ0043898 is presented as cancer inhibitor: it inhibits the cancerous cell in migration and invasion, and induces cell apoptosis and suppresses cell proliferation.
Figure 5. Validation of the capacity of cell proliferation in ECA-109 and Kyse-520 cells. A. The expression level of circ0043898 within different cell lines. B. The performance of the overexpression vector of circ0043898 in ECA-109 and Kyse-520 cells. C. MTS assay determined the capacity of cell proliferation in ECA-109 and Kyse-520 cells. D. Cell colony formation assay proved that circ0043898 can inhibit cell proliferation of ECA-109 and Kyse-520 cells.
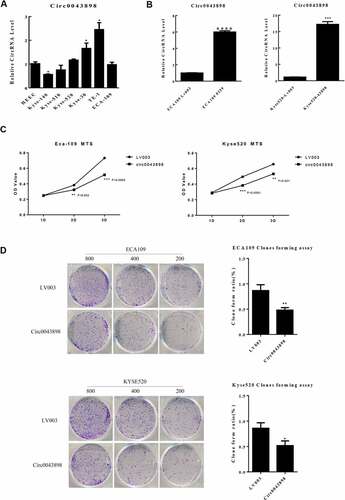
Figure 6. Cell assays to verify the functions of circ0043898 in regulating cell migration and invasion, cell apoptosis and cell cycle. A. Overexpression of circ0043898 inhibited the capacity of cell migration and invasion in ECA-109 and Kyse-520 cells. B. Cell apoptosis assay presented that the overexpression of circ0043898 raised the cell apoptosis rate in ECA-109 and Kyse-520 cells. C. The overexpression of circ0043898 retained the cell in G1 phase.
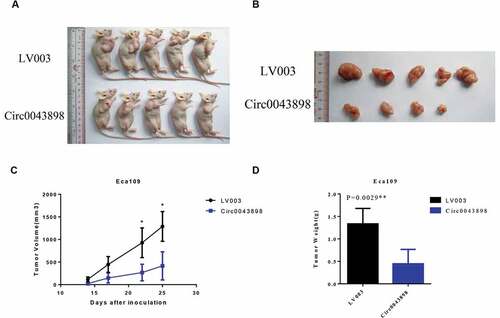
For further validate the conclusion above, the nude mice were used and the in vivo tumor formation assay was conducted by subcutaneous injection. The cancerous cells, ECA-109, were injected to the cheats of the mice. Both cells with and without circ0043898 overexpression treatment were applied. After 25 days, the mice were sacrificed and the tumor tissues were collected. The overexpression of the circ0043898 obviously inhibited the growth of the tumor that the tumor formed by the cells with the treatment was smaller than that in the control group (). By detecting the volume of the tumor, the tumor formed by the cells with circ0043898 overexpression treatment presented in smaller size than that in the control group with the whole time period of the experiment. The difference was showed in 22th day, and became significant at ~ 3-fold in the 25th day (). Meanwhile, the weight of the tumor was showed lower than the control group when circ0043898 was raised in expression (). In a word, circ0043898 was proven to have the functions in inhibiting the cell proliferation in vivo.
Figure 7. In vivo animal studies to verify the functions of circ0043898. A. The cells with/without circ0043898 overexpression were injected to the chest of the mice and thereby tumors were generated. B. The size of the tumor collected from the mice. C. The volumes of the tumors in different period after the cells were injected. D. The comparison of the weight of the tumors formed by the cells with/without circ0043898 overexpression. E. Immunohistochemical staining for the tumor tissues.
Investigating the mechanism of the circ0043898 in inhibition of the oncogenesis process of esophageal carcinoma
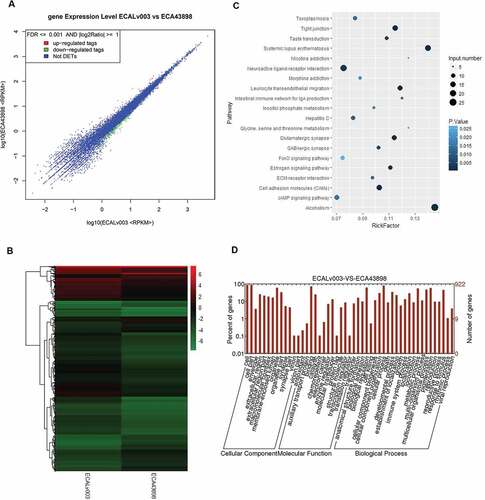
The ECA109 cells with and without circ0043898 overexpression treatment were applied for the transcriptome sequencing study. The sequencing results were summarized and the significantly differential expressed genes were filtered. The scatter graph and cluster graph were presented in and . Both of the results indicated that the overexpression of circ0043898 could affect specific genes’ expression level. Among the genes, most of the differentially expressed genes presented down-regulated expression. The differentially expressed genes were thereby conducted with GO analysis and KEGG pathway analysis ( & ). The KEGG pathway suggested the enriched pathways depending on the gene input number and the rich factor. Therefore, it can be summarized from the graph that the pathways that contained large number of differentially expressed genes and high value of rick factor was focused on systemic lupus erythematosus pathway, neuroactive ligand-receptor interaction pathway, and alcoholism related pathway. On the other hand, the GO analysis presented that the impact of the overexpression of circ0043898 was mainly on cellular components and biological processes as most of the significant differentially expressed genes were concentrated there. However, for the exact genes that regulated by circ0043898 and thereby affect the oncogenesis process of esophageal carcinoma are still not clarified. Similar test has done as well for the Kyse-520 cells. Both KEGG results were showed in Table S1 and S2.
Figure 8. RNA-seq analysis for the ECA-109 cells with/without circ0043898 overexpression treatment. A. Dot graph for the up-regulated and down-regulated differentially expressed genes. B. Cluster graph for the up-regulated and down-regulated differentially expressed genes. C. Pathway enrichment for the significant differentially expressed genes. D. GO functional analysis for the differentially expressed genes.
Discussion
In this study, we found the candidate circRNA, circ0043898, involved in the esophageal carcinoma. Depending on our data, circ0043898 presented down-regulated expression in the tumor, compared with the adjacent tissues. Further study presented that overexpression of the circ0043898 can obviously inhibit the oncogenesis of esophageal carcinoma in terms of inhibition to the cell proliferation, invasion and migration of the cancerous cells. Therefore, circ0043898 should be a tumor inhibitor and presented to be a biomarker in diagnosis of esophageal carcinoma.
Recently, the relevant circRNA studies focused on esophageal carcinoma are rare. Circ_0067934 is proved up-regulated in esophageal carcinoma, and it can promote the tumor differentiation based on the clinical samples.Citation14 circRNA9927-NBEAL1 was filtered from the RNA-seq profile and presented as an important node in regulating the molecules involved in esophageal carcinoma.Citation15 Circ-ITCH can sponge with miR-7, miR-17 and miR-214 and thereby increases the level of ITCH and inhibits the Wnt pathway, which indicates an inhibitory effect on esophageal carcinoma.Citation12 According to these studies, it suggested that circRNAs should have important regulatory effect on esophageal carcinoma, and also have potential value as a biomarker in diagnosis of esophageal carcinoma. For circ0043898, it is currently no report about this specific circRNA regarding its functions and target. Based on the data on our studies, it can inhibit the cell proliferation, migration and invasion, and also induced cell apoptosis and death. Its inhibitive effect on the tumor generation was also proved in the in vivo animal model studies. Therefore, we believe that it functions as a tumor inhibiter of esophageal carcinoma. For validating the regulation mechanism of it, RNA-seq was performed and the mRNA profiles were annotated by GO analysis and KEGG pathway analysis. By analyzing the data, it can be observed that pathway, transcriptional misregulation in cancer, was significantly enriched with the significant differentially expressed genes when circ0067934 was overexpressed in both ECA-109 and Kyse-520 cells. In the pathway, histone H3 and BMI1 were found significant differentially expressed in both cell lines. Both molecules might be the targets of circ0043898. For the histone H3, it involves in the regulation of cell cycle progression by targeting on the gene CCND2. Actually, the effect of histone H3 on esophageal carcinoma was reported in the previous study. Kai et al. proved that the functions of histone H3 should be correlated to the cancer invasive capabilities. It has the functions of methylation, acetylation and butyrylation so that modulate the expression of the downstream genes.Citation16 According to the study from Hu et al., histone H3 locates more at KLF4 promoter region and it is correlated to the cell growth inhibition.Citation17 Therefore, circ0043898 was showed the functions in regulating the level of histone H3, indicating that it might induce the cell arrest and inhibition to cell invasion. It is proven in our study based on the cell invasion assay and flowcytometry assays. For the gene BMI1, it has been reported that BMI1 is overexpressed in a number of malignancies.Citation18,Citation19 And it is also overexpressed in alimentary canal cancers, particularly in esophageal carcinoma, which suggests that BMI1 may confer radioresistance to esophageal carcinoma.Citation20 The study based on the gene chip analysis also showed that BMI1 predicts cancer metastasis,Citation21 promotes cancer cell proliferation and invasion, causes resistance to apoptosis and enhances transfer capabilities.Citation22 In addition, BMI1 might induce the ubiquitination and phosphorylation of H2AX, which is thought to be a critical sensor that can initiate DNA damage response.Citation23–Citation25 Based on the studies, it can be hypothesized that circ004898 might perform a function in inhibiting BMI1 so that to inhibit the cell proliferation and invasion and thereby induced the cell apoptosis in esophageal carcinoma. Except the main findings above, it can be observed individually in the mRNA profile of ECA-109 cell that the Wnt pathway was also under regulation of circ0043898. Therefore, it might also be a potential regulation pathway for circ0043898 to inhibit the cancerous cell proliferation. However, it needs further study to validate it.
In summary, our study proved that circ0043898 has the functions in inhibiting the esophageal carcinoma development and progression. It inhibits the cell proliferation, migration and invasion and also induced the cell death of the cancerous cell so that efficiently inhibits the oncogenesis of esophageal carcinoma. There are some available assays can be performed in order to further validate the conclusion. RT-PCR and western blotting can be conducted to validate the candidate targets of circ0043898 in cell lines and clinical specimens. The amount of the clinical samples is still limited so that large scale of the specimens can be applied to lead the conclusion more convincing. As circ0043898 is proven to be tumor inhibitor and its level was decreased during esophageal carcinoma, it can be used as a biomarker for the therapeutic target and for the diagnosis of esophageal carcinoma.
Availability of data and material
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors’ Contributions
Tingting Xie conceived and designed the study, and critically revised the manuscript. Wei Wang performed the experiments, analyzed the data and drafted the manuscript. Jun Ma, Jianjun Lu, Danqing Fang, Xinming Xiong and Xin Yang participated in study design, study implementation and manuscript revision. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The animal experiment was approved by The Institute Research Medical Ethics Committee of Sun Yat-Sen University.
Consent for publication
Not applicable
Competing interests
The authors declare no potential conflicts of interest.
Supplemental Material
Download ()Acknowledgments
None
Supplemental Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- He C, Wu S, Gao A, Su Y, Min H, Shang ZF, Wu J, Yang L, Ding WQ, Zhou J. Phosphorylation of ETS-1 is a critical event in DNA polymerase iota-induced invasion and metastasis of esophageal squamous cell carcinoma. Cancer Sci. 2017;108:2503–2510. doi:10.1111/cas.13399.
- Chen XX, Zhong Q, Liu Y, Yan SM, Chen ZH, Jin SZ, Xia TL, Li RY, Zhou AJ, Su Z, et al. Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat Commun. 2017;8:524. doi:10.1038/s41467-017-00650-0.
- Hou Z, Abudureheman A, Wang L, Hasim A, Ainiwaer J, Zhang H, Niyaz M, Upur H, Sheyhidin I. Expression, prognosis and functional role of Thsd7a in esophageal squamous cell carcinoma of Kazakh patients, Xinjiang. Oncotarget. 2017;8:60539–60557. doi:10.18632/oncotarget.16966.
- Qi Y, Cao KX, Xing FC, Zhang CY, Huang Q, Wu K, Wen FB, Zhao S, Li X. High expression of MAGE-A9 is associated with unfavorable survival in esophageal squamous cell carcinoma. Oncol Lett. 2017;14:3415–3420. doi:10.3892/ol.2017.6614.
- Chan CM, Lai KKY, Ng EKO, Kiang MN, Kwok TWH, Wang HK, Chan KW, Law TT, Tong DK, Chan KT, et al. Serum microRNA-193b as a promising biomarker for prediction of chemoradiation sensitivity in esophageal squamous cell carcinoma patients. Oncol Lett. 2018;15:3273–3280. doi:10.3892/ol.2017.7698.
- Shang M, Wang X, Zhang Y, Gao Z, Wang T, Liu R. LincRNA-ROR promotes metastasis and invasion of esophageal squamous cell carcinoma by regulating miR-145/FSCN1. Onco Targets Ther. 2018;11:639–649. doi:10.2147/OTT.S157638.
- Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi:10.1186/s12943-017-0663-2.
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi:10.1371/journal.pgen.1001233.
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi:10.1038/emboj.2011.359.
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7:e30733. doi:10.1371/journal.pone.0030733.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:10.1038/nature11928.
- Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi:10.18632/oncotarget.3469.
- Huang SD, Yuan Y, Zhuang CW, Li BL, Gong DJ, Wang SG, Zeng ZY, Cheng HZ. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer. 2012;11:51. doi:10.1186/1476-4598-11-51.
- Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi:10.1038/srep35576.
- Sun J, Yuan X, Li X, Wang D, Shan T, Wang W, Wan Q, Wang X, Yan J, Gao S. Comparative transcriptome analysis of the global circular RNAs expression profiles between SHEE and SHEEC cell lines. Am J Transl Res. 2017;9:5169–5179.
- Zhang K, Li L, Zhu M, Wang G, Xie J, Zhao Y, Fan E, Xu L, Li E. Comparative analysis of histone H3 and H4 post-translational modifications of esophageal squamous cell carcinoma with different invasive capabilities. J Proteomics. 2015;112:180–189. doi:10.1016/j.jprot.2014.09.004.
- Hu C, Liu M, Zhang W, Xu Q, Ma K, Chen L, Wang Z, He S, Zhu H, Xu N. Upregulation of KLF4 by methylseleninic acid in human esophageal squamous cell carcinoma cells: modification of histone H3 acetylation through HAT/HDAC interplay. Mol Carcinog. 2015;54:1051–1059. doi:10.1002/mc.22174.
- Rouhigharabaei L, Ferreiro JF, Put N, Michaux L, Tousseyn T, Lefebvre C, Gardiner A, De Kelver W, Demuynck H, Verschuere J, et al. BMI1, the polycomb-group gene, is recurrently targeted by genomic rearrangements in progressive B-cell leukemia/lymphoma. Genes Chromosomes Cancer. 2013;52:928–944. doi:10.1002/gcc.22088.
- Guo BH, Feng Y, Zhang R, Xu LH, Li MZ, Kung HF, Song LB, Zeng MS. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol Cancer. 2011;10:10. doi:10.1186/1476-4598-10-93.
- Dong Q, Sharma S, Liu H, Chen L, Gu B, Sun X, Wang G. HDAC inhibitors reverse acquired radio resistance of KYSE-150R esophageal carcinoma cells by modulating Bmi-1 expression. Toxicol Lett. 2014;224:121–129. doi:10.1016/j.toxlet.2013.10.014.
- Chen Y, Lian G, Zhang Q, Zeng L, Qian C, Chen S, Huang K. Overexpression of Bmi-1 induces the malignant transformation of gastric epithelial cells in vitro. Oncol Res. 2013;21:33–41. doi:10.3727/096504013X13786659070316.
- Hoenerhoff MJ, Chu I, Barkan D, Liu ZY, Datta S, Dimri GP, Green JE. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28:3022–3032. doi:10.1038/onc.2009.165.
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi:10.1038/nature02985.
- Buchwald G, Van Der Stoop P, Weichenrieder O, Perrakis A, Van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi:10.1038/sj.emboj.7601144.
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi:10.1016/j.molcel.2005.12.002.