ABSTRACT
Background: Substantial progress has been made in metastatic colorectal cancer (mCRC) treatment, but there is still a fraction of patients cannot find any effective therapeutic strategy after guideline-recommended standard chemotherapy and molecular targeted therapy.
Case presentation: Here we present a KRAS/NRAS/BRAF wild-type mCRC patient who has been previously treated with FOLFIRI (fluorouracil, leucovorin, and irinotecan), XELOX (capecitabine and oxaliplatin), cetuximab and bevacizumab, and then received the next generation sequencing (NGS) and whose metastatic subcutaneous nodule was resected to generate patient-derived xenograft (PDX) models. The NGS revealed HER-2 amplification as well as an activating mutation S310F and PDX models tested several drugs finding that afatinib was the optimal agent with notable efficacy and well tolerance among 6 regimens. Therefore, this patient started to take afatinib orally and achieved 3 months progression-free survival (PFS) and relief of clinical symptoms without severe adverse effects.
Conclusions: NGS and PDX models have great significance for precision and individualized medicine in the mCRC treatment, especially for patients whose diseases have been progressed after multiline standard therapies.
Introduction
Colorectal cancer (CRC) is the third most frequent malignancy in both sexes, and in the United States, there are estimated to be 135,430 individuals newly diagnosed with CRC and 50,260 deaths from this disease in 2017. Metastatic colorectal cancer (mCRC) is diagnosed in 21% of patients, the 5-year cancer-specific survival rate of whom is only 14%.Citation1 The conventional approaches to treat mCRC include doublet or triplet cytotoxic drug combinations, such as oxaliplatin-based regimen XELOX and FOLFOX, irinotecan-based regimen FOLFIRI or FOLFOXLIRI.Citation2 Anti-epidermal growth factor receptor (EGFR) and anti-vascular endothelial growth factor (VEGF) drugs, such as cetuximab, panitumumab, bevacizumab and regorafenib, respectively, have markedly prolonged survival for mCRC patients.Citation3,Citation4 However, due to the nature of this disease, many patients would develop drug resistance to the guideline-recommended regimens and suffer from progressive diseases even with a good performance status. For these patients with strong treating willingness, what can help the clinicians to make the treatment decisions? Thanks to the remarkable advances in next generation sequencing (NGS), precision medicine is no longer a faraway dream.
The NGS technology enables to identify gene mutations in patient tumor tissues thoroughly and gives assistance to the scientists for understanding the origin of the disease.Citation5,Citation6 Most importantly, the patients with some given mutations may respond to the corresponding therapies, for example chronic myeloid leukemia (CML) patients with BCR-ABL fusion gene almost all can benefit from imatinib. Unfortunately, this kind of luck only happens to a minority of people, as merely a small fraction of gene variants is considered as functional in the meantime, with food and drug administration (FDA)-approved regimens matched.Citation7 Thus, it is necessary to perform additional functional tests to complement NGS and help oncologists to make the right choice for a greater proportion of patients. The patient-derived xenograft (PDX) mouse model is a novel and reliable approach to test drug sensitivity through implanting the neoplasm from the patient to mice, and imitate the tumor microenvironment in vivo.Citation8 Here we presented a case with HER-2 amplified, RAS-BRAF-wild type mCRC, whose disease progressed on several standard lines of systemic treatment and responded to afatinib as an off-label use according to the results from the NGS and PDX mouse models.
Case presentation
A 64-year-old man was admitted to our hospital in October 2015 due to difficult defecation. Enhanced abdominal CT revealed thickening in the sigmoid colon along with multiple liver metastases. Colonoscope found a mass and pathology showed moderately differentiated adenocarcinoma. In the November, the colon mass was completely removed with lymph node dissection. Postoperative pathological findings: a stage IV (pT3N1M1) moderately-poorly differentiated adenocarcinoma with RAS and BRAF wild-type and then the systemic therapy of FOLFIRI-cetuximab was performed from Dec 2015 to Feb 2016. However, the disease progressed with a subcutaneous nodule in the abdominal wall, which was considered as metastasis. Consequently, the patient converted to administer XELOX plus bevacizumab for 8 cycles, and xeloda plus bevacizumab as maintenance treatment from Oct 2016, with a mix response that stable hepatic foci but progressive disease of the subcutaneous nodule.
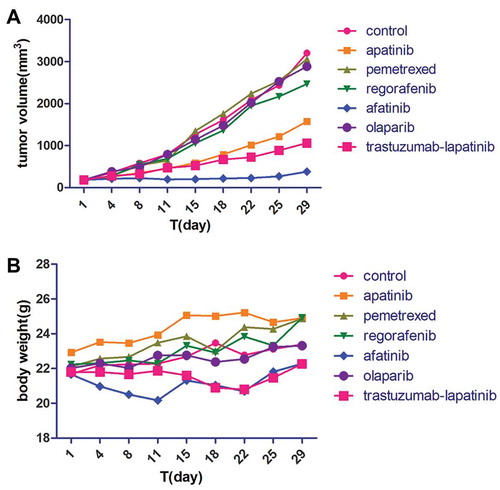
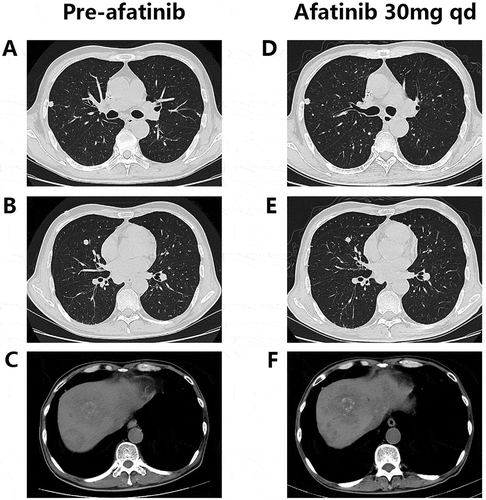
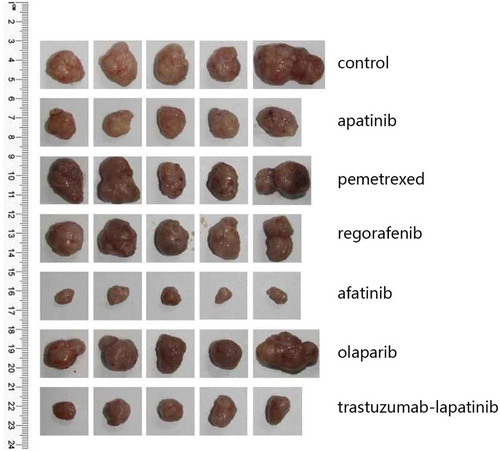
In Feb 2017, the NGS covering 45 genes showed HER2 amplification and an activating mutation S310F in the patient tumor tissue inferring that several HER2-targeted agents aiming at it may have certain effects on the disease control, including trastuzumab and T-DM1. To choose the most appropriate therapy, the subcutaneous nodule in the abdominal wall was resected for the generation of PDX mouse models to test the following 6 regimens in vivo, including apatinib, pemetrexed, regorafenib, afatinib, olaparib and trastuzumab plus lapatinib. And as xeloda – bevacizumab failed, the patient began receiving the triplet drug combination (oxaliplatin, raltitrexed plus bevacizumab), however, the disease was still not controlled effectively with multiple lung metastases occurring. Then according to the NGS and literature about the relativity between HER2-amplification and the activating mutation S310F,Citation9 we had intended to recommend him to receive trastuzumab plus lapatinib. Yet in July 2017, the PDX models indicated that in the drug sensitivity test, apatinib, trastuzumab-lapatinib and afatinib could inhibit tumor growth, and unexpectedly, afatinib had the more significant effect than trastuzumab-lapatinib “30 days after modeling, the tumors removed from each group are seen in and the change of tumor volume during the medication can be seen in ” in terms of adverse events, which was evaluated via weight changes of tumor-bearing mice, a visible loss of weight was seen in the mice from the afatinib and trastuzumab-lapatinib group (). So, from August 2017, after failure of multiline therapies, the patient ultimately orally took afatinib with a reduced dose of 30mg per day due to limited fitness and maintained stable disease for 3 months with well tolerance (). During the medication, hepatalgia and pelvic pain was obviously relieved. In January 2018, the patient died of severe lung infection and kidney failure.
Figure 1. The PDX models from top to bottom are tumors of the control group and 6 drug sensitivity test groups (n = 5 per group), which were removed 30 days after modeling.
Discussion
In recent years, standard lines of systemic therapy, including chemotherapy, targeted therapy and immunotherapy, have potently improved survival for mCRC patients, but a high proportion of them are incapable of benefitting in the long term. The key lies in the incompleteness of precision medicine. Combination NGS with PDX models provides an opportunity for the selection of rational and individualized agents based on the tumor molecular features. With increased efficiency and decreased expenses, the NGS has been a widespread approach to evaluate the patient genomes in clinical settings. Besides the innovation in understanding tumorigenesis, NGS also plays an important role in cancer prevention, screening, diagnosis, and management.Citation10 Whole genome sequencing is able to reveal 3 to 4 million variants for a typical person,Citation11 but because of insufficient knowledge of genetics and the enormous imbalance between substantial mutations and limited available antitumor agents, NGS only succeeded to guide clinical decisions in some cases and small studies to date.Citation12
It is thought that performing additional functional tests, like the PDX models can help clinicians to better interpret and utilize the genetic information of patients. According to the research gains from case reports and pilot clinical studies, the prospects of PDX mouse models are very cheerful: a. Hidalgo et al generated models for 14 patients with advanced cancer and tested 63 drugs in 232 therapeutic regimens, then 11 (79%) patients achieved durable partial remissions with the treatments deemed effective in mouse models;Citation13 b. in the PDX-directed sarcoma treatment, clinical outcome was observed in 13 of 16 (81%) patients.Citation14 However, there exist some limitations in the clinical application of this technology. First of all, the whole process of building PDX models requires 6 to 8 months,Citation13 so some patients have died before receiving the experimental results, and this situation has been seen in both above two clinical researches. What’s more, the number of regimens tested in the PDX models is quite limited, as in this case, it took over 5 months to complete the whole experiment and only 6 regimens were tested. Thus, there is a high probability that the patient expends a great deal of time and money but fails to find the optimal therapeutic regimen. As for this, the clinicians may consider recommending the sufferers to receive the drug susceptibility study in vitro, that use cancer cell line from the tumor tissue instead of the mouse model as “avatar” of the patient tumor. The patient in this case also tried this method in December 2017 as the disease progressed again, and 11 drug options were tested in less than a month with a higher efficiency than PDX models, but he died before taking the prescription (osimertinib, cobimetinib plus palbociclib). Secondly, tumor material needs to be fresh, adequate and sterile as possible which is difficult to reach, especially for CRC patients with primary lesion resected.Citation15 Finally, the results of the drug sensitive test would become unreliable due to the heterogenicity between the patient tumor tissue and the third-generation PDX models.Citation16 Even so, the promising data gained from some PDX platforms encourages researchers to conduct large-scale prospective randomized clinical trials for this fresh therapeutic selection strategy.
Previous researches reveal that HER2 amplification is implicated in resistance to anti-EGFR therapy, which explains the reason why the PFS of this patient was only 3 months. In breast cancer and gastric cancer treatment, HER2-targeted therapy has been the standard treatment for the patients with HER2 amplification,Citation17,Citation18 which are also found in 4.9% mCRC patients.Citation19 As for this CRC subgroup, preclinical studies have suggested that dual blockade of HER2 and EGFR with trastuzumab, a HER2 human monoclonal antibody and lapatinib, a tyrosine kinase inhibitor (TKI) targeting HER2 and EGFR, enables remarkably inhibit colorectal cancer growth in vivo and in vitro, but either drug alone is of no effect.Citation20 The phase II clinical trial HERACLES (HER2 Amplification for Colon-Rectal Cancer Enhanced Stratification) evaluates the efficacy of combination trastuzumab with lapatinib in treating HER2-amplified CRC determined by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). In the HERACLES, a total of 24 cetuximab-pretreated mCRC patients enrolled. 34.7% patients responded to this therapy (complete and partial response), with an 8.5-month median duration of response and 5.5-month median progression-free survival (PFS).Citation21 Therefore, trastuzumab plus lapatinib is generally recommended in HER2-positive mCRC treatment. In the drug sensitive test of PDX models, afatinib, the irreversible HER2-EGFR-directed TKI, is significantly better than trastuzumab plus lapatinib, so a reduced dose of afatinib was used in the third-line treatment of this patient, considering his limited physical score. Afatinib has been shown to lead an inhibitory effect on CRC cell lines with HER2 activating mutations.Citation9 In this case, the patient experienced disease progression during the first-line FOLFIRI plus cetuximab treatment, and after taking afatinib, achieved 3-month progression free. Compared with the median PFS of other molecule-targeting drugs for mCRC third-line treatment, like regorafenib (1.9months)Citation22, trifluridine-tipiracil (TAS-102, 2.0months)Citation23 and fruquintinib (3.7months)Citation24, afatinib brought more impressive benefit for this HER2-amplified mCRC patient.
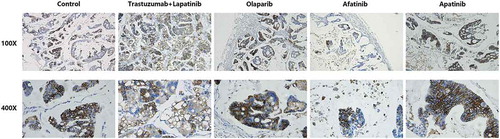
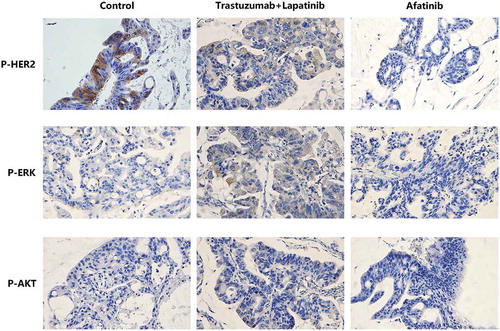
To explore the molecular mechanism underlying the different effects between afatinib and trastuzumab-lapatinib on PDX models, we performed IHC staining on the tumors from each group of PDX models to determine the expression of HER2. It is shown that all tumors strongly express HER2, which infers that in the process of PDX-model generation, the heterogeneity between the primary lesion and tumors in each group is quite small and the predictive value of this drug-sensitive test is correspondingly high (). Since homo- or heterodimerization of the HER receptors modifies tumor cell proliferation, survival and migration through Ras/MAPK and PI3K/AKT pathways,Citation25 we also used IHC staining to confirm the phosphorylation of HER2, ERK1/2 and AKT T308 (). We found that both afatinib and trastuzumab-lapatinib inhibit the phosphorylation of HER2, and the former has a stronger inhibitory effect. As for the Ras/MAPK pathway, there is no positive signal detected in tumors of the control and afatinib group, but we found this pathway is unusually activated in the tumor exposed to trastuzumab-lapatinib. We suppose that there exists a bypass activating ERK1/2 with HER2 inhibited. In addition, PI3K/AKT pathway is not activated in any group as we haven’t seen any tumor expressing phosphorylated AKT T308. In short, in this case, afatinib could effectively interrupt HER2 activating as well as its downstream oncogenic pathways to promote tumor development, but due to some unknown cause, Ras/MAPK pathway in the tumor of trastuzumab-lapatinib group is abnormally activated. That is the reason why afatinib can produce much more significant effects than conventional trastuzumab-lapatinib.
Conclusions
In summary, it is the first clinical evidence of a mCRC patient on afatinib who achieved 3-month PFS, based on the results from NGS and PDX models. It highlighted the practicability of combination these two technologies in precision medicine for advanced colorectal cancer patients to select individualized regimens who were extensively pretreated with several standard treatments. In addition, this case provided clinical evidence supporting the relativity between the resistance to cetuximab and HER2 amplification. For these special mCRC patients, we propose that besides the guideline-recommended regimen, NGS technology and PDX models could assist clinicians to choose optimal agents for the patients with good physical fitness. Finally, this case reminds us that afatinib which is usually used for the treatment of advanced non-small cell lung cancer (NSCLC) with EGFR sensitive mutations, could be an option for these RAS-BRAF-wild type, HER2-positive mCRC patients as an off-label use.
Disclosure statement
Disclosure of potential conflicts of interest: No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics. CA Cancer J Clin. 2017;67:177–193. Internet. doi:10.3322/caac.21395.
- Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol. 2015;33:1809–1824. Internet. doi:10.1200/JCO.2014.59.7633.
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. Internet. doi:10.1056/NEJMoa033025
- Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. Internet. doi:10.1056/NEJM200103153441101
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. Internet. doi:10.1038/nature12912.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: figure 1. Cancer Discov. 2012;2:401–404. Internet. doi:10.1158/2159-8290.CD-12-0095
- Dienstmann R, Jang IS, Bot B, Friend S, Guinney J. Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov. 2015;5:118–123. Internet. doi:10.1158/2159-8290.CD-14-1118.
- Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15:311–316. Internet. doi:10.1038/nrc3944.
- Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, Searleman AC, Shen W, Monsey J, Trusolino L, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5:832–841. Internet. doi:10.1158/2159-8290.CD-14-1211
- Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275. Internet. doi:10.1001/jamainternmed.2013.12048.
- Biesecker LG. Opportunities and challenges for the integration of massively parallel genomic sequencing into clinical practice: lessons from the ClinSeq project. Genet Med Internet. 2012;14:393–398. http://www.nature.com/articles/gim201178.
- Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer Internet. 2015;15:747–756. http://www.nature.com/articles/nrc4015.
- Hidalgo M, Bruckheimer E, Rajeshkumar NV, Garrido-Laguna I, De Oliveira E, Rubio-Viqueira B, Strawn S, Wick MJ, Martell J, Sidransky D. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther. 2011;10:1311–1316. Internet. doi:10.1158/1535-7163.MCT-11-0233.
- Stebbing J, Paz K, Schwartz GK, Wexler LH, Maki R, Pollock RE, Morris R, Cohen R, Shankar A, Blackman G, et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer. 2014;120:2006–2015. Internet. doi:10.1002/cncr.28696
- Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, Shi C, Danenberg K, Danenberg PV, Kuramochi H, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. Internet 2006;12:4652–4661. doi:10.1158/1078-0432.CCR-06-0113.
- Majumder B, Baraneedharan U, Thiyagarajan S, Radhakrishnan P, Narasimhan H, Dhandapani M, Brijwani N, Pinto DD, Prasath A, Shanthappa BU, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun. 2015;6:6169. Internet. doi:10.1038/ncomms7169
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. Internet. doi:10.1056/NEJM200103153441101
- Bang Y-J, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. Internet. doi:10.1016/S0140-6736(10)61121-X
- Ross JS, Ali SM, Elvin JA, Schrock AB, Suh J, Vergilio J-A, Ramkissoon S, Severson EA, Daniel S, Fabrizio D, et al. Targeted therapy for HER2 driven colorectal cancer. J Clin Oncol. 2017;35:3583. Internet. doi:10.1200/JCO.2017.35.15_suppl.3583
- Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti C, et al. A molecularly annotated platform of patient- derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi:10.1158/2159-8290.CD-11-0109.
- Siena S, Sartore-Bianchi A, Lonardi S, Trusolino L, Martino C, Bencardino K, Leone F, Zagonel V, Valtorta E, Torri V, et al. Trastuzumab and lapatinib in HER2-amplified metastatic colorectal cancer patients (mCRC): the HERACLES trial. J Clin Oncol. 2015;33:3508. Internet. doi:10.1200/jco.2015.33.15_suppl.3508
- Grothey A, Van CE, Sobrero A, Siena S, Falcone A, Ychou M, Humblet Y, Bouché O, Mineur L, Barone C, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303–312. Internet. doi:10.1016/S0140-6736(12)61900-X
- Mayer RJ, Van Cutsem E, Falcone A, Yoshino T, Garcia-Carbonero R, Mizunuma N, Yamazaki K, Shimada Y, Tabernero J, Komatsu Y, et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med. 2015;372:1909–1919. Internet. doi:10.1056/NEJMoa1414325
- Li J, Qin S, Xu R-H, Shen L, Xu J, Bai Y, Yang L, Deng Y, Chen Z, Zhong H, et al. Effect of fruquintinib vs placebo on overall survival in patients with previously treated metastatic colorectal cancer. JAMA. 2018;319:2486. Internet. doi:10.1001/jama.2018.7855
- Citri A, Yarden Y. EGF–ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505. Internet. doi:10.1038/nrm1962.