ABSTRACT
Background: Gastric cancer (GC) is a serious threat for public health worldwide. Long non-coding RNA (lncRNA) linc00152 has been well reported to be an oncogene and a potential biomarker in multiple cancers including GC. However, the molecular mechanisms of linc00152 in GC development need to be further investigated.
Methods: RT-qPCR assay was employed to detect the levels of linc00152, microRNA-193b-3p (miR-193b-3p) and ETS1 mRNA. ETS1 protein level was measured by western blot assay. Cell proliferative, migratory and invasive capacities were assessed by colony formation together with CCK-8 assays, transwell migration and invasion assays, respectively. Bioinformatics analyses and luciferase reporter assay were used to explore whether miR-193b-3p could interact with linc00152 or ETS1 3ʹUTR. The roles and molecular basis of linc00152 silence on the growth of GC xenograft tumors were tested in vivo.
Results: Linc00152 expression was notably upregulated in GC tissues and cells. The proliferative, migratory and invasive abilities of GC cells were weakened by linc00152 depletion, miR-193b-3p overexpression or ETS1 knockdown. Linc00152 upregulation inhibited miR-193b-3p expression by direct interaction and abolished miR-193b-3p-mediated anti-proliferation, anti-migration and anti-invasion effects in GC cells. ETS1 was a target of miR-193b-3p and linc00152 could promote ETS1 expression by downregulating miR-193b-3p. In vivo experiments further validated that linc00152 knockdown inhibited the growth of GC xenograft tumors by upregulating miR-193b-3p and downregulating ETS1.
Conclusion: Knockdown of linc00152 inhibited GC progression by sequestering miR-193b-3p from ETS1 in vitro and in vivo, elucidating a novel molecular mechanism of linc00152 in promoting GC carcinogenesis.
Introduction
Gastric cancer (GC) is a major public health problem worldwide especially in China with about 50% of global GC cases and deaths occurring in China.Citation1 Despite the great advances in the diagnosis, staging, image and treatment, GC is still the fifth most frequently diagnosed cancer and the third cause of cancer deaths globally.Citation2–Citation5 GC is usually diagnosed at an advanced stage Citation3 and the percentage of patients with metastatic GC is around 40%.Citation6,Citation7 Moreover, the prognosis for patients with metastatic GC is poor with the median survival time of about 3 months to 1 year.Citation8,Citation9 Recently, numerous researchers paid much attentions to the search of potential tumor markers and targets, which might can contribute to the early detection, monitoring, prognosis and treatment of patients with GC.Citation10
Long noncoding RNAs (lncRNAs), a group of transcripts longer than 200 nucleotides (nt) without protein-coding potential, are emerging as positive or negative regulators and possible biomarkers in various cancers including GC.Citation11,Citation12
For instance, lncRNA XIST level was associated with larger tumor size and advanced pathological status for GC patients, and the silence of XIST1 suppressed GC cell proliferation, migration and invasion in vitro as well as curbed growth and metastasis of GC xenograft tumors in vivo by regulating microRNA-101/EZH2 axis.Citation13 Also, lncRNA linc00628 exerted anti-tumor effect by long-range regulating cell cycle related genes in GC.Citation14 Linc00152 has been well documented as an oncogene in multiple malignancies such as hepatocellular cancer, colon cancer and renal cell cancer.Citation15 Moreover, some evidences indicated that linc00152 has potential diagnostic and prognostic values in plenty of cancers.Citation15 For instance, linc00152 overexpression facilitated cell proliferation, migration, invasion in vitro and accelerated tumor growth in vivo in gallbladder cancer.Citation16 Additionally, linc00152 level was positively associated with lymph node metastasis and higher TNM stage in gallbladder cancer.Citation16
Also, previous studies showed that linc00152 could promote the tumorigenesis and metastasis of GC in vitro and in vivo.Citation17–Citation20 Additionally, linc00152 expression was positively correlated with advanced pathological phenotypes and poor prognosis in GC.Citation19–Citation21 However, the pathogenesis of linc00152 in the development of GC has not been well featured till now.
In the present study, we demonstrated that linc00152 knockdown suppressed GC cell proliferation, migration and invasion in vitro and hampered GC tumor growth in vivo by elevating miR-193b-3p expression and reducing ETS1 (a target of miR-193b-3p) expression.
Materials and methods
Clinical samples and cell culture
Thirty-six pairs of GC tissues and adjacent normal tissues were obtained from primary GC patients undergoing radical gastrectomy at Beijing University of Chinese Medicine Third Affiliated Hospital. Prior to surgery, all enrolled patients did not receive any therapy including chemotherapy and radiotherapy. After surgical resection, these specimens were immediately frozen in liquid nitrogen and then stored at −80°C. Patients were staged following the 2010 UICC/AJCC classification criteria for GC. Our project was performed with the written informed consents from all patients and the approval from the Ethics Committee of our hospital.
GC cell lines AGS and SGC-7901 were purchased from cell bank of China Academy of Sciences (Shanghai, China). Human normal gastric mucosal epithelial cell line (GES-1) was purchased from Shanghai Yubo Biological Technology Co., ltd (Shanghai, China). AGS cells were cultured in F-12K medium (Thermo Fisher Scientific, Rockford, IL, USA) containing 10 % fetal bovine serum (FBS, Thermo Fisher Scientific). SGC-7901 cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with 10 % FBS (Thermo Fisher Scientific).
Reagents and cell transfection
Small interference RNA (siRNA) targeting ETS (si-ETS) or linc00152 (si-linc00152) and their negative control (si-NC), miR-193b-3p mimic and its negative control (miR-NC), miR-193b-3p inhibitor (in-miR-193b-3p) and its negative control (in-miR-NC) were obtained from GenePharma Co., Ltd. (Shanghai, China). Linc00152 overexpression plasmid (pcDNA-linc00152) and its empty vector (pcDNA3.1) were purchased from Sangon Biotech Co., Ltd (Shanghai, China). All siRNAs, miRNAs, miRNA inhibitors and plasmids, alone or in combination, were transfected into AGS and SGC-7901 cells using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) referring to the instructions of manufacturer. The sequences of si-ETS1 and si-linc00152 were listed as follows: si-linc00152, 5ʹ-GGAAUGCAGCUGAAAGAUUTT-3ʹ (sense) and 5ʹ-AAUUUUCAGCUGCAUUCCTT-3ʹ (antisense) .Citation16; si-ETS1, 5ʹ-GCAUUAAAAGCUACUUUCATT-3ʹ (sense) and 5ʹ-UGAAAGUAGCUUUUAAUGCTT −3ʹ (antisense).Citation22
Reverse transcription-quantitative PCR (rt-qpcr) assay
Total RNA was extracted from clinical samples and cells using Trizol reagent (Thermo Fisher Scientific) following the protocols of manufacturer. The expression analysis of miR-193b-3p was conducted using TaqMan MicroRNA Assays (Thermo Fisher Scientific) with U6 snRNA as the endogenous control. For the measurement of linc00152 and ETS1 mRNA levels, RNA was reversely transcribed into cDNA using M-MLV Reverse Transcriptase (Thermo Fisher Scientific) together with random primers and then cDNA was used for qPCR detection by PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific) along with specific quantitative primers. The quantitative PCR primers were presented as follows: 5ʹ-GAAGGTGTCGGCAAGATC-3ʹ (forward) and 5ʹ-TCGGTGTCTGTCATATTCG-3ʹ (reverse) for linc00152, 5ʹ-CTCAGATATGGAATGTGCAG-3ʹ (forward) and 5ʹ-TGCTGTTCTTTAGTGAAACC-3ʹ (reverse) for ETS1, 5ʹ-AAAGACCTGTACGCCAACAC-3ʹ (forward) and 5ʹ-GTCATACTCCTGCTTGCTGAT-3ʹ (reverse) for β-actin. β-actin was employed to normalize the expression of linc00152 and ETS1.
Western blot assay
Cell and tissue samples were lysed with cell lysis solution (RIPA buffer, Cell Signaling Technology, Danvers, MA, USA) supplemented with protease inhibitor cocktail (Thermo Fisher Scientific). Next, the samples were incubated on ice for 30 min and then subjected to high-speed centrifugation in order to obtain cell supernatant containing protein. Subsequently, protein concentration was measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Following this, an equal amount of protein (45 μg/samples) was separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA), and blocked with 5% skimmed milk. Then, the membranes were incubated overnight at 4°C with primary antibody for ETS1 (ab186844, 1/1000 dilution, Abcam, Cambridge, UK) or β-actin (ab8227, 1/2000 dilution, Abcam). Next, the membranes were probed for 2 h at room temperature with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (ab6721, 1/2000 dilution, Abcam).Finally, Immobilon ECL Ultra Western HRP Substrate (Millipore) was used to detect protein signals. And, the gray values of protein bands were quantified using Quantity One Software Version 4.1.1 (BIO-Rad Laboratories, Hercules, CA, USA).
Luciferase reporter assay
The fragment from linc00152 or ETS1 3ʹUTR containing predicted miR-193b-3p binding sites was constructed into psiCHECK-2 luciferase vector (Promega, Madison, WI, USA) to generate wide type linc00152 reporter (linc00152-WT) or ETS1 3ʹUTR reporter (ETS1 3ʹUTR-WT), respectively. Also, linc00152-MUT and ETS1 3ʹUTR-MUT reporters carrying mutant miR-193b-3p binding sites were also constructed using KOD-plus-mutagenesis kit (Toyobo, Osaka, Japan). Then, linc00152-WT, linc00152-MUT, ETS1 3ʹUTR-WT, or ETS1 3ʹUTR-MUT reporter was transfected into AGS and SGC-7901 cells together with miR-NC, miR-193b-3p mimic, in-miR-NC or in-miR-193b-3p. After 48 h of transfection, luciferase activities of linc00152-WT, linc00152-MUT, ETS1 3ʹUTR-WT, or ETS1 3ʹUTR-MUT reporter were detected using a dual luciferase reporter assay kit (Promega) with the renilla luciferase activity as the internal control.
Colony formation assay
Transfected AGS and SGC-7901 cells were seeded into 6-well plates and then incubated for 2 weeks at the incubator. Then, cells were fixed with methanol for 20 min and then stained with 0.1% crystal violet solution (Sigma-Aldrich, St. Louis, MO, USA) for 20 min. Next, the number of colonies containing over 50 cells was counted.
Cell counting kit-8 (CCK-8) assay
Cell proliferative ability was assessed by CCK-8 assay kit (Dojindo Molecular Technologies, Rockville, MD, USA) referring to the protocols of manufacturer. Briefly, after transfected with corresponding siRNAs, miRNAs or plasmids for 24 h, cells were plated into 96-well plates. Then, 10 μl of CCK-8 solution was added into corresponding wells every 24 h for a total of 96 h. At 2 h after CCK-8 treatment, cell absorbance was determined at the wavelength of 450 nm.
Transwell migration and invasion assays
The migratory and invasive abilities of AGS and SGC-7901 cells were evaluated by transwell migration and invasion assays. Briefly, transfected AGS and SGC-7901 cells in serum-free medium were seeded into the upper chamber of 24-well transwell chambers with 8 μm pore size membranes (Costar, Lowell, MA, USA). Moreover, for cell invasion assay, the membranes were precoated with 1 mg/ml matrigel (BD Bioscience, San Jose, CA, USA). Also, the low chamber was filled with 600 μl of medium containing 20% FBS. At 24 h upon incubation, cells at the upper side of membranes were removed, and cells attached to the lower side of membranes were fixed with methanol for 30 min and stained with 0.1 % crystal violet solution for 20 min (Sigma-Aldrich). Then, cells adhering to the undersurface of membranes were photographed and counted in 10 random fields.
Tumorigenesis assay
All animal experiments were approval by the Institutional Animal Care and Use Committee of Beijing University of Chinese Medicine Third Affiliated Hospital. Female BABL/c athymic nude mice (n = 10, 6–8 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) and treated following the national standard for the care and use of laboratory animals. Mice were randomly divided into sh-NC group and sh-linc00152 group with 5 mice in each group. SGC-7901 cells (1 × 107 cells) with the stable knockdown of linc00152 (sh-linc00152) and negative control cells (Hanbio Biotechnology Co., Ltd, Shanghai, China) were subcutaneously injected into the right flanks of sh-linc00152 group and sh-NC group mice, respectively. Tumor volume was monitored every 7 days and calculated referring to the formula: V = (shortest diameter)2 × (longest diameter) × 0.5. Mice were killed on day 35 after injection and xenograft tumors were resected and weighted. Also, expression levels of linc00152, miR-193b-3p and ETS1 mRNA in tumors were measured by RT-qPCR assay and ETS1 protein level in tumors was detected by western blot assay.
Statistical analysis
Results were presented as means ± standard deviations (SD). Student’s t-test or one-way ANOVA was used to conduct difference analysis with Student’s t-test for the comparison of two group data and one-way ANOVA for the comparison of more than two group data. Also, P value below 0.05 was denoted as statistically significant. Kaplan-Meier curve and log-rank test was used to explore the effect of linc00152 on overall survival of GC patients.
Results
Linc00152 expression was notably upregulated in GC tissues and cells
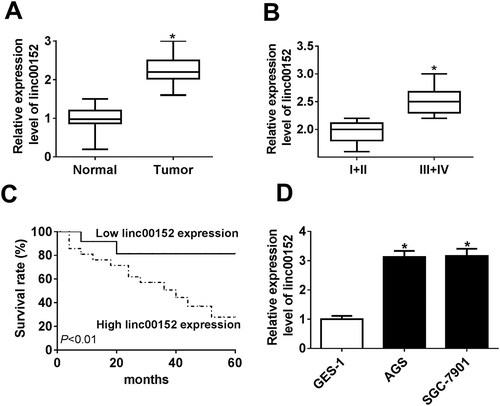
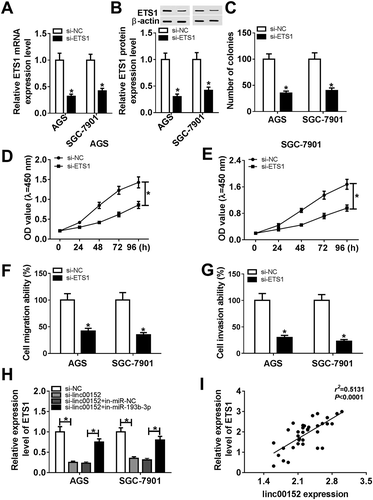
Firstly, we demonstrated that linc00152 was highly expressed in 36 cases of GC tissues than that in adjacent normal tissues (). Moreover, linc00152 level was markedly upregulated in the tumor tissues of patients with advanced GC (III + IV) as compared to that of patients with early GC (I + II) (). Additionally, to further test the association between linc00152 expression and prognosis of GC patients, GC patients were divided into low (n = 14) and high (n = 22) linc00152 expression groups with the median value as a cutoff point (linc00152 level in high linc00152 expression group ≥ median value, linc00152 level in low linc00152 expression group < median value). Following Kaplan-Meier analysis showed that the overall survival rate and survival time of GC patients in high linc00152 expression group was relatively lower than that in low linc00152 expression group, indicating that higher linc00152 expression was associated with poor prognosis for GC patients (). Moreover, a notable increase in linc00152 level was observed in two types of GC cell lines (AGS and SGC-7901) as compared to that in normal gastric epithelial cell line (GES-1) (). In a word, these data indicated that linc00152 might be involved in the development of GC.
Figure 1. Linc00152 expression was notably upregulated in GC tissues and cells. (A, B, D) RT-qPCR assay was performed to examine the level of linc00152 in 36 pairs of GC tissues and adjacent normal tissues (A), different pathological stages (n (I +Ⅱ) = 16, n (Ⅲ + Ⅳ) = 20) (B), different cell lines (GES-1, AGS and SGC-7901) (D). (C) Kaplan-Meier analysis of overall survival for GC patients based on the difference of linc00152 level. *P < 0.05.
The knockdown of linc00152 hampered GC cell proliferation, migration and invasion
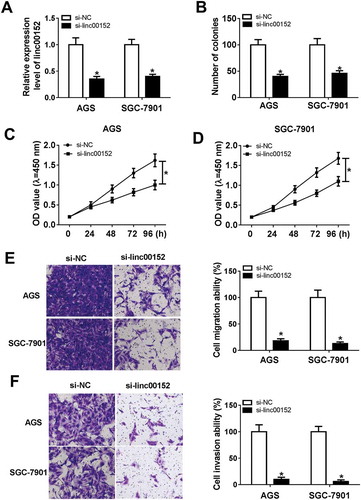
To further explore the roles and molecular mechanisms of linc00152 in the development of GC, si-linc00152 and its negative control (si-NC) were transfected into GC cells. RT-qPCR assay testified that the introduction of si-linc00152 resulted in the remarkable reduction in linc00152 level in AGS and SGC-7901 cells compared with cells transfected with si-NC (), meaning that si-linc00152 could be used for ensuing loss-of-function investigations. Next, colony formation assay showed that the number of colonies was notably reduced in linc00152-depleted AGS and SGC-7901 cells than that in si-NC-transfected cells (). Also, CCK-8 assay further confirmed that the knockdown of linc00152 suppressed the proliferation of AGS and SGC-7901 cells ( and ). Transwell migration and invasion assays also showed that the linc00152 silence resulted in the obvious downregulation of cell migratory and invasive abilities in AGS and SGC-7901 cells ( and ). Taken together, these results showed that linc00152 loss hindered the development of GC cells.
Figure 2. The knockdown of linc00152 hampered GC cell proliferation, migration and invasion. (A-F) AGS and SGC-7901 cells were transfected with si-NC or si-linc00152. (A) At 48 h after transfection, linc00152 level was measured by RT-qPCR assay. (B) At 2 weeks post transfection, the number of cell colonies was determined by colony formation assay. (C and D) Cell proliferation ability was assessed by CCK-8 assay at 0, 24, 48, 72, 96 h after transfection. (E and F) Cell invasion capacity was evaluated by transwell migration and invasion assays at 24 h upon incubation, respectively. *P < 0.05.
Linc00152 inhibited mir-193b-3p expression by direct interaction in GC cells
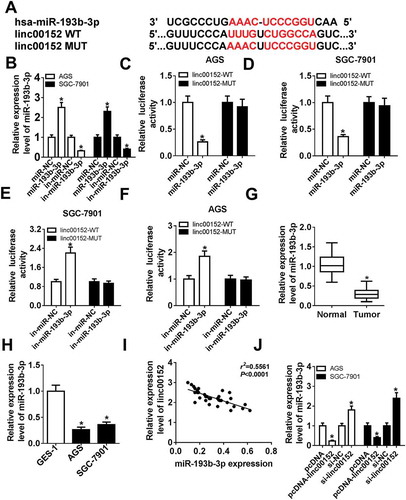
To have a deep insight into the molecular mechanisms of linc00152 in GC progression, starBase online website (http://starbase.sysu.edu.cn/mirLncRNA.php) was used to predict potential miRNAs that have a chance to interact with linc00152. Results showed that there existed some complementary sites between linc00152 and miR-193b-3p (), hinting the possible interaction of linc00152 and miR-193b-3p. To further substantiate this prediction, miR-193b-3p mimic (miR-193b-3p), miR-193b-3p inhibitor (in-miR-193b-3p) and their corresponding controls were synthesized and then transfected into AGS and SGC-7901 cells, followed by the detection of miR-193b-3p level. Results showed that miR-193b-3p level was strikingly increased in AGS and SGC-7901 cells transfected with miR-193b-3p mimic, but was notably reduced in in-miR-193b-3p-transfected AGS and SGC-7901 cells (). Next, the effect of miR-193b-3p increase or reduction on luciferase activity of linc00152-WT or linc00152-MUT reporter was determined by luciferase reporter assay. As presented in -, the overexpression of miR-193b-3p resulted in the noticeable downregulation in the luciferase activity of linc00152-WT reporter, whereas the depletion of miR-193b-3p enhanced the luciferase activity of linc00152-WT reporter. Moreover, the increase or reduction of miR-193b-3p had no much influence on the luciferase activity of linc00152-MUT reporter (-). In other words, the results in - showed that linc00152 could interact with miR-193b-3p by putative binding sites in GC cells. Subsequently, RT-qPCR assay further revealed that miR-193b-3p level was remarkably downregulated in GC tissues () and cell lines () as compared to that in matching controls. Moreover, linc00152 level was inversely associated with miR-193b-3p level in 36 cases of GC tissues (). Additionally, the enforced expression of linc00152 resulted in the noticeable reduction of miR-193b-3p level in AGS and SGC-7901 cells (). Conversely, the depletion of linc00152 promoted miR-193b-3p expression in AGS and SGC-7901 cells (). Taken together, these results showed that linc00152 inhibited miR-193b-3p expression by direct interaction in GC.
Figure 3. Linc00152 suppressed miR-193b-3p expression by direct interaction in GC cells. (A) The complementary sites between linc00152 and miR-193b-3p and mutant sites in linc00152-MUT reporter. (B) AGS and SGC-7901 cells were transfected with miR-NC, miR-193b-3p mimic, in-miR-NC, or in-miR-193b-3p, followed by the measurement of miR-193b-3p level at 48 h upon transfection. (C-F) AGS and SGC-7901 cells with the transfection of linc00152-WT or linc00152-MUT reporter were also transfected with miR-NC, miR-193b-3p mimic, in-miR-NC, or in-miR-193b-3p. At 48 h post transfection, luciferase activities of linc00152-WT or linc00152-MUT reporter were detected by dual luciferase reporter assay. (G and H) MiR-193b-3p level was measured by RT-qPCR assay in 36 pairs of GC tissues and adjacent normal tissues and different cell lines (GES-1, AGS and SGC-7901). (I) Correlation analysis between linc00152 and miR-193b-3p in 36 cases of GC tissues. (J) AGS and SGC-7901 cells were transfected with pcDNA3.1 empty vector, pcDNA-linc00152 overexpression plasmid, si-NC, or si-linc00152, followed by the determination of miR-193b-3p level at 48 h after transfection. *P < 0.05.
The overexpression of linc00152 abrogated the inhibitory effects of mir-193b-3p on cell proliferation, migration and invasion in GC
Next, RT-qPCR assay further demonstrated that the transfection of pcDNA-linc00152 overexpression plasmid induced the prominent upregulation of linc00152 level in AGS and SGC-7901 cells (). Also, functional analyses disclosed that the ectopic expression of miR-193b-3p curbed cell colony formation (), proliferation ( and ), migration () and invasion () in AGS and SGC-7901 cells, while these effects of miR-193b-3p were notably abolished by linc00152 upregulation.
Figure 4. The overexpression of linc00152 weakened the inhibitory effects of miR-193b-3p on cell proliferation, migration and invasion in GC. (A) AGS and SGC-7901 cells were transfected with pcDNA3.1 empty vector or pcDNA-linc00152 overexpression plasmid, followed by the measurement of linc00152 level at 48 h upon transfection. (B-F) AGS and SGC-7901 cells were transfected with miR-NC, miR-193b-3p mimic, miR-193b-3p mimic + pcDNA3.1 empty vector, or miR-193b-3p mimic + pcDNA-linc00152. (B) Colony formation assay was performed at 2 weeks after transfection. (C and D) Cell proliferative capacity was evaluated by CCK-8 assay at 0, 24, 48, 72, 96 h after transfection. (E and F) Cell migration and invasion capacities were examined by transwell migration and invasion assays at 24 h upon incubation, respectively. *P < 0.05.

ETS1 was a target of mir-193b-3p
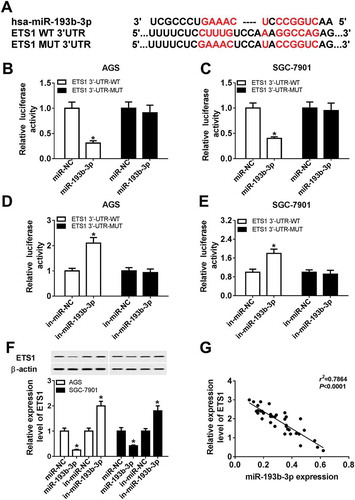
Next, miRTarBase online analysis (http://mirtarbase.mbc.nctu.edu.tw/php/search.php) was performed to search for possible targets of miR-193b-3p. Among numerous candidate targets of miR-193b-3p, ETS1 was chosen due to its significant roles in the development of various cancers.Citation23,(. Following luciferase reporter assay further showed that the luciferase activity of ETS1 3ʹUTR-WT reporter was notably reduced in miR−193b−3p-overexpressed AGS and SGC−7901 cells, but was markedly increased in AGS and SGC−7901 cells transfected with miR−193b−3p inhibitor (-. However, upregulation or downregulation of miR−193b−3p had no much impact on the luciferase activity of ETS1 3ʹUTR-MUT reporter in AGS and SGC−7901 cells (-, denoting that miR−193b−3p could interact with ETS1 3ʹUTR by putative binding sites. Also, western blot assay further revealed that the ectopic expression of miR−193b−3p resulted in the reduction of ETS1 protein level in AGS and SGC−7901 cells (. Conversely, the depletion of miR−193b−3p induced ETS1 expression in AGS and SGC−7901 cells (. Additionally, correlation analysis showed that ETS1 level was negatively associated with miR−193b−3p level in 36 cases of GC tissues (). In a word, these data showed that ETS1 was a target of miR−193b−3p in GC.
Figure 5. ETS1 was a target of miR-193b-3p. (A) The complementary sites between miR-193b-3p and ETS1 3ʹUTR and mutant sites in ETS1 3ʹUTR-MUT reporter. (B-E) AGS and SGC-7901 cells were co-transfected with ETS1 3ʹUTR-WT or ETS1 3ʹUTR-MUT reporter and miR-NC, miR-193b-3p mimic, in-miR-NC, or in-miR-193b-3p. Then, luciferase activities of ETS1 3ʹUTR-WT or ETS1 3ʹUTR-MUT reporter were determined at 48 h post transfection. (F) AGS and SGC-7901 cells were transfected with miR-NC, miR-193b-3p mimic, in-miR-NC, or in-miR-193b-3p, followed by the measurement of ETS1 protein level at 48 h post transfection. (G) Correlation analysis of ETS1 and miR-193b-3p expressions in 36 cases of GC tissues. *P < 0.05.
ETS1 knockdown inhibited GC cell proliferation, migration and invasion
Next, si-ETS1 and its negative control si-NC were synthesized and transfected into AGS and SGC-7901 cells. RT-qPCR assay firstly confirmed that the transfection of si-ETS1 resulted in the remarkable downregulation of ETS1 expression at mRNA () and protein () levels in AGS and SGC-7901 cells, manifesting that si-ETS1 could be used for subsequent loss-of-function explorations. Colony formation assay showed that ETS1 silence resulted in the noticeable reduction of colony number in AGS and SGC-7901 cells (). CCK-8 assay further confirmed that ETS1 knockdown inhibited the proliferation of AGS and SGC-7901 cells ( and ). Moreover, a conspicuous downregulation of cell migratory and invasive capacities was observed in ETS1-silenced AGS and SGC-7901 cells ( and ). In summary, these outcomes disclosed that ETS1 knockdown hampered the development of GC. Moreover, we further demonstrated that si-linc00152-mediated linc00152 depletion triggered the prominent downregulation of ETS1 mRNA level, whereas the loss of miR-193b-3p abrogated the inhibitory effect of linc00152 knockdown on ETS1 mRNA expression in AGS and SGC-7901 cells (), suggesting that linc00152 could promote ETS1 expression by sequestering miR-193b-3p from ETS1 like a “sponge”. Additionally, ETS1 expression was positively associated with linc00152 expression in 36 cases of GC tissues ().
Figure 6. ETS1 knockdown inhibited GC cell proliferation, migration and invasion. (A-G) AGS and SGC-7901 cells were transfected with si-NC or si-ETS1. (A and B) ETS1 mRNA and protein levels were determined at 48 h post transfection. (C) The number of colonies was measured by colony formation assay at 2 weeks after transfection. (D and E) Cell proliferative capacity was detected at 0, 24, 48, 72, 96 h post transfection. (F and G) Cell migratory and invasive abilities were examined at 24 h upon incubation. (H) AGS and SGC-7901 cells were transfected with si-NC, si-linc00152, si-linc00152 + in-miR-NC, or si-linc00152 + in-miR-193b-3p, followed by the measurement of ETS1 mRNA level at 48 h after transfection. (I) Correlation analysis between ETS1 and linc00152 levels in 36 cases of GC tissues. *P < 0.05.
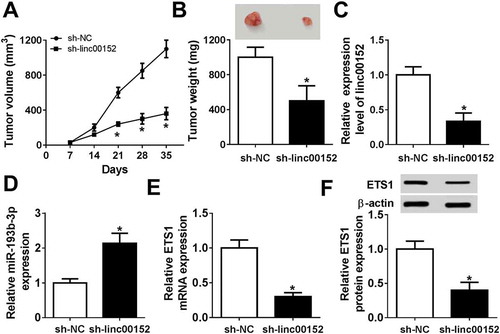
The knockdown of linc00152 inhibited the growth of GC xenograft tumors by upregulating mir-193b-3p and downregulating ETS1 in vivo
Next, we further demonstrated that the depletion of linc00152 inhibited the growth of GC xenograft tumors, as evidenced by the reduction of tumor volume () and weight () in linc00152-slienced mouse xenograft models of GC. Moreover, linc00152 level was markedly reduced in the tumors with the stable knockdown of linc00152 (). Also, the knockdown of linc00152 resulted in the obvious upregulation of miR-193b-3p level () and the remarkable downregulation of ETS1 mRNA () and protein () levels in xenograft tumors of GC.
Figure 7. The knockdown of linc00152 inhibited the growth of GC xenograft tumors by upregulating miR-193b-3p and downregulating ETS1 in vivo. SGC-7901 cells (1 × 107 cells) with the stable knockdown of linc00152 (sh-linc00152) and negative control cells (sh-NC) were subcutaneously injected into the right flanks of sh-linc00152 group and sh-NC group mice, respectively. (A) Tumor volume was measured every 7 days upon injection. (B) At 35 day after injection, tumors were excised and weighted. (C-E) Expression levels of linc00152, miR-193b-3p and ETS1 mRNA in tumors were measured by RT-qPCR assay on day 35 after injection. (F) ETS1 protein level in tumors was determined by western blot assay on day 35 upon injection. *P < 0.05.
Discussion
Over the past decades, emerging evidence shows that non-coding RNAs (ncRNAs) including lncRNAs and microRNAs (miRNAs) function as critical players in various cellular processes relevant to human health and disease including cancers.Citation24,Citation25 Moreover, mounting lncRNAs and miRNAs have been reported to be implicated in the tumorigenesis, diagnosis and treatment of GC.Citation26,Citation27
Linc00152 has been reported to be highly expressed in GC tissues and cells.Citation17,Citation18,Citation20,Citation28 Also, subcellular location analysis showed that linc00152 existed in the cytoplasm.Citation18 Moreover, prior studies showed that the knockdown of linc00152 suppressed GC cell proliferation and the growth of GC xenograft tumors by regulating miR-193a-3p/MCL1 axis and EGFR-mediated PI3K/AKT pathway.Citation17,Citation18 Additionally, the depletion of linc00152 repressed cell proliferation, epithelial-to-mesenchymal transition (EMT), migration and invasion and induced apoptosis in GC.Citation20 Consistent with earlier reports Citation17,Citation18,Citation20,Citation28, our data showed that linc00152 expression was markedly upregulated in GC tissues and cells. Also, higher linc00152 expression was associated with advanced pathological phenotypes and poor overall survival of GC patients, which was in line with prior data .Citation17,Citation19. Functional analysis showed that the silence of linc00152 resulted in the marked downregulation of cell proliferative, migratory and invasive capacities and the notable reduction of xenograft tumor size and weight in GC, which was in accordance with previous findings.Citation17,Citation18,Citation20
It has been well documented that lncRNAs can exert “sponge-like” effects on many specific miRNAs, resulting the downregulation of miRNA levels and inhibition of miRNA-mediated functions.Citation24,Citation29 In the present study, we further demonstrated that linc00152 could function as a molecular sponge of miR-193b-3p, resulting in the reduction of miR-193b-3p level in GC.
MiR-193b-3p, a mature miRNA derived from the 3ʹ arm of miR-193b precursor (pre-miR-193b), has been widely reported as a tumor suppressor in multiple cancers such as ovarian cancerCitation30, neuroblastoma.Citation31 and acute lymphoblastic leukemia.Citation32 For instance, the enforced expression of miR-193b-3p suppressed cell proliferation and promoted cell apoptosis by targeting KRAS in pancreatic cancer.Citation33 The upregulation of miR-193b-3p inhibited cell proliferation and invasion in colorectal cancer by targeting stathmin 1.Citation34 However, the carcinogenic effect of miR-193b-3p also has been reported in some cancers. For instance, Zhong et al. pointed out that miR-193b-3p facilitated cell proliferation by targeting smad3 in glioma.Citation35 Also, Lenarduzzi et al.disclosed that miR-193b silence suppressed cell proliferation, migration and invasion in vitro and hampered tumor formation in vivo by targeting neurofibromin 1 in head and neck squamous cell carcinomas (HNSCC).Citation36 Additionally, Wang et al. showed that miR-193b level was strikingly reduced in GC tissues and lower miR-193b level was associated with more aggressive GC phenotypes.Citation37 Also, the ectopic expression of miR-193b could hamper cell proliferation, migration and invasion in GC.Citation37 Our study demonstrated that miR-193b-3p was low expressed in GC tissues and cells. Also, the level of miR-193b-3p was negatively associated with linc00152 level in GC tissues. Moreover, the overexpression of miR-193b-3p suppressed proliferation, migration and invasion in GC cells. Further analyses revealed that linc00152 upregulation could weaken miR-193b-3p-mediated anti-proliferation, anti-migration and anti-invasion effects in GC cells.
Next, we further confirmed that E26 oncogene homolog 1 (ETS1) was a target of miR-193b-3p in GC by bioinformatics test, luciferase reporter assay, western blot assay and correlation analysis. Also, ETS1 as a target of miR-193b has been reported in hepatocellular cancer and urothelial cancer cells.Citation38,Citation39
ETS1, a member of ETS family, can functionally and physically interact with many proteins including transcriptional factors, kinases and acetyl transferases.Citation40 Also, the dysregulation of ETS1 plays vital roles in numerous biological processes such as haematopoietic development, angiogenesis and apoptosis.Citation40 Moreover, ETS1 is abnormally expressed in lots of cancers and is implicated in tumour progression and malignant transformation (including proliferation, invasion, and metastasis).Citation23 Additionally, ETS1 has been widely reported to be an oncogene in multiple cancers. For instance, ETS1overexpression induced cell migration and invasion in vitro and liver metastases in vivo in non-small cell lung cancer.Citation41 The knockdown of ETS1 inhibited cell proliferation, migration and invasion in ovarian cancer.Citation42 Also, prior studies manifested that ETS1 expression was notably upregulated in GC tissues and cells .Citation22,Citation43 and associated with aggressive tumor behaviors and poor prognosis in GC.Citation44,Citation45 Moreover, Taniguchi et al. substantiated that the decoy of ETS1 markedly inhibited GC cell proliferation and GC peritoneal dissemination.Citation46 ETS1 knockdown resulted in the reduction of cell proliferative and invasive abilities in AGS.Citation22 However, Zheng et al. showed that ETS1 silence suppressed cell migration and invasion, but had no effect on miR-9-mediated proliferation inhibition in GC.Citation43
Our data further unveiled that cell proliferative, migratory and invasive capacities were markedly attenuated in ETS1-silenced GC cells. Moreover, linc00152 could sequester miR-193b-3p from ETS1 and relieve the inhibitory effect of miR-193b-3p on ETS1 expression, resulting in the elevation of ETS1 level in GC cells. Moreover, ETS1 level was positively associated with linc00152 level in GC tissues. In vivo experiments further demonstrated that the knockdown of ETS1 inhibited tumor growth by upregulating miR-193b-3p expression and downregulating ETS1 expression in mouse xenograft models of GC.
Conclusions
Collectively, our data showed that linc00152 inhibited the development of GC by regulating miR-193b-3p/ETS1 axis, deepening our understanding on pathogenesis of GC and providing some possible targets or biomarkers for the diagnose, treatment and prognosis of GC. To further confirm our conclusion, the effect of linc00152, miR-193b-3p and ETS1, alone or in combination, on GC progression need to further investigated in other GC cells including undifferentiated or low-differentiated GC cells. Also, the effect of linc00152, miR-193b-3p and ETS1, alone or in combination, on GC tumor growth and metastasis remains to be deeply explored in vivo.
Abbreviations
Gastric cancer (GC); Long non-coding RNA (lncRNA); microRNA; 193b-3p (miR-193b-3p); nucleotides (nt); Small interference RNA (siRNA); Linc00152 overexpression plasmid (pcDNA-linc00152); standard deviations (SD); non-coding RNAs (ncRNAs); miR-193b precursor (pre-miR-193b); head and neck squamous cell carcinomas (HNSCC)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
Not applicable
References
- Nie Y, Wu K, Yu J, Liang Q, Cai X, Shang Y, Zhou J, Pan K, Sun L, Fang J. A global burden of gastric cancer: the major impact of China. Expert Rev Gastroenterol Hepatol. 2017;11:651–661. doi:10.1080/17474124.2017.1312342.
- Balakrishnan M, George R, Sharma A, Graham DY. Changing trends in stomach cancer throughout the world. Curr Gastroenterol Rep. 2017;19:36. doi:10.1007/s11894-017-0575-8.
- Ajani JA, D’Amico TA, Almhanna K, Bentrem DJ, Chao J, Das P, Denlinger CS, Fanta P, Farjah F, Fuchs CS. Gastric cancer, version 3.2016; clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2016;14:1286–1312.
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin D, Forman D, Bray F. Globocan 2012 v1.1, cancer incidence and mortality worldwide: IARC cancerbase No. 11. Intl J Cancer. 2014;136:E359–E86. doi:10.1002/ijc.29210.
- De ML, Lardiã¨Re-Deguelte S, Volet J, Kianmanesh R, Bouchã© O. Recent insights in the therapeutic management of patients with gastric cancer. Dig Liver Dis. 2016;48:984–994. doi:10.1016/j.dld.2016.04.010.
- Bernards N, Creemers GJ, Nieuwenhuijzen GA, Bosscha K, Pruijt JF, Lemmens VE. No improvement in median survival for patients with metastatic gastric cancer despite increased use of chemotherapy. Ann Oncol. 2013;24:3056–3060. doi:10.1093/annonc/mdt401.
- Thomassen I, van Gestel YR, Van RB, Luyer MD, Bosscha K, Nienhuijs SW, Lemmens VE, de Hingh IH. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Intl J Cancer. 2013;134:622–628. doi:10.1002/ijc.28373.
- Van CE, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;57:2654–2664.
- Riihimäki M, Hemminki A, Sundquist K, Sundquist J, Hemminki K. Metastatic spread in patients with gastric cancer. Oncotarget. 2016;7:52307–52316. doi:10.18632/oncotarget.10740.
- Sawaki K, Kanda M, Kodera Y. Review of recent efforts to discover biomarkers for early detection, monitoring, prognosis, and prediction of treatment responses of patients with gastric cancer. Expert Rev Gastroenterol Hepatol. 2018;12:657–670. doi:10.1080/17474124.2018.1489233.
- Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi:10.1186/s12943-016-0530-6.
- Yang Z, Guo X, Li G, Shi Y, Li L. Long noncoding RNAs as potential biomarkers in gastric cancer: opportunities and challenges. Cancer Lett. 2016;371:62–70. doi:10.1016/j.canlet.2015.11.011.
- Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Feng W, Qiu MZ, Wang DS. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi:10.1186/s13046-016-0444-6.
- Zhang ZZ, Zhao G, Zhuang C, Shen YY, Zhao WY, Xu J, Wang M, Wang CJ, Tu L, Cao H. Long non-coding RNA LINC00628 functions as a gastric cancer suppressor via long-range modulating the expression of cell cycle related genes. Sci Rep. 2016;6:27435. doi:10.1038/srep27435.
- Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC00152: A pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50:e12349.
- Cai Q, Wang ZQ, Wang SH, Li C, Zhu ZG, Quan ZW, Zhang WJ. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am J Transl Res. 2016;8:4068–4081.
- Huang Y, Luo H, Li F, Yang Y, Ou G, Ye X, Li N. LINC00152 down-regulated miR-193a-3p to enhance MCL1 expression and promote gastric cancer cells proliferation. Biosci Rep. 2018;38:BSR20171607. doi:10.1042/BSR20171607.
- Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J, Wang J, Zhang Q, Xu Z. Linc00152 promotes proliferation in gastric cance r through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135. doi:10.1186/s13046-015-0250-6.
- Chen WM, Huang MD, Sun DP, Kong R, Xu TP, Xia R, Zhang EB, Shu YQ. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7:9773–9787. doi:10.18632/oncotarget.6949.
- Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi:10.1080/15384101.2015.1078034.
- Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumor Biol. 2014;35:5441–5447. doi:10.1007/s13277-014-1709-3.
- Chou NH, Lo YH, Wang KC, Kang CH, Tsai CY, Tsai KW. MiR-193a-5p and −3p play a distinct role in gastric cancer: miR-193a-3p suppresses gastric cancer cell growth by targeting ETS1 and CCND1. Anticancer Res. 2018;38:3309–3318. doi:10.21873/anticanres.12596.
- Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38. doi:10.1016/j.semcancer.2015.09.010.
- Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H, Tang YL, Kim IM. Crosstalk between long noncoding RNAs and MicroRNAs in health and disease. Int J Mol Sci. 2016;17:356. doi:10.3390/ijms17030356.
- Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi:10.1152/physrev.00041.2015.
- Yang Q, Zhang RW, Sui PC, He HT, Ding L. Dysregulation of non-coding RNAs in gastric cancer. World J Gastroenterol. 2015;21:10956–10981. doi:10.3748/wjg.v21.i39.10956.
- Xie SS, Jin J, Xu X, Zhuo W, Zhou TH. Emerging roles of non-coding RNAs in gastric cancer: pathogenesis and clinical implications. World J Gastroenterol. 2016;22:1213–1223. doi:10.3748/wjg.v22.i3.1213.
- Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658–3664. doi:10.3748/wjg.v19.i23.3658.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi:10.1038/nature12986.
- Zhang J, Qin J, Su Y. miR-193b-3p possesses anti-tumor activity in ovarian carcinoma cells by targeting p21-activated kinase 3. Biomede Pharmacother. 2017. doi:10.1016/j.biopha.2017.11.086.
- Roth SA, Hald ØH, Fuchs S, Løkke C, Mikkola I, Flægstad T, Schulte J, Einvik C. MicroRNA-193b-3p represses neuroblastoma cell growth via downregulation ofCyclin D1,MCL-1andMYCN. Oncotarget. 2018;9:18160–18179. doi:10.18632/oncotarget.24793.
- Mets E, Meulen JVD, Peer GV, Boice M, Mestdagh P, Walle IVD, Lammens T, Goossens S, Moerloose BD, Benoit Y. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:798–806. doi:10.1038/leu.2014.276.
- Jin X, Sun Y, Yang H, Li J, Yu S, Chang X, Lu Z, Chen J. Deregulation of the MiR-193b-KRAS axis contributes to impaired cell growth in pancreatic cancer. PLoS One. 2015;10:e0125515. doi:10.1371/journal.pone.0125515.
- Guo F, Luo Y, Mu YF, Qin SL, Qi Y, Qiu YE, Zhong M. miR-193b directly targets STMN1 and inhibits the malignant phenotype in colorectal cancer. Am J Cancer Res. 2016;6:2463–2475.
- Zhong Q, Wang T, Lu P, Zhang R, Zou J, Yuan S. miR-193b promotes cell proliferation by targeting Smad3 in human glioma. J Neurosci Res. 2014;92:619–626. doi:10.1002/jnr.23338.
- Lenarduzzi M, Hui AB, Alajez NM, Shi W, Williams J, Yue S, O’Sullivan B, Liu FF. MicroRNA-193b enhances tumor progression via down regulation of neurofibromin 1. PLoS One. 2013;8:e53765. doi:10.1371/journal.pone.0053765.
- Wang L, Zhang Y, Zhao L, Liu S, Yu S, Ma Y, Sun G. MicroRNA-193b inhibits the proliferation, migration and invasion of gastric cancer cells via targeting cyclin D1. Acta Histochem. 2016;118:323–330. doi:10.1016/j.acthis.2016.02.001.
- Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, Xing R, Jin Y, Sun Z, Zheng X. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;46:2828–2836. doi:10.1016/j.ejca.2010.06.127.
- Lin SR, Yeh HC, Wang WJ, Ke HL, Lin HH, Hsu WC, Chao SY, Hour TC, Wu WJ, Pu YS. MiR‐193b mediates CEBPD‐induced cisplatin sensitization through targeting ETS1 and cyclin D1 in human urothelial carcinoma cells. J Cell Biochem. 2016;118:1563–1573. doi:10.1002/jcb.25818.
- Dittmer R. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi:10.1186/1476-4598-2-29.
- Zhou X, Zhou R, Zhou H, Li Q, Hong J, Meng R, Zhu F, Zhang S, Dai X, Peng G. ETS-1 induces endothelial-like differentiation and promotes metastasis in non-small cell lung cancer. Intl J Exp Cell Physiol Biochem Pharmacol. 2018;45:1827–1839. doi:10.1159/000487874.
- Tomar S, Plotnik JP, Haley J, Scantland J, Dasari S, Sheikh Z, Emerson R, Lenz D, Hollenhorst PC, Mitra AK. ETS1 induction by the microenvironment promotes ovarian cancer metastasis through focal adhesion kinase. Cancer Lett. 2018;414:190–204.
- Zheng L, Qi T, Yang D, Qi M, Li D, Xiang X, Huang K, Tong Q. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS One. 2013;8:e55719. doi:10.1371/journal.pone.0055719.
- Tsutsumi S, Kuwano H, Nagashima N, Shimura T, Mochiki E, Asao T. Ets-1 expression in gastric cancer. Hepatogastroenterol. 2005;52:654–656.
- Yu Y, Zhang YC, Zhang WZ, Shen LS, Hertzog P, Wilson TJ, Xu DK. Ets1 as a marker of malignant potential in gastric carcinoma. World J Gastroenterol. 2003;9:2154–2159.
- Taniguchi H, Fujiwara Y, Doki Y, Sugita Y, Sohma I, Miyata H, Takiguchi S, Yasuda T, Tomita N, Morishita R. Gene therapy using ets‐1 transcription factor decoy for peritoneal dissemination of gastric cancer. Intl J Cancer. 2007;121:1609–1617. doi:10.1002/ijc.22870.
