ABSTRACT
Eosinophilia is a severe disease with increased eosinophil count. The transcript of FIP1L1-PDGFRA fusion gene is a genetic biomarker of clonal eosinophilia screened routinely by reverse transcript PCR (RT-PCR) during diagnosis. Another significant genetic biomarker is the PDGFRA gene alone as some of its mutations are targets of imatinib. In this study, we identified a patient who had typical symptoms of Eosinophilia but had no response to the first-line treatment of hormonotherapy. This patient also showed bone rupture and eosinophil bone infiltration, which are extremely rare among all known eosinophilia patients. We identified the FIP1L1-PDGFRA fusion gene via RT-PCR and Sanger sequencing. Using next generation sequencing (NGS), we detected point mutations in PDGFRA, MYOM2, and ASXL3. The patient then received imatinib therapy, leading to the complete disappearance of FIP1L1-PDGFRA fusion gene and mutated MYOM2. The level of PDGFRA point mutation was also decreased from pre-treatment: 57.86% down to 42.99% at 6 months and to 38.80% at one-year after treatment. The level of ASXL3 mutations did not change significantly. To the best of our knowledge, this is the first case in which the point mutation of PDGFRA has been identified at p.P6L in exon 2, likely making it sensitive to imatinib and thus should be further studied as a potential new molecular target of imatinib therapy.
Introduction
Hypereosinophilic syndromes (HES) is a group of rare and heterogeneous disorders, characterized with sustained and significant peripheral eosinophilia (> 1.5 × 109/L for more than six months) for unknown reasons, eosinophil-mediated tissue injury and organ damage, and lacking any other causes of eosinophilia such as malignant, allergic or parasitic disorders.Citation1 HES mainly involves the skin, lungs, heart and nervous system. Other reported complications include hepatitis-splenomegaly, eosinophilic gastroenteritis, and coagulation disorders.Citation2-Citation4 The FIP1L1-PDGFRA fusion gene (F/P+ variant) is considered the benchmark molecular biomarker and has recently proven to be sensitive to treatment with tyrosine-kinase inhibitor drugs, such as imatinib (Gleevec and other brand names).Citation5,Citation6 Since it has been difficult to identify a gene mutation or to prove eosinophil clonality in association with peripheral eosinophilia, it has been difficult to diagnose HES accurately.Citation7
Clonal eosinophilia, a subgroup of HES, mainly results from PDGFRA, PDGFRB or FGFR1 gene rearrangements or point mutations.Citation8 The most common cause is the formation of a FIP1L1-PDGFRA fusion gene caused by the deletion of genetic material between FIP1L1 and PDGFRA genes.Citation9 It encodes the FIP1L1-PDGFRA fusion protein, which is a receptor of platelet-derived growth factor.Citation9 The fusion receptor is constantly active even without the presence of platelet-derived growth factor since the first 29 amino acids of the FIP1L1 protein are able to activate the kinase domain of PDGFRα. Activation leads to constant phosphorylation of the receptor on a tyrosine that causes the activation of the entire subsequent signal pathway, resulting in the transformation of hematopoietic cells to a continuous growth state.Citation10 Fusion of these two genes also induces the overexpression of PDGFRA, which also causes hematopoietic cell over proliferation.Citation11 One of the consequences of such a modification is eosinophilia, which makes FIP1L1-PDGFRA fusion gene a diagnostic biomarker of eosinophilia.Citation9,Citation10
The most common front-line chemotherapy to treat eosinophilia uses the drugs prednisone and hydroxyurea.Citation9 Second-line drugs including vincristine, pulsed chlorambucil, cyclophosphamide, and etoposide also have shown therapeutic effectiveness, but these drugs are also cytotoxic to normal cells and sometimes cause severe side effects poorly tolerated by patients.Citation9 The PDGFRA gene encodes one of the family of PDGF kinases, PDGFRα, which is a known target of imatinib.Citation10,Citation12 Targeted therapy drugs including imatinib have higher efficacy towards eosinophilia patients with specific mutations and much milder side effects during long term treatments.Citation9
In this study, we report a HES patient suffering primarily from bone pain and bone lesions, which is very rare in HES. Also, for the first time, we report that a PDGFRA point mutation was detected at p.P6L in exon 2 in this patient. We also demonstrated for the first time that this mutation is sensitive to imatinib therapy. The identification of this novel molecular target may provide important insights to the personalized therapy of eosinophilia and related hematological diseases.
Clinical case report
A 28-year-old male who had experienced sore throat and repetitive cough as well as slight fever, fatigue and night sweating for half a year went to a local hospital for medical attention. His peripheral blood eosinophilia count reached as high as 12.5 × 109/L, which is significantly higher than normal eosinophil counts in peripheral blood that range between 0.05 and 0.5 × 109/L. For the next three months, his peripheral blood eosinophilia count was sustained at about 12 × 109/L, and he also started to feel sternocostal pain in addition to the symptoms described above. Prednisone at 60 mg/day (1 mg/kg/day) was administrated for 3 months, but resulted in no improvement (, ). Six months after initial diagnosis, he was transferred to our hospital in December 2016. At the time of arrival, he was found with enlargement of superficial lymph nodes, sternal tenderness and multiple areas of rib bone bumps. Two of the rib bone bumps showed red and itchy inflammation. No obvious abnormal signs were found in the lungs or with cardiac auscultation. Laboratory examination showed no exact evidence of allergic disorders, autoimmune disease or parasites.
Table 1. Hematology test results of peripheral blood prior to and after treatments.
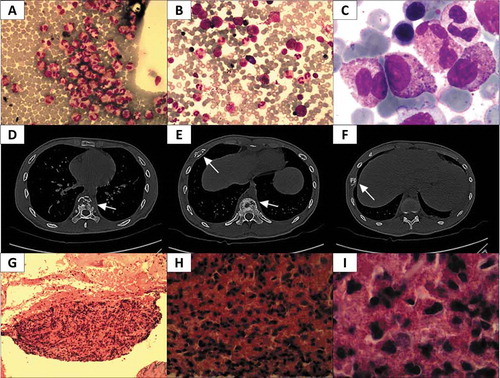
Figure 1. Summary of CT scan and pathological assessment. A Image of Wright stained peripheral blood smear (200x); a large number of eosinophils (red) were observed. B-C Wright-Giemsa stained bone marrow smear (B, 200x; C, 1000x); a large amount of orange eosinophil was observed. D-F Pictures of CT scans at different heights. In D, osteolytic lesions were observed in vertebrae (arrow); E reveals osteolytic lesions in a rib and the vertebrae (arrows); in F, osteolytic lesions were found in another rib (arrow). G-I Pictures of tenderness area rib biopsy stained with hematoxylin/eosin (G, 100x; H, 400x; I, 1000x), numerous orange or red eosinophils were observed.
Color doppler ultrasound showed enlargement of cervical, axillary, and inguinal lymph nodes. Splenomegaly was found with a size of 16.2 cm × 5.7 cm. Liver, gallbladder, pancreas and lymph nodes in the abdominal cavity and behind the peritoneum were normal. Echocardiography indicated no abnormality of form, structure, functionality or valve activities.
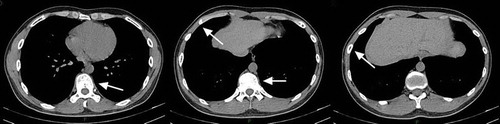
A computed tomography (CT) scan showed the manifestations of bilateral bronchitis, subpleural proliferation in the lower lobe of the right lung, moderate pericardial effusion, splenomegaly (greater than 7 ribs), and the bone window showed multiple areas of bone damage. Positron emission tomography–computed tomography (PET-CT) showed that bone metabolism increased, partly due to bone changes and destruction. No obvious malignant tumor lesions were observed in the rest of the body. A CT scan with 3D reconstruction showed multiple sites of osteoid destruction in ribs, thoracic vertebrae, sternum, right clavicle, right shoulder peak, shoulder blade, left scapula and bilateral humeral head, and indicated pathologic fracture of multiple ribs ().
Hematology testing, including peripheral blood smear, bone marrow smear, bone marrow biopsy and analysis of bone marrow specimens, all indicated a predominant eosinophilic component without blast cells or monoclonal lymphocytes. Subsequently, tissue samples were taken from ribs with an osteolytic lesion area and evaluated by a pathologist. The biopsy revealed apparent fibrous proliferation between bone trabeculae and plenty of diffuse eosinophils (). No lymphocyte count increase was observed during the pathological evaluation. It is likely that the bone lesions were caused by eosinophil infiltration into the bones.
Testing by fluorescence in situ hybridization (FISH) for PDGFRA, PDGFRB and FGFR1 rearrangement in bone marrow specimens was negative. Further molecular biological detection for rearrangements and point mutations in BCR-ABL, FIP1L1-PDGFRA, TEL-PDGFRB, JAK2, MPL, CALR, and c-KIT were performed by real-time fluorescent quantitative PCR and Sanger sequencing. Our results showed that all were negative except FIP1L1-PDGFRA fusion, which was detected at a low level (3.84%).
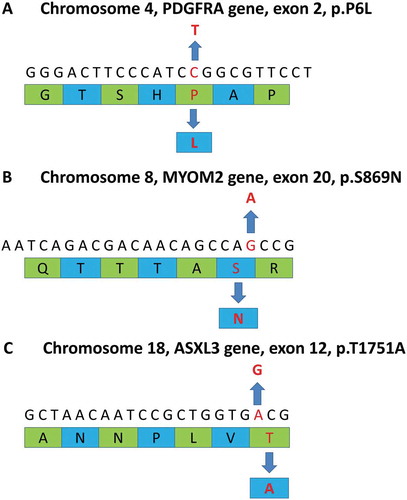
To obtain a comprehensive genetic profile, whole-genome sequencing was carried out on this patient’s bone marrow sample that was collected prior to treatment at the Shanghai Ositai Medical Technology Center. By this method, we detected a PDGFRA mutation at p.P6L in exon 2 and the rate of the mutation was 57.86% (). Point mutations at p.S869N in exon 20 of the MYOM2 gene were also found with a frequency of 100% mutation (). Another point mutation was also detected on p.T1751A in exon 12 of the ASXL3 gene at the rate of 49.02% (). All these mutations are missense mutations which lead to alterations in their corresponding amino acid sequences. None of these mutations have been reported in any literature related to hematological or lymphatic system disorders.
Figure 2. Summary of whole exome sequencing. The upper sequences in each diagram represent gene sequences and lower green and blue strips are amino acid sequences. A The diagram reveals a point mutation (C→T) at p.P6L in exon 2 of the PDGFRA gene. The point mutation is a missense mutation in chromosome 4 which alters its corresponding amino acid (P→L). B The diagram indicates a point mutation (G→A) at p.S869N in exon 20 of the MYOM2 gene. The point mutation is a missense mutation in chromosome 8 that leads to a change in the corresponding amino acid (S→N). C The diagram shows a point mutation (A→G) at p.T1751A in exon 12 of the ASXL3 gene. This is a missense mutation on chromosome 18 causing a substitution of its corresponding amino acid (T→A).
Based on the clinical manifestations and the results of laboratory examinations, this patient was diagnosed with clonal eosinophilia. The patient was then treated with imatinib at 100 mg/day (1.67 mg/kg/day)accompanying a decreased dose of prednisone at 30 mg/day (0.5 mg/kg/day). Four days later, the eosinophil level was normal. One week after treatment, the symptoms that had persisted for 6-months, including cough, slight fever, night sweating, fatigue and sternocostal pain, were all relieved and the eosinophil level had been maintained at a normal level. The patient recovered quickly and was soon discharged with a prescription for continued imatinib at 100 mg/day without any other medicine.
This patient was monitored routinely and his once-a-month blood examinations showed stable and normal eosinophil counts. Six months after discharge, he underwent a return examination and his hematology tests showed that all his original abnormal parameters had recovered or decreased to a level close to reference ranges (). His CT scan also revealed that all his osteolytic lesions and pathological fractures had recovered () and his spleen size returned to normal. Bone marrow RT-PCR showed no FIP1L1-PDGFRA fusion gene expression. Whole-genome sequencing during his 6-month follow-up visit indicated that the rate of PDGFRA mutation at p.P6L in exon 2 decreased from 57.86% (pre-treatment) to 42.99% (post-treatment). At one-year follow-up, his blood cell count and spleen size were normal, his osteolytic lesions and pathological fractures were gone, and finally the rate of PDGFRA mutation at p.P6L in exon 2 had dropped to 38.80% (post-treatment). Moreover, MYOM2 mutations had vanished completely after one year of treatment. However, the ASXL3 mutation rate was not changed significantly, only dropping slightly from 49.02% to 42.76%. Currently, the patient feels perfectly normal without any discomfort from treatment with imatinib.
Discussion
Hypereosinophilic syndrome (HES) is a rare disorder characterized by chronic eosinophilia with tissue damage, excluding eosinophilia associated with parasitic, allergic and neoplastic diseases. Although almost any tissue or organ can be involved in HES, clinical complications in more than 50% of patients were found in skin, heart, lung, digestive tract and nervous system. In this study, we detected a novel PDGFRA point mutation (p.P6L in Exon 2) along with a FIP1L1-PDGFRA fusion gene in a HES patient who showed osteolytic lesions as the dominant clinical feature. We demonstrated that this patient was sensitive to treatment with imatinib.
In this study, we have shown that bone damage could be a cardinal symptom associated with eosinophilia. As indicated by our bone biopsy data, bone damage in this case was likely caused by eosinophil infiltration. Thus, examination of eosinophil infiltration of bone and the resulting osteolytic lesions and pathological fractures could provide valuable support for the diagnosis of HES. It is important for a clinician to make a proper diagnosis by including pathological examination of specific organ or tissue biopsies if that organ or tissue is likely the target of eosinophilic infiltration.
The PDGFRA gene is a target of imatinib since it encodes a cell surface receptor of the platelet-derived growth factor (PDGF) family.Citation13 The receptor is essential to cell proliferation and survival.Citation13 While bound to PDGF, platelet-derived growth factor receptor alpha is activated as a tyrosine kinase and undergoes phosphorylation that triggers its downstream signaling pathways.Citation14 PDGFRA gene rearrangements forming fusions with other genes only occur in 15% of eosinophilic patients.Citation15 In our case, FIP1L1-PDGFRA fusion gene expression was confirmed by RT-PCR to be positive but at an extraordinarily low level (3.84%). A variety of point mutations had been reported in the PDGFRA gene from patients without PDGFRA fusion genes via Sanger sequencing, including R481G, I562M, H570R, M628T, L705P, G729D, H650Q, L507P, N659S, Y849S and R748G.Citation15 Over proliferation, led by PDGFRA point mutations in eosinophilia, has been shown to be alleviated by imatinib.Citation15 It has also been shown that PDGFRA point mutations are associated with other types of leukemia. N870S and F808L point mutations in the PDGFRA gene were detected in two acute myeloid leukemia patients via Sanger sequencing.Citation16 In the current study, we detected a PDGFRA mutation at p.P6L in the patient’s bone marrow via WES. We showed that the mutation rate in this patient dropped from a pre-treatment high of 57.86% to 42.99% after 6-months of imatinib treatment, and later down to 38.80% at the one-year follow-up, indicating a positive effect of imatinib. The efficiency of imatinib treatment on this mutation warrants additional studies to verify our result. However, while prednisone is used as part of the common first-line treatment in eosinophilia, it was ineffective in our case presented here. This is likely due to the presence of the FIP1L1-PDGFRA fusion gene and/or PDGFRA mutations in this patient.
Prednisone was ineffective in our case, although it can be effective towards other types of eosinophilia, such as necrotizing eosinophilic myocarditis.Citation17 Imatinib is a widely used target therapy drug designated to treat multiple carcinomas and other diseases.Citation12 The drug is able to bind to different types of mutated tyrosine kinases, including PDGF receptor kinases, and inhibits their related signaling pathways.Citation12 While these signaling pathways are blocked, cancer cell proliferation will be inhibited and apoptosis will be induced.Citation12 Some leukemia patients who carried a FIP1L1-PDGFRA fusion gene and were resistant to chemotherapy drugs were responsive to imatinib, achieving complete recovery with no subsequent relapse.Citation18 Consistent with this line of evidence, we have shown that no FIP1L1-PDGFRA fusion gene was detected after 6 months of imatinib treatment, in addition to a significant drop in the level of mutated PDGFRA. Imatinib was also found to be particularly effective towards eosinophilia secondary to myeloproliferative leukemia.Citation19
Accompanying the FIP1L1-PDGFRA fusion gene and PDGFRA gene point mutation in our patient, MYOM2 and ASXL3 gene point mutations were also identified by NGS, suggesting his cancer was heterogeneous. The MYOM2 gene plays a significant role in recurrent diffuse large B-cell lymphoma.Citation20 The ASXL3 gene is an epigenetic modifier which was confirmed as associated to natural killer/T-cell lymphoma relapse,Citation21 but rarely detected in acute myeloid leukemia.Citation22 Although there were no reports on the effects of these 2 gene mutations in eosinophilia, they might contribute to the unique clinical features (bone damage and eosinophil infiltration in bones) in this patient, including his abnormal hematology parameter changes. Interestingly, these two mutations showed opposite effects toward imatinib treatment. It is possible that the MYOM2 mutation effect was associated with the FIP1L1-PDGFRA gene translocation in this patient, as both showed complete response to the drug. Similarly, the effects of the ASXL3 mutation were likely associated with PDGFRA point mutations as both showed poor responses to imatinib treatment. It is also possible that the limited drop in PDGFRA gene point mutation rate after imatinib treatment may be attributed to the presence of ASXL3 in the same tumor cells. Further studies should be conducted to determine whether these two mutations are concurrent in other patients and their roles in eosinophilia.
In conclusion, as shown in this case, our patient presented osteolytic lesions as his dominant clinical abnormality, which is rare in eosinophilia. While showing no response to hormone (prednisone) therapy, he responded well to imatinib, likely owing to the presence of both a FIP1L1-PDGFRA fusion gene and a novel PDGFRA point mutation at p.P6L in exon 2. In addition, the levels of MYOM2 and ASXL3 mutations were altered at variable levels upon imatinib treatment and their pathological effects were likely associated with the FIP1L1-PDGFRA fusion gene and PDGFRA point mutation, respectively. These results further indicate the heterogeneity of clonal eosinophilia in our patient, which may contribute to his unique clinical features and treatment outcome. Further studies are necessary to confirm the occurrence of this novel PDGFRA mutation and the efficacy of an imatinib regimen in other patients.
Methods
Detection of FIP1L1-PDGFRA fusion gene through RT-PCR
RNA was extracted from the patient’s bone marrow using an RNeasy Kit manufactured by Qiagen (CA, USA) and remaining DNA was removed by DNase I (Ambion, Applied Biosystems, TX, USA). The purified RNA was then used for determination of FIP1L1-PDGFRA fusion gene by RT-PCR using the Leukemia Related Fusion Gene Detection Kit (Cat# C10021, Yuanqi Bio-Pharmaceutical Co., Shanghai, China). RT-PCR was performed on an ABI 7300 Real Time PCR System (ABI Corporation, USA).
Detection of PDGFRA mutation via whole exome sequencing (WES) on a NGS platform
Prior to sequencing, genomic DNA was isolated from patient’s bone marrow sample using QIAmp DNA Blood Mini kit (Qiagen, Valencia, California) per manufacturer’s protocol. The quality of DNA was determined using Qubit 2.0 fluorometer and electrophoresis using 1% (w/v) agarose gel. Library was prepared in accordance with the procedure of the Kapa Library Preparation Kit for Illumina Sequencing System (Illumina, San Diego, CA). Exome capture was prepared by SureSelect Human All Exon V6 Kit (Agilent Technologies, Palo Alto, CA, USA). Qualification of Library was performed using the Agilent DNA 1000 Kit (Agilent Corporation, USA). Finally, WES was performed on a HiSeq 3000 (Illumina, San Diego, CA) NGS platform per manufacturer’s instructions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Titlic M, Kodzoman K, Loncar D. Neurologic manifestations of hypereosinophilic syndrome–review of the literature. Acta Clin Croat. 2012;51:65–69.
- Inayat F, Hurairah A. Gastrointestinal and Hepatic Involvement in Hypereosinophilic Syndrome. Cureus. 2016;8:e760. doi:10.7759/cureus.760.
- Ortiz C, Jimenez M, Matos NA. Chronic eosinophilic leukemia: a rare cause of hypereosinophilic syndrome. Bol Asoc Med P R. 2014;106:33–36.
- Park SM, Park JW, Kim SM, Koo EH, Lee JY, Lee CS, Choi DC, Lee BJ. A case of hypereosinophilic syndrome presenting with multiorgan infarctions associated with disseminated intravascular coagulation. Allergy Asthma Immunol Res. 2012;4:161–164. doi:10.4168/aair.2012.4.3.161.
- Klion AD. How I treat hypereosinophilic syndromes. Blood. 2015;126:1069–1077. doi:10.1182/blood-2014-11-551614.
- Fraticelli P, Kafyeke A, Mattioli M, Martino GP, Murri M, Gabrielli A. Idiopathic hypereosinophilic syndrome presenting with severe vasculitis successfully treated with imatinib. World J Clin Cases. 2016;4:328–332. doi:10.12998/wjcc.v4.i10.328.
- Roufosse FE, Goldman M, Cogan E. Hypereosinophilic syndromes. Orphanet J Rare Dis. 2007;2:37. doi:10.1186/1750-1172-2-37.
- Savage N, George TI, Gotlib J. Myeloid neoplasms associated with eosinophilia and rearrangement of PDGFRA, PDGFRB, and FGFR1: a review. Int J Lab Hematol. 2013;35:491–500. doi:10.1111/ijlh.12057.
- Gotlib J, Cools J, Malone JM 3rd, Schrier SL, Gilliland DG, Coutre SE. The FIP1L1-PDGFRalpha fusion tyrosine kinase in hypereosinophilic syndrome and chronic eosinophilic leukemia: implications for diagnosis, classification, and management. Blood. 2004;103:2879–2891. doi:10.1182/blood-2003-06-1824.
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–1214. doi:10.1056/NEJMoa025217.
- Metzgeroth G, Walz C, Score J, Siebert R, Schnittger S, Haferlach C, Popp H, Haferlach T, Erben P, Mix J, et al. Recurrent finding of the FIP1L1-PDGFRA fusion gene in eosinophilia-associated acute myeloid leukemia and lymphoblastic T-cell lymphoma. Leukemia. 2007;21:1183–1188. doi:10.1038/sj.leu.2404662.
- Waller CF. Imatinib mesylate. Recent Results Cancer Res. 2014;201:1–25. doi:10.1007/978-3-642-54490-3_1.
- Sihto H, Sarlomo-Rikala M, Tynninen O, Tanner M, Andersson LC, Franssila K, Nupponen NN, Joensuu H. KIT and platelet-derived growth factor receptor alpha tyrosine kinase gene mutations and KIT amplifications in human solid tumors. J Clin Oncol. 2005;23:49–57. doi:10.1200/jco.2005.02.093.
- Qian C, Wong CWY, Wu Z, He Q, Xia H, Tam PKH, Wong KKY, Lui VCH. Stage specific requirement of platelet-derived growth factor receptor-alpha in embryonic development. PLoS One. 2017;12:e0184473. doi:10.1371/journal.pone.0184473.
- Elling C, Erben P, Walz C, Frickenhaus M, Schemionek M, Stehling M, Serve H, Cross NC, Hochhaus A, Hofmann WK, et al. Novel imatinib-sensitive PDGFRA-activating point mutations in hypereosinophilic syndrome induce growth factor independence and leukemia-like disease. Blood. 2011;117:2935–2943. doi:10.1182/blood-2010-05-286757.
- Hiwatari M, Taki T, Tsuchida M, Hanada R, Hongo T, Sako M, Hayashi Y. Novel missense mutations in the tyrosine kinase domain of the platelet-derived growth factor receptor alpha(PDGFRA) gene in childhood acute myeloid leukemia with t(8;21)(q22;q22) or inv(16)(p13q22). Leukemia. 2005;19:476–477. doi:10.1038/sj.leu.2403638.
- Watanabe N, Nakagawa S, Fukunaga T, Fukuoka S, Hatakeyama K, Hayashi T. Acute necrotizing eosinophilic myocarditis successfully treated by high dose methylprednisolone. Jpn Circ J. 2001;65:923–926.
- Shah S, Loghavi S, Garcia-Manero G, Khoury JD. Discovery of imatinib-responsive FIP1L1-PDGFRA mutation during refractory acute myeloid leukemia transformation of chronic myelomonocytic leukemia. J Hematol Oncol. 2014;7:26. doi:10.1186/1756-8722-7-26.
- Kumar AN, Sathyanarayanan V, Devi VL, Rajkumar NN, Das U, Dutt S, Chinnagiriyappa LK. FIP1L1-PDGFRA-positive chronic eosinophilic leukemia: a low-burden disease with dramatic response to imatinib - a report of 5 cases from South India. Turk J Haematol. 2014;31:56–60. doi:10.4274/Tjh.2013.0086.
- Camicia R, Winkler HC, Hassa PO. Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: a comprehensive review. Mol Cancer. 2015;14:207. doi:10.1186/s12943-015-0474-2.
- Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG, Pan CM, Hu Y, Cai CP, Dong Y, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47:1061–1066. doi:10.1038/ng.3358.
- Duployez N, Micol JB, Boissel N, Petit A, Geffroy S, Bucci M, Lapillonne H, Renneville A, Leverger G, Ifrah N, et al. Unlike ASXL1 and ASXL2 mutations, ASXL3 mutations are rare events in acute myeloid leukemia with t(8;21). Leuk Lymphoma. 2016;57:199–200. doi:10.3109/10428194.2015.1037754.