ABSTRACT
We aimed at the effects of long non-coding RNA (lncRNA) SNHG5 on proliferation, metastasis and migration of colorectal cancer (CRC) cells. We also investigated regulatory relationships among miR-132-3p, SNHG5 and CREB5 and their roles in CRC. 25 pairs of samples containing CRC tissues and matched para-tumor tissues were obtained to examine SNHG5, miR-132-3p and CREB5 expression by qRT-PCR or Western blot. The targeted relationship between miR-132-3p and SNHG5 or CREB5 was confirmed by dual luciferase report assay as well as RNA pull down assay. The expression of SNHG5, miR-132-3p and CREB5 in CRC cells were regulated by cell transfection. CRC cellular proliferation was assayed by CCK-8 and meanwhile flow cytometry was adopted to observe apoptosis. Metastasis and migration of CRC cells were determined respectively by means of Transwell assay and scratch test. The effects of SNHG5 on CRC were researched in vivo, too. SNHG5 or CREB5 was up-regulated in CRC tissues and cells, whereas miR-132-3p was down-regulated. Overexpression of SNHG5 and CREB5 resulted in the enhancement of proliferation, metastasis, migration and the inhibition of apoptosis in CRC cells, while miR-132-3p led to the opposite result. LncRNA SNHG5 promoted proliferation, migration and metastasis of CRC cells but inhibited apoptosis by modulating miR-132-3p/CERB5.
Introduction
All over the world, colorectal cancer (CRC) has become the second most common cancer type in women and the third in men, which shows a rapid progression. More pressingly, CRC is becoming more and more prevalent in developing nations, especially in China.Citation1 Substantial development in the tools to prevent, diagnose and treat CRC has been gained, and as a result the survival of CRC patients at early stage has improved in the last decades.Citation2 However, the overall five-year survival rate of patients with CRC remains low because of the fact that the molecular nosogenesis of this carcinoma remains unclear to a great extent.Citation3 Our study aimed to draw a clearer map of molecular mechanisms of the proliferation and the metastasis of CRC.
Long non-coding RNAs (lncRNAs) are a class of mRNA-like transcripts, which all are longer than 200 nucleotides. They lack protein coding ability and are universally acknowledged to be involved in various kinds of biological processes.Citation4 Increasing evidence has indicated that lncRNAs are commonly aberrantly expressed in plenty of cancers such as CRC,Citation4 breast cancer, gastric cancer,Citation5 bladder cancer, esophageal cancer,Citation6 and so on. SNHG5 is a member of the non-coding multiple small nucleolar RNA host gene family. The SNHG5 gene is 524 bp in size and located on chromosome 6q15 at the breakpoint of chromosomal translocation and this site is involved in human B-cell lymphoma.Citation7 SNHG5 was reported as a translocation partner with BCL6 in the patient suffering from diffuse large B-cell lymphoma. Besides, SNHG5 encodes the Small Nucleolar RNAs (snoRNAs) U50 and U500 in introns 4 and 5, respectively. Though snoRNAs of the C/D box family are well-known to guide fibrillarin mediated 200 methylation of the ribosomal RNA,Citation8 reduced expression and both somatic and germ line mutations in U50 have been reported from breast cancerCitation9 and prostate cancer.Citation10 Despite of the above review and summary, the mechanisms of SNHG5 have not previously been characterized in CRC.
Micro RNAs (miRNAs) are noncoding RNA molecules of 19–25 nucleotides that potentially mediate protein expressions accounting for 20–30 %.Citation11 For the past few years, many miRNAs are believed to play a large part in carcinogenesis via regulating mRNA expression.Citation12 Both downregulation of tumor suppressive miRNAs targeting proto-oncogenes and upregulation of oncogenic miRNAs targeting tumor suppressor genes can lead to the tumorigenesis and tumor progression.Citation13 MiR-132, which has been vigorously studied, is located in chromosome 17p13.3. As a possible target of SNHG5, it has exhibited correlation with various kinds of malignancies including glioma,Citation14 CRC,Citation15 breast cancer,Citation16 prostate cancer and pancreatic cancer.Citation12 Some studies showed that miR-132 could promote proliferative capacity of cancer cells.Citation17,Citation18 Most of the reports about the biological functions and throttling capability of miR-132 have emerged on the basis of the studies performed in the neuronal context. Additionally, miR-132 has also been described in a number of other fields, such as inflammation and cell transformation.Citation17
CAMP-responsive element binding protein 5 (CREB5), the product of which belongs to the CRE (cAMP response element)-binding protein family, which contain zinc-finger and bZIP DNA-binding domains.Citation19 A previous research revealed that higher expressions of CREB5, PTPRB and COL4A3 predicted favorable disease free survival in patients with pulmonary carcinoid tumors.Citation20 The biological function of CREB5 in CRC has not been characterized in previous studies.
Our objective of this report was to conduct the thorough researches into the biological function of lncRNA SNHG5 in CRC and the exact functional mechanisms. What’s more, it was demonstrated by luciferase reporter assay that lncRNA SNHG5 targeted binding miR-132-3p and CREB5 was a downstream target of miR-132-3p in CRC. Furthermore, by multiple bioinformatics techniques and in vitro cell researches such as RNA pull-down, transwell assay and so on, we finally reach a conclusion that lncRNA SNHG5 affected cell proliferation, migration and metastasis of CRC through regulating miR-132-3P/CREB5.
Results
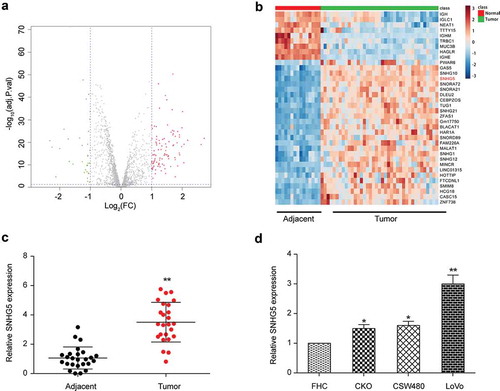
Lncrna SNHG5 was overexpressed in CRC tissues and cells
GSE18105 microarray from GPL96 platform was analyzed using the R project for statistical computing. The lncRNAs with different expression were screened out by Fold change greater than 2 and P < 0.05. Statistical analysis revealed that SNHG5 was up-regulated in human CRC tissues ()). Compared with paracancerous tissues, there were 98 up-regulated lncRNAs in CRC tissues, whereas 35 lncRNAs were down-regulated. The expression level of SNHG5 in CRC tissues increased by 1.79 times (P < 0.001, ). The expression of SNHG5 in CRC and adjacent tissues was detected. The qRT-PCR results revealed that SNHG5 expression in CRC tissues was markedly higher than that in adjacent tissues (P < 0.01, ). The expression of SNHG5 in CRC cell lines RKO, SW480, LoVo and normal colorectal mucosa cell FHC was detected, and results by qRT-PCR revealed SNHG5 expression in CRC cells was markedly up-regulated (P < 0.01, ). Up-regulation in LoVo cell line was the most obvious, so we selected LoVo cells for the follow-up experiments.
Figure 1. SNHG5 was overexpressed in colorectal cancer tissues and cells. (a) Volcano plot. (b) Heat map. (c) qRT-PCR results of SNHG5 in colorectal cancer tissues and adjacent normal tissues. (d) qRT-PCR results of SNHG5 in colorectal cancer cell lines RKO, SW480, LoVo and normal colorectal mucosa cell line FHC. * P<0.05, ** P<0.01, compared with control group.
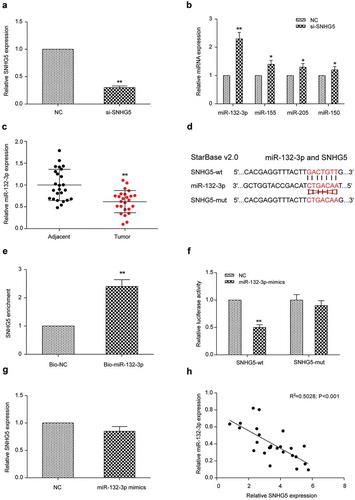
Relationship between miR-132-3p and SNHG5
LoVo cells were transfected with si-SNHG5 and revealed a remarkable reduction in SNHG5 expression determined by qRT-PCR (P < 0.01, ). By using the Starbase database, we found that miR-132-3p, miR-155, miR-205 and miR-150 could interact with SNHG5. QRT-PCR results revealed the expression of miR-155, miR-205 and miR-150 in cells transfected with si-SNHG5 was remarkably higher than that in cells of NC group (P < 0.05, ), among which the up-regulation level of miR-132-3p was the highest. Therefore, we selected miR-132-3p for the follow-up experiments. qRT-PCR revealed that miR-132-3p expression in CRC tissues was remarkably lower than that in adjacent tissues (P < 0.01, ). According to the binding-site predictions from the Starbase database, SNHG5 could target miR-132-3p (). By using biotinylated miR-132-3p probes for pulling down lncRNA SNHG5, it was found SNHG5 expression detected by miR-132-3p probes was remarkably increased compared with that of the control group (P < 0.01, ). The addition of miR-132-3p mimics was able to inhibit luciferase activity of cells transfected with SNHG5-wt, but not that of cells transfected with SNHG5-mut, indicating that SNHG5 could directly bind to the 3ʹUTR region of miR-132-3p and inhibit its expression (). In order to further demonstrate the interactions between SNHG5 and miR-132-3p at the transcriptional level, we used qRT-PCR to detect expression of SNHG5 in LoVo cells transfected with miR-132-3p mimics or NC group. SNHG5 expression in miR-132-3p mimics group didn’t change remarkably compared with NC group (P > 0.05, ), indicating that this was a unidirectional adjustment. MiR-132-3p was negatively correlated with SNHG5 expression (). Based on all the experimental data, we reached a conclusion that miR-132-3p was a downstream target of SNHG5 in CRC cells.
Figure 2. SNHG5 regulated the expression of miR-132-3p in LoVo cells (a) Transfection efficiency of si-SNHG5 was verified by qRT-PCR. (b) qRT-PCR results of miR-132-3p, miR-155, miR-205 and miR-150 in si-SNHG5 group and NC group. (c) qRT-PCR results of miR-132-3p in colorectal cancer tissues and adjacent normal tissues. (d) Bioinformatics predicted the binding sites of SNHG5 and miR-132-3p. (e) RNA pull down experiments demonstrated SNHG5 bound to miR-132-3p. (f) Dual luciferase assay detected the relationship between SNHG5 and miR-132-3p. (g) qRT-PCR detected the expression of SNHG5 in LoVo cells transfected with NC or miR-132-3p mimics. (h) The correlation between SNHG5 and miR-132-3p. * P<0.05, ** P<0.01, compared with NC group.
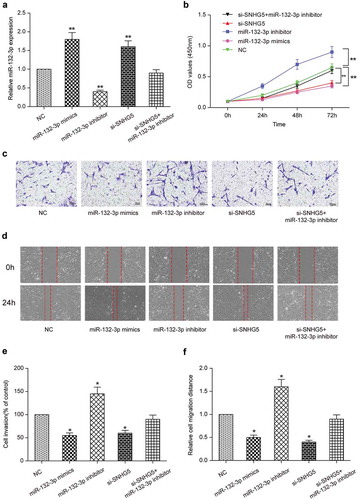
Effect of SNHG5 on proliferation, metastasis and migration of CRC cells by regulating miR-132-3p
To determine the effect of SNHG5 on tumorigenesis, we divided cells into five groups: miR-132-3p mimics group, miR-132-3p inhibitor group, si-SNHG5 group, si-SNHG5 + miR-132-3p inhibitor group and NC group. QRT-PCR results displayed miR-132-3p expression was significantly up-regulated in miR-132-3p mimics group (P < 0.01, ). On the contrary, miR-132-3p was significantly down-regulated in miR-132-3p inhibitor group (P < 0.05, ). CCK-8 assay displayed that the multiplication capacity of LoVo cells transfected with miR-132-3p inhibitor was dramatically higher than that of NC group (P < 0.01, ), whereas the multiplication capacity of LoVo cells transfected with miR-132-3p mimics or si-SNHG5 was decreased in comparison to that of NC group (P < 0.01, ). Proliferation of LoVo cells transfected with si-SNHG5 + miR-132-3p inhibitor was comparable to that of NC group (P > 0.05, ). The metastasis and migration ability of LoVo cells were detected by transwell assay and wound healing assay respectively. Compared with NC group, the metastasis and migration ability of LoVo cells transfected with miR-132-3p inhibitor were significantly enhanced (P < 0.01, ), the metastasis and migration ability of LoVo cells transfected with miR-132-3p mimics or si-SNHG5 were inhibited (P < 0.05, ), while the metastasis and migration ability of LoVo cells transfected with si-SNHG5 + miR-132-3p inhibitor was comparable to that of NC group (). Our results indicated that SNHG5 regulated miR-132-3p to promote cell proliferation, metastasis and migration in CRC cells.
Figure 3. SNHG5 regulated miR-132-3p to promote colorectal cancer cell proliferation, migration and invasion. (a) Transfection efficiency was verified by qRT-PCR. (b) CCK-8 detected the proliferation of cells in miR-132-3p mimics group, miR-132-3p inhibitor group, si-SNHG5 group, si-SNHG5 + miR-132-3p inhibitor group and NC group. (c) (e) Transwell detected the relationship between SNHG5 and miR-132-3p and their effects on colorectal cancer cell invasion (× 200). (d) (f) Wound healing assay detected the relationship between SNHG5 and miR-132-3p and their effects on colorectal cancer cell migration (× 50). * P<0.05, ** P<0.01, compared with NC group.
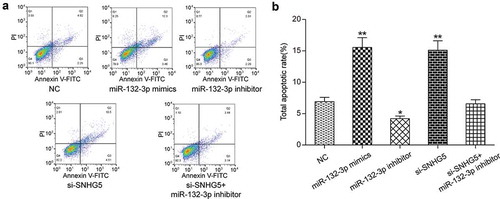
Effects of SNHG5 on apoptosis of CRC cells by regulating miR-132-3p
Flow cytometry was employed and its results displayed the apoptosis rate of miR-132-3p mimics group and si-SNHG5 group were dramatically increased (P < 0.01, ), the apoptosis rate of miR-132-3p inhibitor group was significantly inhibited (P < 0.05, ), while the apoptosis rate of si-SNHG5 + miR-132-3p inhibitor group was comparable to that of NC group (P > 0.05, ). Our results indicated that SNHG5 regulated miR-132-3p to reduce apoptosis in CRC cells.
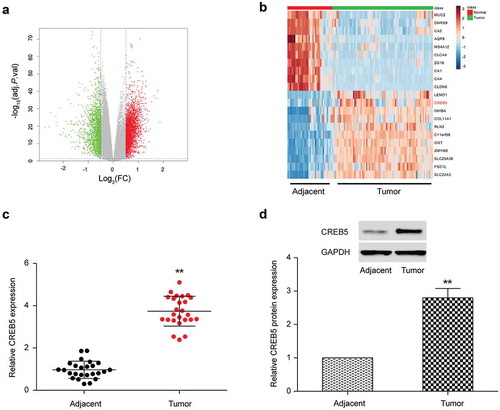
CREB5 was differentially expressed in CRC tissues and cells
The differently expressed mRNAs were screened by Fold change value greater than 2 and P < 0.01. Results proved there were 158 mRNAs that were up-regulated in CRC cells and 175 mRNAs that were down-regulated in CRC cells (). We selected mRNAs that had significant difference in expression levels to carry out the heat map (). Statistical analysis proved that CREB5 was up-regulated in human CRC and qRT-PCR results confirmed CREB5 expression in CRC tissues was dramatically higher than that in adjacent tissues (). Results of Western blot were consistent with the results of qRT-PCR ().
Figure 5. CREB5 was under-expressed in colorectal cancer tissues and cells. (a) Volcano plot. (b) Heat map. (c) qRT-PCR results of CREB5 in colorectal cancer tissues and adjacent normal tissues. (d) Western blot results of CREB5 in colorectal cancer tissues and adjacent normal tissues. ** P<0.01, compared with Adjacent group.
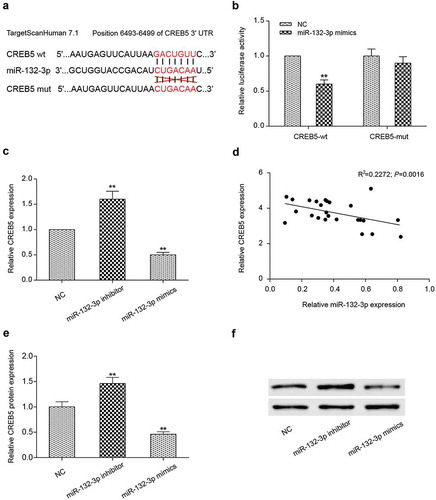
Relationship between miR-132-3p and CREB5
The bioinformatics software (Targetscan 7.1) predicted miR-132-3p could target to CREB5 (). The addition of miR-132-3p mimics was able to inhibit luciferase activity of cells transfected with CREB5-wt, but not that of cells transfected with CREB5-mut, indicating that miR-132-3p could directly bind to the 3ʹUTR region of CREB5 and inhibit its expression (). In order to further demonstrate that miR-132-3p could inhibit CREB5 expression, we detected the mRNA and protein expression level of CREB5 by qRT-PCR and Western blot in LoVo cells that were transfected with miR-132-3p inhibitor or miR-132-3p mimics. Results proved compared with NC group, expression of CREB5 in miR-132-3p inhibitor group was up-regulated (P < 0.01, ), while expression of CREB5 in miR-132-3p mimics group was down-regulated (P < 0.05, ). MiR-132-3p was negatively correlated with CREB5 expression (). In addition, CREB5 protein expression level changes showed the same trend with mRNA (). Based on the experimental data, we came to a conclusion that CREB5 was a downstream target of miR-132-3p in CRC cells.
Figure 6. MiR-132-3p targeted the expression of CREB5 in LoVo cells. (a) Bioinformatics predicted the binding sites of miR-132-3p and CREB5. (b) The combination of miR-132-3p and CREB5-wt was detected by dual-luciferase reporter system. (c) The mRNA expression of CREB5 in miR-132-3p-mimics, miR-132-3p-inhibitor LoVo cells was detected by qRT-PCR. (d) The correlation between miR-132-3p and CREB5. (e,f) The CREB5 protein expression level in miR-132-3p-mimics, miR-132-3p-inhibitor LoVo cells was tested by Western blot. ** P<0.01, compared with NC group.
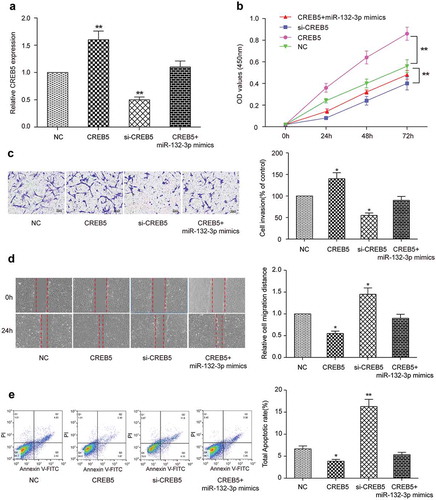
Effects of miR-132-3p on proliferation, metastasis, migration and apoptosis of CRC cells by targeting CREB5
To verify the effect of miR-132-3p on CRC by targeting CREB5, we divided the cells into four groups: CREB5 group, si-CREB5 group, CREB5 + miR-132-3p mimics group and NC group. Results of qRT-PCR exhibited CREB5 group promoted expression of CREB5 (P < 0.01, ), si-CREB5 group inhibited expression of CREB5 (P < 0.01, ), and the expression level of CREB5 in CREB5 + miR-132-3p mimics group was similar to that of control group (P > 0.05, ). CCK-8 results exhibited that compared with NC group, transfection of CREB5 promoted the proliferation ability of LoVo cells (P < 0.01, ), transfection of si-CREB5 inhibited the proliferation ability of LoVo cells (P < 0.01, ), while the proliferation level of cells in CREB5 + miR-132-3p mimics group was similar to that of control group (P > 0.05, ). Transwell migration assay () and wound healing assay () also yielded similar results. The results of Flow cytometry exhibited that the apoptosis rate of CREB5 group was significantly decreased (P < 0.05, ), the apoptosis rate of si-CREB5 group was observably increased (P < 0.01, ) and there was no significant difference between CREB5 + miR-132-3p mimics group and NC group (P > 0.05, ). These results illustrated that miR-132-3p inhibited cell proliferation, metastasis and migration, and promoted apoptosis of CRC cells by targeting CREB5.
Figure 7. Effects of miR-132-3p on the proliferation, migration, invasion and apoptosis of colorectal cancer cells by targeting CREB5. (a) The expression of CREB5 in CREB5 group, si-CREB5 group, CREB5 + miR-132-3p-mimics group and NC group was detected by qRT-PCR in LoVo cells. (b) CCK-8 was used to detect the proliferation of four groups of cells. (c) Transwell was used to detect the invasion ability of four groups of cells (× 200). (d) The effects of miR-132-3p on cell migration through targeting CREB5 were detected by wound healing assay (× 50). (E) The apoptosis ability of four groups of cells was detected by flow cytometry. * P<0.05, ** P<0.01, compared with NC group.
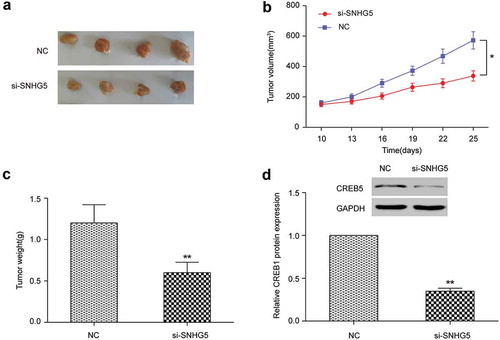
In vivo experiments verified the effect of lncrna SNHG5 on CRC
The experiments were divided into two groups: si-SNHG5 group and NC group. The growth of subcutaneous tumor in nude mice was measured by the vernier caliper. Compared with NC group, si-SNHG5 group could observably inhibit the growth of subcutaneous tumor in nude mice (). The volume of tumor in si-SNHG5 group was observably lower than that in control group (P < 0.01, ). Nude mice was sacrificed at day 25 and tumor mass was removed. The tumor mass of si-SNHG5 group was smaller than that of NC group (P < 0.01, ). Western blot exhibited that si-SNHG5 group significantly inhibited the expression of CREB5 (P < 0.01, ). These results exhibited in vivo that SNHG5 could affect the multiplication capacity of CRC by regulating CREB5.
Figure 8. In vivo experiments verified the effect of lncRNA SNHG5 on colorectal cancer. (a) Subcutaneous tumors were removed from nude mice. (b) Changes in the volume of subcutaneous tumors of nude mice. (c) Changes in the weight of tumors extracted from nude mice. (d) Western blot detected CREB5 expression in the tumor tissues. * P<0.05, ** P<0.01, compared with NC group.
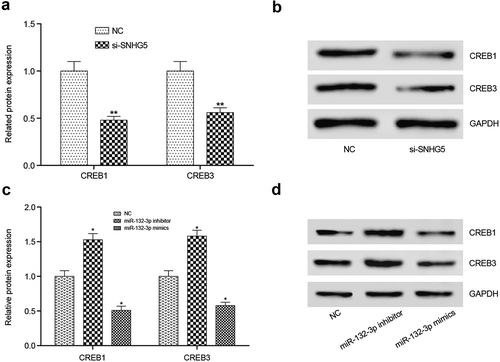
Effects of lncRNA SNHG5 or miR-132-3p on CREB family protein in CRC cells
Subsequently, Western blot experiments was used to verified the effect of lncRNA SNHG5 and miR-132-3p on the change of CREB family protein expression levels. The results displayed that CREB1 and CREB3 proteins were decreased in si-SNHG5 group compared with NC group (). While after treated with miR-132-3p inhibitor or miR-132-3p mimics, the variation trends of CREB1 and CREB3 proteins level were similar to CREB5 ().
Figure 9. Effects of lncRNA SNHG5 or miR-132-3p on CREB family protein in CRC cells. (a,b) CREB1 and CREB3 protein expression levels decreased observably in si-SNHG5 group compared with NC group. (c,d) CREB1 and CREB3 protein expression levels were prominently increased in miR-132-3p inhibitor compared with NC group, while decreased in miR-132-3p mimics group. *P < 0.05, **P < 0.01, compared with NC group.
Discussion
In this study, we identified the lncRNA SNHG5 in CRC cells and found that it was dramatically up-regulated in CRC cells using the qRT-PCR assay, indicating the potential function of lncRNA SNHG5 in CRC. A series of experiments illustrated the correlation among lncRNA SNHG5, miR-132-3p and CREB5, concluding that lncRNA SNHG5 regulated CRC progression by regulating miR-132-3p and CREB5. Our findings indicated the important role of lncRNA SNHG5 during CRC tumorigenesis.
Along with small RNAs and proteins, lncRNAs have broad applications in CRC diagnosis and treatment.Citation21 SNHG5 belongs to a large family of non-coding genes hosting small RNAs, such as snoRNAs and microRNAs, most often residing in the introns of the host genes. It has been identified in previous study that SNHG5 was significantly up-regulated in CRC cells with respect to normal tissues. And SNHG5 has been identified and characterized as a stable cytoplasmic lncRNA with up-regulated expression in CRC.Citation8 As in our study, SNHG5 expression in CRC tissues was observably higher than that in adjacent tissues. And by scratch test and transwell array, we also confirmed overexpression of SNHG5 could promote metastasis and migration in CRC. By CCK-8 and flow cytometry we demonstrated that SNHG5 could reduce apoptosis and promote cell multiplication capacity in CRC cells.
MiR-132 has been reported as a tumor suppressive miRNA targeting proto-oncogenes in a variety of cancers, such as HCC (hepatocellular carcinoma)Citation22 and prostate cancer.Citation23 Zheng et al. found that miR-132 inhibited the capacity to migrate around and invade other tissues of CRC cells via directly targeting ZEB2 using 62 CRC cells samples.Citation11 A recent report indicated that miR-132 was markedly downregulated in CRC tissues, leading to the marked inhibition of cell metastasis and epithelial-mesenchymal transition (EMT) in CRC cell lines.Citation3 Our results found miR-132-3p was significantly downregulated in CRC cells which was the same in previous studies. And we also confirmed that miR-132-3p functioned as a tumor suppressor in CRC. Expression of miR-132-3p could significantly inhibit proliferation, metastasis and migration and promote apoptosis of CRC cells.
The CREB5 gene belongs to the CRE (cAMP response element)-binding protein family. In previous studies, both knockdown and overexpression of CREB5 demonstrated that it was a negative regulator of hepatitis B virus (HBV) replication.Citation24 There were also functional studies showed that knockdown of CREB1/CREB5 increased tumor necrosis factor alpha (TNF-α) level and enhanced expression of phospho–NF-κB p65 and NF-κB p65 in human monocytes.Citation25 In our present study, CREB5 was markedly overexpressed in CRC cells, and overexpression of CREB5 promoted cell proliferation, migration and metastasis, reduced apoptosis in CRC cells. Colorectal cancer-related lncRNAs have been demonstrated to regulate the genes by various mechanisms, including epigenetic modifications, lncRNA-miRNA and lncRNA-protein interactions, and by their actions as miRNA precursors or pseudogenes.Citation21 Based on our laboratory finding, we came to a conclusion that miR-132-3p was a downstream target of SNHG5 and CREB5 was a downstream target of miR-132-3p in CRC cells.
There appears to be some deficiencies in the current study. For example, we studied only 25 samples in this study, and therefore more samples should be included to make our conclusion more accurate and reliable. Further studies are needed to focus on the regulation of SNGH5 in normal and cancer tissues at different stages. With the development of research and technology, SNHG5 will facilitate the diagnosis and treatment of CRC in the near future.
Basically, our study demonstrated that expression of SNHG5 or CREB5 was up-regulated in CRC cells, whereas expression of miR-132-3p was down-regulated. This differential expression resulted in the enhancement of proliferation, metastasis, migration in CRC cells and the inhibition of apoptosis. Finally we reached a conclusion that lncRNA SNHG5 regulates CREB5 by sponging miR-132-3p. Our findings have achieved a clearer understanding of how lncRNA SNHG5 is connected with the molecular pathogenesis of CRC, which could offer a potential path for treatment of this disease.
Materials and methods
Patients or participants
25 couples of tumor and adjacent tissues were collected from CRC patients who had surgical treatment in The Second Hospital of Shandong University from May 2014 to June 2015. All patients were treated firstly neither radiotherapy nor chemotherapy before surgical treatment. All the specimens were made a confirmed diagnosis according to the pathological examination, and all the experiments were approved by The Second Hospital of Shandong University medical ethics committee and informed consent from patients were obtained. The CRC cell line RKO, SW480, LoVo and normal colorectal mucosa cells FHC were purchased from GEFAN BIOTECHNOLOGY.CO., LTD (Shanghai, China).
Microarray analysis
The expression data was analyzed from GSE18105 microarray in Gene Expression Omnibus (GEO) public database including 34 normal samples and 77 CRC samples. The expression of differently expressed lncRNAs and mRNAs was screened with Fold change> 2 and P < 0.05 as filter criteria.
Cell culture and transfection
The CRC cell line RKO was cultured in Eagle’s minimal essential medium (EMEM) with 10 % fetal bovine serum (FBS) (Halingbio, Shanghai, China). The CRC cell lines SW480 and FHC were cultured in RPMI-1640 medium containing 10 % FBS (Biosun, Shanghai, China). LoVo cell line was cultured in Ham’s F12K medium (Shanghai Haoran Biotechnology CO. LTD, Shanghai, China) with 10 % FBS. All CRC cell lines were subcultured in an incubator with an appropriate growth environment for the cells. LoVo cells were selected for transfection because of the highest expression level of SNHG5 in this cell line. All recombined plasmid vectors used for transfection were extracted via the plasmid Midiprep kit (Shanghai Haoran Biotechnology CO. LTD). MiR-132-3p mimics, miR-132-3p inhibitor, si-CREB5, si-SNHG5, pcDNA3.1-CERB5 and pcDNA3.1-SNHG5 were purchased from Biovector Co., LTD (Beijing, China). Vectors were transferred into cells using LipofectaminTM 3000 (Life Technologies, Gaithersburg, MD, USA). Cell grouping: miR-132-3p mimics group, miR-132-3p inhibitor group, si-SNHG5 group, si-SNHG5 + miR-132-3p inhibitor group and NC group; CREB5 group, si-CREB5 group, CREB5 + miR-132-3p mimics group and NC group; si-SNHG5 group and NC group.
QRT-PCR
Total RNA from tissues and cell lines was extracted and purified with Trizol reagent (Yeasen, Shanghai, China). After the quality and quantity of the extracted total RNA were confirmed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc), complementary DNA (cDNA) was synthesized by 200 ng total RNA using ReverTra Ace qPCR RT Kit (Toyobo life science, Osaka, Japan) following the product’s instructions for use label. The product obtained by reverse transcription was analyzed by real-time PCR via THUNDERBIRD SYBR® qPCR Mix (Toyobo). 2−ΔΔCt method was used to calculate the expression levels of miRNAs and mRNAs relative to GAPDH and U6 respectively. The primer sequences used for PCR were listed in .
Table 1. Primers for qRT-PCR.
Western blot
Total proteins from CRC tissues and cells were prepared using RIPA buffer (Yeasen). BCA method was employed to measure the total protein concentration. Proteins were electrophoresed in SDS-PAGE, and then blotted on a PVDF membrane, which was then blocked in 5 % fat-free milk. The membrane was co-incubated with the primary antibody mouse polyclonal to human CREB5 (ab168928, 1: 500) (Abcam, Cambridge, MA, USA), anti-CREB antibody [E306] (ab32515, 1:500–1:1000, Abcam) and anti-CREB3 antibody [EPR14215(B)] (ab180119, 1:2000, Abcam). The PVDF membrane was washed with Tris Buffered Saline Tween (TBST) by three times. HRP-labeled goat anti-mouse IgG secondary antibody (ab6782, Abcam) was prepared with 1 % fat-free milk by 1: 10,000. Then it was added and incubated, followed by being washed three times by TBST. Semi-quantitative analysis was performed using ImageJ software to analyze the cumulative optical density of the bands, and finally the relative expression of each protein was figured out.
CCK-8 assay
The ability of cell proliferation was detected using CCK-8 reagent (Well Biotech, Shanghai, China). Logarithmically growing LoVo cells were picked out, digested with 0.25 % trypsin, and made into single-cell suspensions with F12K medium. Transfected cells were seeded at a density of 8 × 10Citation3 cells/well in 96-well flat-bottom plates and cultured in a suitable circumstance. Afterwards, culture medium was substituted by F12K medium without FBS, and cells were cultured followed by discarding the supernatant after 24 h. After cells being cultured for 24 h, 48 h and 72 h, CCK-8 reagent was added (10 μl/well) and incubated for 4 h. The optical density was determined at 450 nm by the microplate reader.
Transwell assay
Transfected LoVo cells were harvested, trypsin digested, and resuspended. Then the mixture was seeded into the upper chamber at a density of 5 × 103 per well. 500 µL F12K medium was added into the lower chamber. After one day incubation, the chambers were washed and cells were fixed in 4% paraformaldehyde, and subsequently stained with 0.1 % crystal violet. At last, the images were shot under a microscope to observe the metastasis of cells (× 200).
Wound healing assay
Cells were incubated with normal cell growth medium in six-well plates. Once the cell adherence reached 90 %, the cell layer was scratched with a 10μl sterile pipette tip. The old F12K medium was discarded, after which the shed cells were washed with sterile PBS. At different time points (0, 24 h), observation of the cells in plates were acquired using a microscope. Images were opened and edited by Image J, and then the mean of the migration cells was calculated.
Flow cytometry assay
Transfected LoVo cells were harvested after trypsin digestion without ethylene diamine tetraacetic acid (EDTA). After that, the mixture were centrifuged at speed of 1000rpm for 5 min and washed with cold PBS twice after removing the upper clear liquid. 5μl Annexin V-FITC was added into 200μl binding buffer and reacted for 15 min. Then 5μl PI was added and reacted for 15 min. BD FACS Calibur Flow Cytometer was used to assay the cell apoptosis and BD FACS Diva was used for data analysis.
RNA pull-down assay
The biotinylated SNHG5 and miR-132-3p probes (Sangon Biotech, Shanghai, China) were transfected into LoVo cells. Cells were lysed (Lysis Buffer: 40mmol/L Tris-HCL (PH = 7.5), 150mmol/L NaCl, 1mmol/L EDTA, 0.5% NP-40, 10 % glycerinum, 1mmol/L DTT, 0.4mmol/L PMSF, 2μg/mL leupeptin hemisulfate salt, 2μg/mL aprotinin, 5mg/L lysozyme) and centrifuged. The biotinylated probes were added to the supernatant for co-incubation. Then cells were incubated with m-280 streptavidin beads (Invitrogen). The beads were then washed twice with lysis buffer. At last, qRT-PCR was performed on the supernatant.
Dual-luciferase reporter assay
The cells were seeded in 24-well plates at a density of 1 × 105 and transfected after logarithmic growth phase. The fragments of SNHG5 and CREB5 were inserted into the restriction enzyme cutting site of psiCHECK TM-2 (Promega, Madision, WI, USA) and luciferase reporter plasmids and psiCHECK empty vector, miR-132-3p mimics, miR-132-3p inhibitor were transfected. 48 h after being transfected, each well was added with 80μl of 1× Passive Lysis Buffer and placed on a shaker for 15 min to lyse cells. Cell lysate were centrifuged at 12000rpm, and then the supernatant was saved in the refrigerator at −80°C. 10μl of lysed cells was added to each well of a 96-well plate, and 30μl of Luciferase Assay Reagent II (LAR II, Promega) was added. Then 30μl of the stop solution was added to terminate the LAR II reaction. Finally, the fluorescence photometer was set to measure the luciferase activity.
Tumor formation assay in nude mice
Cells in well-grown state were digested by trypsin into single cell suspension, washed three times with PBS buffer, and then resuspended in serum-free medium. LoVo cells transfected with si-SNHG5 and NC were subcutaneously inoculated in the back of 5 nude mice, with the density of inoculated cells 1 × 106/200μl. From the 10th day, the long and short diameters of the tumor were measured every 3 days with a vernier caliper. The tumor volume was calculated making use of the formula V = (D× dCitation2)/2, where D is on behalf of the longer diameter and d is on behalf of the shorter diameter. The animals were sacrificed and the tumor tissues were removed and weighed at the 25th day. The animal tissue samples were fixed with 4 % formalin and embedded in paraffin, to be prepared for the follow-up immunohistochemistry assay. All the experimental procedures were approved by the Animal Care Use Committee of The Second Hospital of Shandong University
Statistical analysis
Independent experiments were performed in triplicate. GraphPad Prism 6.0 was used to analyze the data expressed as mean ± SD to detect the statistical significance. The relationship between miR-132-3p and SNHG5 or CREB5 expression was calculated by Pearson’s correlation analysis. Quantitative data of at least three experiments were compared using one-way analysis of variance and that of two below experiments were compared using the Student’s t-test. Data with P < 0.05 were considered to have statistically significant difference.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest were disclsoed.
Ethics approval
This study was authorized by The Second Hospital of Shandong University.
Consent for publication
This manuscript has been approved by all authors for publication.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request
References
- Yu B, Ye X, Du Q, Zhu B, Zhai Q, Li XX. 2017. The long non-coding RNA CRNDE promotes colorectal carcinoma progression by competitively binding miR-217 with TCF7L2 and enhancing the Wnt/beta-Catenin signaling pathway. Cell Physiol Biochem. 41:2489–2502. doi:10.1159/000475941.
- Xu J, Zhang R, Zhao J. 2017. The novel long noncoding RNA TUSC7 inhibits proliferation by sponging miR-211 in colorectal cancer. Cell Physiol Biochem. 41:635–644. doi:10.1159/000457938.
- Qin J, Ke J, Xu J, Wang F, Zhou Y, Jiang Y, Wang Z. 2015. Downregulation of microRNA-132 by DNA hypermethylation is associated with cell invasion in colorectal cancer. Onco Targets Ther. 8:3639–3648. doi:10.2147/OTT.S91560.
- Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F, Yan D. 2015. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 106:1323–1332. doi:10.1111/cas.12759.
- Shao Y, Chen H, Jiang X, Chen S, Li P, Ye M, Li Q, Sun W, Guo J. 2014. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 35:9591–9595. doi:10.1007/s13277-014-2243-z.
- Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–1710. doi:10.1136/gutjnl-2013-305806.
- Ichigozaki Y, Fukushima S, Jinnin M, Miyashita A, Nakahara S, Tokuzumi A, Yamashita J, Kajihara I, Aoi J, Masuguchi S, et al. Serum long non-coding RNA, snoRNA host gene 5 level as a new tumor marker of malignant melanoma. Exp Dermatol. 2016;25:67–69. doi:10.1111/exd.12868.
- Damas ND, Marcatti M, Come C, Christensen LL, Nielsen MM, Baumgartner R, Gylling HM, Maglieri G, Rundsten CF, Seemann SE, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. doi:10.1038/ncomms13875.
- Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT. 2009. Implication of snoRNA U50 in human breast cancer. J Genet Genomics. 36:447–454. doi:10.1016/S1673-8527(08)60134-4.
- Dong XY, Rodriguez C, Guo P, Sun X, Talbot JT, Zhou W, Petros J, Li Q, Vessella RL, Kibel AS, et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet. 2008;17:1031–1042. doi:10.1093/hmg/ddm375.
- Mokutani Y, Uemura M, Munakata K, Okuzaki D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K, Takemasa I, et al. Down-regulation of microRNA-132 is associated with poor prognosis of colorectal cancer. Ann Surg Oncol. 2016;23:599–608. doi:10.1245/s10434-016-5133-3.
- Zhang X, Tang W, Chen G, Ren F, Liang H, Dang Y, Rong M. 2016. An encapsulation of gene signatures for hepatocellular carcinoma, MicroRNA-132 predicted target genes and the corresponding overlaps. PLoS One. 11:e0159498. doi:10.1371/journal.pone.0159498.
- Kaur S, Lotsari JE, Al-Sohaily S, Warusavitarne J, Kohonen-Corish MR, Peltomaki P. 2015. Identification of subgroup-specific miRNA patterns by epigenetic profiling of sporadic and Lynch syndrome-associated colorectal and endometrial carcinoma. Clin Epigenetics. 7:20. doi:10.1186/s13148-015-0059-3.
- Liu Q, Liao F, Wu H, Cai T, Yang L, Wang ZF, Zou R. 2014. Upregulation of miR-132 expression in glioma and its clinical significance. Tumour Biol. 35:12299–12304. doi:10.1007/s13277-014-2541-5.
- Zheng YB, Luo HP, Shi Q, Hao ZN, Ding Y, Wang QS, Li SB, Xiao GC, Tong SL. 2014. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J Gastroenterol. 20:6515–6522. doi:10.3748/wjg.v20.i21.6515.
- Zhang ZG, Chen WX, Wu YH, Liang HF, Zhang BX. 2014. MiR-132 prohibits proliferation, invasion, migration, and metastasis in breast cancer by targeting HN1. Biochem Biophys Res Commun. 454:109–114. doi:10.1016/j.bbrc.2014.10.049.
- Liu X, Yu H, Cai H, Wang Y. 2014. The expression and clinical significance of miR-132 in gastric cancer patients. Diagn Pathol. 9:57. doi:10.1186/1746-1596-9-57.
- Wang J, Xu G, Shen F, Kang Y. 2014. miR-132 targeting cyclin E1 suppresses cell proliferation in osteosarcoma cells. Tumour Biol. 35:4859–4865. doi:10.1007/s13277-014-1637-2.
- Bhardwaj A, Singh H, Rajapakshe K, Tachibana K, Ganesan N, Pan Y, Gunaratne PH, Coarfa C, Bedrosian I. 2017. Regulation of miRNA-29c and its downstream pathways in preneoplastic progression of triple-negative breast cancer. Oncotarget. 8:19645–19660. doi:10.18632/oncotarget.14902.
- Deng B, Molina J, Aubry MC, Sun Z, Wang L, Eckloff BW, Vasmatzis G, You M, Wieben ED, Jen J, et al. Clinical biomarkers of pulmonary carcinoid tumors in never smokers via profiling miRNA and target mRNA. Cell Biosci. 2014;4:35. doi:10.1186/2045-3701-4-35.
- Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong H, Zhou JY, Yang SM. 2016. Long non-coding RNAs in colorectal cancer. Oncotarget. 7:5226–5239. doi:10.18632/oncotarget.6446.
- Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, Croce N, Vandesompele J, Mestdagh P, Finazzi-Agro E, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–134. doi:10.1038/onc.2012.14.
- Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, Liu R, Wu Z. 2013. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal. 25:1037–1043. doi:10.1016/j.cellsig.2013.01.019.
- Zhang X, Liu H, Xie Z, Deng W, Wu C, Qin B, Hou J, Lu M. 2016. Epigenetically regulated miR-449a enhances hepatitis B virus replication by targeting cAMP-responsive element binding protein 5 and modulating hepatocytes phenotype. Sci Rep. 6:25389. doi:10.1038/srep25389.
- Long X, Li Y, Qiu S, Liu J, He L, Peng Y. 2016. MiR-582-5p/miR-590-5p targeted CREB1/CREB5-NF-kappaB signaling and caused opioid-induced immunosuppression in human monocytes. Transl Psychiatry. 6:e757. doi:10.1038/tp.2016.4.