ABSTRACT
Objective: To investigate the role and mechanism of action of nicotinamide nucleotide transhydrogenase antisense RNA 1 (NNT-AS1) in osteosarcoma (OS).
Methods: Bioinformatic analysis suggested miR-320a as potential target of NNT-AS1. Influence of NNT-AS1 overexpression or knockdown on OS cell proliferation, colony-formation, apoptosis, migration and invasion capacity was first investigated. Expression levels of NNT-AS1, miR-320a, beta-catenin, RUNX2, IGF-1R, c-Myc, Cyclin D1 and MMP13 were also evaluated by RT-qPCR and western blotting accordingly. Xenograft models using U2OS and OS-732 cells with different NNT-AS1 gene modifications were constructed for tumor formation assay as well as evaluation of miR-320a, beta-catenin and RUNX2 expression in primary lesion. NNT-AS1-overexpressing U2OS cells and NNT-AS1-knockdown OS-732 cells were subject to miR-320a mimic and inhibitor transfection, respectively, to investigate the miR-320a dependency of the osteosarcoma-promoting role of NNT-AS1.
Results: NNT-AS1 overexpression significantly increased proliferation, survival and mobility of U2OS cells in vitro as well as its tumor formation ability in vivo, while NNT-AS1 knockdown showed opposite effect on OS-732 cells. In both in vitro and in vivo model, NNT-AS1 expression level significantly correlated with that of beta-catenin, RUNX2, IGF-1R, c-Myc, Cyclin D1 and MMP13 as well as Akt phosphorylation level, and inversely correlated with miR-320a expression. Transfection of miR-320a mimic significantly inhibiter the promoting effect of NNT-AS1 on cell proliferation, survival and mobility of U2OS cells, while miR-320 inhibitor partially rescued that of OS-732 cells.
Conclusion: NNT-As1 functions as a cancer-promoting lncRNA by downregulating miR-320a, thus increasing the protein expression level of beta-catenin, RUNX2 and IGF-1R as well as activation of Akt in osteosarcoma.
Introduction
Osteosarcoma is the most prevalent bone tumor and is often highly malignant.Citation1 Although the overall survival of patients with osteosarcoma (OS) has improved to a five-year survival of 70%, the outcomes of patients with metastatic or recurrent OS remain poor.Citation2 Development of new and more effective treatment against OS is obstacle by our lack of understanding of the molecular mechanisms in detail of pathogenesis and progression of OS.Citation1 Several key oncogenic genes and pathways have been identified in OS, such as genes belonging to the NOTCH and WNT/beta-catenin signaling pathway as well as transcription factor RUNX2 and Osterix, and amplification or upregulation of these genes are frequently reported in OS.Citation3–Citation6
Long noncoding RNAs (lncRNAs) can regulate gene expression by interacting with microRNAs (miRNAs) or mRNAs. The best understood function of miRNAs is blocking mRNA transcription by direct binding, and lncRNA can rescue the mRNA translation by “sponging” the targeting miRNA and inhibit its function. lncRNA can function as either cancer-promoting or suppressing in OS oncogenesis and development.Citation7,Citation8 PI3K/Akt, NF-kappaB and HIPPO signaling pathways are frequently potentiated by upregulation of lncRNAs in OS, such as MALAT1, HOTTIP and HULC, while decrease in MEG3 has been shown to facilitate OS development.Citation8–Citation11 Nicotinamide Nucleotide Transhydrogenase-antisense RNA1 (NNT-AS1) is a recently identified lncRNA with its function largely underexplored. Wang et al’s research showed that NNT-AS1 is upregulated in colorectal cancer (CRC) and promote CRC development possibly by activating MAPK/Erk signaling pathway with no detectable protein-coding potential.Citation12 Upregulation of NNT-AS1 and its cancer-promoting role in cervical and liver cancer have been demonstrated by Hua et al and Lu et al.Citation13,Citation14
The present research aimed to explore the role of NNT-AS1 in OS. Our preliminary research using DIANA-LncBase V2 found that NNT-AS1 might promote cancer development by sponging miR-320a and other miR-320 family miRNAs. Although the role of miR-320a in OS is undetermined before, its cancer-suppressing role in solid tumors such as liver, lung, breast and pancreatic cancer has been well documented.Citation15–Citation18 Notably, miR-320a is known for its negative regulation of transcription factor RUNX2 and beta-catenin as well as insulin growth factor 1 receptor (IGF-1R).Citation19–Citation22 In the present research we tested our hypothesis that NNT-AS1 promote OS development by sponging miR-320a, thus facilitating the expression of beta-catenin and RUNX2 that functions as oncogenes.
Materials and methods
Cell culture and preparation
U2OS and OS-732 human bone osteosarcoma epithelial cells (referred to as “OS cells” in the present manuscript) were purchased from Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). OS cells were cultured in DMEM with 4500 mg/L D-Glucose (STEMCELL Technologies, Vancouver, Canada) supplemented with 10% FBS (Cell Applications, San Diego, USA) and 100 U/ml of penicillin-streptomycin (Thermo Fisher Scientific, Waltham, USA) in a cell incubator with an humidified atmosphere at 37°C with 5% CO2. U2OS cells with stable NNT-AS1 overexpression and OS-732 cells with stable NNT-AS1 knockdown were constructed by Fulengen (Guangzhou, China). MiR-320a overexpression in NNT-AS1-overexpressing U2OS cells and MiR-320a inhibition in NNT-AS1-knockdown OS-732 cells were achieved by lentiviral transfection of miR-320a precursor and inhibitor, respectively, using a customized kit purchased from Genecopoeia (Guangzhou, China) following manufacturer’s instructions.
RT-qPCR and western blot
For qPCR, total RNA from cell lysate was extracted using a total RNA isolation kit (EpiGentek, Farmingdale, USA) following manufacturer’s instructions. Extracted RNAs were reversely transcribed into cDNAs using a cDNA synthesis kit purchased from Bio-Rad (Hercules, USA) following manufacturer’s instructions. qPCR was performed using FAST-SYBR Green Master Mix and StepOne Plus Real-Time PCR Detection System (Applied Biosystems) following manufacturer’s instructions. GAPDH and U6 snRNA were used as housekeeping genes for determining the relative expression level of NNT-AS1 and miR-320a by 2−ΔΔCt method, respectively. Primer pairs for RT-qPCR evaluating the transcription level of NNT-AS1 and miR-320a were purchased from Genecopoeia. Primer sequences are listed in .
Table 1. Sequences of primer pairs used for RT-qPCR.
For western blot evaluating protein expression, cells were lysed using home-made RIPA buffer, and tumor tissue samples were homogenized by tissue grinder with pre-chilled RIPA buffer. Cell or tissue lysate were obtained by harsh centrifugation, and protein expression level of beta-catenin, RUNX2, IGF-1R, c-MYC, cyclin D1, MMP13, Akt as well as Akt phosphorylation at Ser473 in lysate samples were evaluated by western blot using GAPDH as housekeeping gene. Relative expression was determined by comparing the gray scale of each protein band to that of GAPDH using ImageJ software.
Cell functional assays
Cell proliferation assay was performed using CCK-8 cell counting kit (Dojindo, absorbance, Japan) following manufacturer’s instructions. Briefly, cells cultured on 96-well-plate were incubated with CCK-8 solution for 1 hour before measuring the absorbance at 450 nm (O.D 450) using a microplate reader. Colony formation assay of OS cells were performed as described by Wang et al.Citation23 Cell apoptosis were evaluated by cell surface staining with FITC-conjugated Annexin V/PI probes (BD Biosciences, Franklin USA) followed by flow cytometry. Annexin V positive cells were considered as apoptotic cells, while Annexin V negative, PI positive cells being necrotic. Trans-well assay was performed on a 24-well-plate using Matrigel pre-coated chamber inserts (Costar, Washington DC, USA). OS cells were added in the chamber in serum free condition, and the chamber were inserted in culture medium with 10% FBS. Cells migrated to the bottom of the chamber were stained with 1% crystal violet for 15 min in room temperature before being photographed under magnified view. Scratch assay was performed by scratching the OS cell culture on 12-well plate with a sterile 1ml pipette tip, followed by incubation for 24 hours. Cell mobility were evaluated by determining the migration rate as described previously.Citation24
Tumor formation assay
NU/NU nude mice for tumor formation assay were purchased from Charles River (Wilmington, USA). Xenograft model were constructed by subcutaneous injection of OS cells on the left back of the mice. Tumor volume were determined by measuring the size of tumor with a caliper. At the endpoint of the tumor formation assay, mice were euthanized, and tumor were dissected for weighting and evaluating the expression level of NNT-AS1, miR-320a as well as proteins mentioned above by western blot. Laboratory animal use in this research was approved by the ethical review committee of People’s Hospital of Rizhao.
Statistical analysis
Statistical analysis was performed using SPSS software (Ver. 17). Data in each group represents three replicates and were presented as mean ± SD, unless otherwise indicated. Students’ t test and Sidak’s multiple comparisons test were used for significance test, and statistical significance threshold was set as p < 0.05.
Results
NNT-AS1 promotes cell proliferation, survival and mobility of osteosarcoma cells in vitro
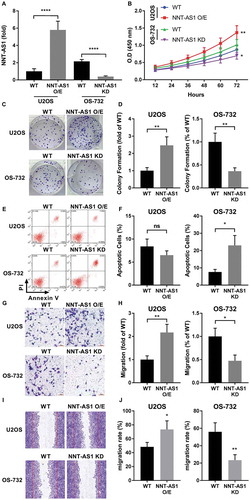
The present study aimed to investigate the role and mechanism of action of NNT-AS1 in OS. Preliminary research analyzing NNT-AS1 expression level in various of OS cell lines in vitro by RT-qPCR suggested that NNT-AS1 expression is relatively high in OS-732 cells and low in U2OS cells (data not shown). Therefore, we chose U2OS and OS-732 cells for gain-of-function and loss-of-function assays, respectively. To investigate the role of NNT-AS1 in OS, we constructed NNT-AS1 overexpressing (O/E) U2OE cells and NNT-AS1 knockdown OS-732 cells ()). Cell functional assays suggested that, after NNT-AS1 overexpression, cell proliferation and colony formation capacity of U2OS cells were significantly increased in vitro, while NNT-AS1 knockdown showed opposite effect on OS-732 cells (~). Apoptosis assay suggested that NNT-AS1 overexpression moderately (not statistically significant) decreased cell apoptosis in U2OS cells, and NNT-AS1 knockdown in OS-732 significantly raised apoptosis rate (,). These results suggested that NNT-AS1 is involved in cell proliferation and survival of OS cells in vitro. Previous research also suggested that increase in NNT-AS1 expression may promote cancer metastasis.Citation12–Citation14 In the present research we investigated the influence of NNT-AS1 overexpression or knockdown on mobility of OS cells in vitro by trans-well assay and scratch assay. We found that NNT-AS1 overexpression significantly increased cell migration rate and trans-well invasion capacity of U2OS cells, while NNT-AS1 knockdown in OS-732 cells showed opposite effects (~). These results suggested that NNT-AS1 is also involved in migration and invasion of OS cells.
Figure 1. NNT-AS1 is involved in osteosarcoma cell proliferation, survival and metastasis in vitro. Cells were assayed 24 hours after transfection for NNT-AS1 overexpression (O/E) or knockdown (KD). A, evaluation of NNT-AS1 in U2OS and OS-732 OS cells with NNT-AS1 overexpression or knockdown by transfection. Data of each group was normalized to that of wildtype (WT) U2OS cells transfected with control vector and presented as fold change. B, cell proliferation evaluated by CCK-8 method. Density of cells in each well on a 96-well plate correlated to absorbance at 450 nm (O.D 450 nm) after incubation with CCK-8 for 1 hour. C and D, colony formation assay evaluating clonogenicity of OS cells with or without NNT-AS1 overexpression or knockdown. C is a representative result of colony formation assay. E and F, flow cytometry evaluating Annexin V/PI surface staining of each group of cells after NNT-AS1 overexpression or knockdown. E is a representative result of flow cytometry. G and H, trans-well assay evaluating the invasiveness of OS cells with different NNT-AS1 expression. G is a representative result of trans-well assay. I and J, scratch assay evaluating cell mobility. I is a representative result of scratch assay. Vertical lines in each image indicate the width of the initial scratch at 0 hour. Data in panel D, H and J were normalized to WT group in the same panel and presented as fold change. Students’ t test was used for significance test in panel A, D, F, H and J, and two-way ANOVA was used for significance test in panel B. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
NNT-AS1 increased OS promoting gene expression while inhibiting miR-320a
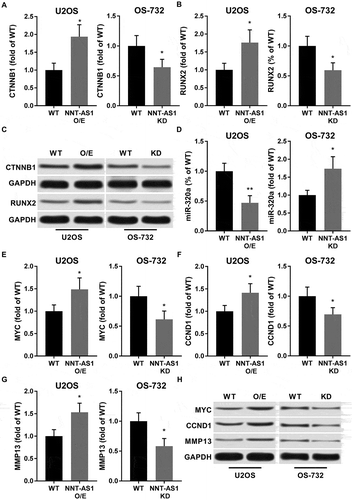
To test our hypothesis that NNT-AS1 promotes OS development by inhibiting miR-320a, we analyzed the expression level of miR-320a as well as its known target genes beta-catenin and RUNX2 that have been determined as important OS-promoting genes.Citation3,Citation19,Citation20 Western blot results suggested that beta-catenin and RUNX2 protein expression level in U2OS cells were significantly increased after NNT-AS1 overexpression along with a significant decrease in miR-320a expression level, and NNT-AS1 knockdown in OS-732 cells showed opposite effect (~). beta-catenin and RUNX2 as two transcription factors promote OS development by increasing the transcription of other cancer promoting genes, such as MYC, cyclin D1 (CCND1) and MMP-13. Our western blot results suggested that protein expressions of these genes were significantly increased by NNT-AS1 overexpression in U2OS cells and significantly decreased in OS-732 cells by NNT-AS1 knockdown, which was in accordance with beta-catenin and RUNX2 protein expression level (~).
Figure 2. NNT-AS1 increased protein expression level of beta-catenin, RUNX2 as well as their down-stream genes MYC, Cyclin D1 and MMP-13 in OS cells in vitro, while inhibiting miR-320a. Cells were assayed 24 hours after transfection. A, B and C, western blot evaluating protein expression level of beta-catenin (CTNNB1) and RUNX2 in different group of cells. GAPDH was used as housekeeping genes. C is a representative result of western blot. D, RT-qPCR evaluating miR-320a expression level in each group of cells. E ~ H, western blot evaluating protein expression level of c-Myc (MYC), cyclin D1 (CCND1) and MMP-13 (MMP13). GAPDH was used as housekeeping genes. F is a representative result of western blot. Data normalized to the WT group and presented as fold change. Students’ t test was used for significance test. *, p < 0.05; **, p < 0.01.
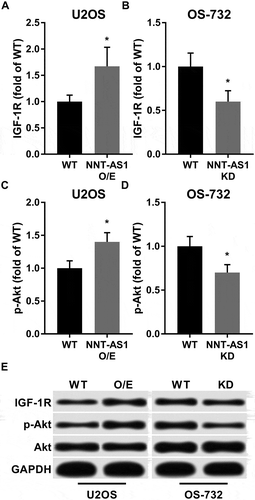
IGF-1R is another previously reported miR-320a target gene, and IGF-1R promotes OS development by activating the PI3K/AKT signaling pathway. Our western blot results showed that protein expression level of IGF-1R as well as Ser473 phosphorylation level of Akt were significantly increased in U2OS cells by NNT-AS1 and decreased in OS-732 cells by NNT-AS1 knockdown in consistent with that of beta-catenin and RUNX2 (). Collectively, these results suggested that NNT-AS1 liberates the expression of these OS promoting genes while inhibiting miR-320a.
Figure 3. Western blot showed that NNT-AS1 increased protein expression level of IGF-1R and Ser473 phosphorylation level of Akt in OS cells in vitro. Cells were treated and analyzed as described in . Total protein expression level of Akt was intact under NNT-AS1 overexpression or knockdown conditions. E is a representative result of western blot. Data in panel A ~ D were normalized to the WT in the same panel group and presented as fold change. Students’ t test was used for significance test. *, p < 0.05.
MiR-320a antagonized the OS-promoting effect of NNT-AS1 in vitro
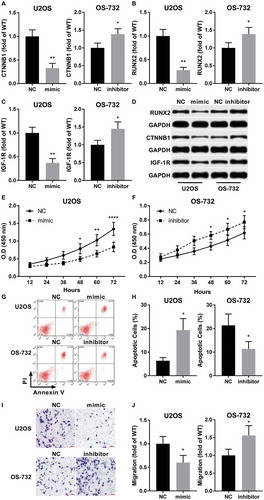
To further investigate the role of miR-320a in NNT-AS1 promoting OS development, we transfected miR-320a precursor into U2OS cells with NNT-AS1 overexpression and miR-320a inhibitor into NNT-AS1 knockdown OS-732 cells. Western blot results suggested that beta-catenin, RUNX2 and IGF-1R protein expression levels in NNT-AS1 overexpressing U2OS cells were dramatically decreased by miR-320a precursor transfection, while transfection of miR-320a inhibitor significantly increased the expression level of these proteins (~). Cell functional assays suggested that mir-320a precursor transfection significantly decreased cell proliferation and increased apoptosis in U2OS cells with NNT-AS1 overexpression, while transfection of mir-320a inhibitor in OS-732 cells showed opposite effects (~). Correspondingly, trans-well assay showed that the invasiveness of NNT-AS1 overexpressing U2OS cells were significantly decreased by miR-320a precursors transfection while that of NNT-AS1 knockdown OS-732 cells significantly increased by miR-320a inhibitor transfection (~). Collectively, these results suggested that NNT-AS1 facilitates OS cell proliferation, survival and invasion in vitro by inhibiting miR-320a, thus increasing the expression of OS-promoting genes targeted by this OS-inhibiting miRNA.
Figure 4. MiR-320a antagonized the OS-promoting role of NNT-AS1 in vitro. Cells were assayed 24 hours after transfection. A ~ D, western blot evaluating protein expression level of beta-catenin (CTNNB1), RUNX2 and IGF-1R in different group of cells. GAPDH was used as housekeeping genes. D is a representative result of western blot. E and F, cell proliferation assay by CCK-8 (as described in ), G and H, evaluation of cell apoptosis by flow cytometry. Cell apoptosis was determined by Annexin V positive staining. G is a representative result of flow cytometry. I and J, tran-well assay evaluating the invasiveness of each group of cells. I is a representative result of trans-well assay. Sidak’s multiple comparisons test was used for significance test in panel E and F, while students’ t test was used for significance test panel A, B, C, H and J. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
NNT-AS1 promote OS tumor development in vivo by inhibiting miR-320a
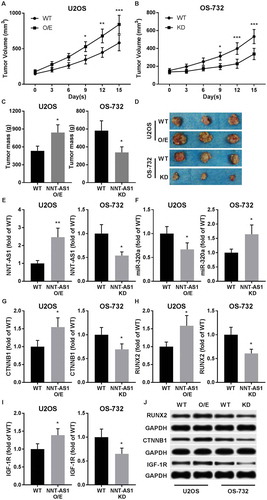
To verify whether NNT-AS1 inhibits miR-320a and promote OS development in vivo, xenograft model was established by subcutaneous injection of U2OS or OS-732 cells with different NNT-AS1 gene modifications in NU/NU nude mice. Our results showed that tumor formation of U2OS cells were significantly facilitated by NNT-AS1 overexpression, while NNT-AS1 knockdown significantly inhibited the tumor formation capacity of OS-732 cells. (~). RT-qPCR results suggested that miR-320a expression was significantly inhibited in U2OS cells vivo by NNT-AS1 overexpression, while its expression in OS-732 cells were significantly increased after NNT-AS1 knockdown (~). Western blot results suggested that protein expression level of beta-catenin, RUNX2 and IGF-1R were significantly increased in xenograft by NNT-AS1 overexpression and decreased by NNT-AS1 knockdown, in consistent with in vitro assay results (~). These data suggested that NNT-AS1 promotes OS development in vivo by inhibiting miR-320a and increasing beta-catenin and RUNX2 protein expression.
Figure 5. NNT-AS1 facilitated OS development in xenograft model. A and B, tumor formation assay evaluating tumor volume in tumor bearing nude mice. OS cells of different groups were subcutaneously injected on the left back of mice. Assay began when initial tumor volume formed by WT cells reached 150 mm3. C and D, tumor weight at the end-point of tumor formation assay. E and F, RT-qPCR evaluating NNT-AS1 and miR-320a in dissected tumor tissue. G ~ J, western blot evaluating protein expression level of beta-catenin (CTNNB1), RUNX2 and IGF-1R in dissected tumor tissues using GAPDH as housekeeping gene. J is a representative result of western blot. Data in panel C, E ~ I were normalized to WT group in the same graph. Sidak’s multiple comparisons test was used for significance test in panel A and B, while students’ t test was used for significance test in panel C, E ~ I. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
In the present research, we discovered for the first time that NNT-AS1 functions as a promoting lncRNA in OS cells in vitro and in vivo by inhibiting miR-320a and increasing the protein expression of beta-catenin and RUNX2. Currently the study on the expression and function of NNT-AS1 is still in its infancy. Three previous study based on clinical data and laboratory findings reported the upregulation of NNT-AS1 in colorectal, cervical and liver cancer in addition to the cancer-promoting role of NNT-AS1 in these three cancer types.Citation12–Citation14 A lncRNA influences gene transcription in cells primarily by interacting with miRNA or mRNA in a complementary base pairing manner.Citation25 We started our investigation by exploring potential NNT-AS1-interacting miRNAs using DIANA-LncBase V2, an online platform that provide lncRNA-miRNA interaction with either experimental evidence basis or high prediction score.Citation26,Citation27 By sorting the experimental evidence (mostly from CLIP-seq data)-supported miRNAs interacting with NNT-AS1 and cross-referencing previous reports regarding these miRNAs, we found that NNT-AS1 might interact with miR-320a, which has been suggested as a cancer-suppressing miRNA in various cancer types such as liver, lung, and breast cancer.Citation15–Citation17 The role of miR-320a in OS cells hasn’t been explored before, but downregulation of beta-catenin and RUNX2 by miR-320a has already been reported in other cell types, and these two transcription factors play vital role in OS pathogenesis and progression.Citation3,Citation20,Citation28 Targeting of RUNX2 by other miRNAs has also been demonstrated as OS suppressive.Citation29,Citation30 Similarly, IGF-1R has been well demonstrated to promote OS development by activating the PI3K/AKT signaling pathway when activated by its ligand, IGF-1, and the direct targeting of IGF-1R by miR-320a has been confirmed by luciferase reporter asay.Citation21,Citation22,Citation31,Citation32
Based on these findings we hypothesized that NNT-AS1 might promote OS development by sponging and functionally inhibiting miR-320a, unleashing the expression of OS-promoting genes such as beta-catenin and RUNX2. Our western blot and RT-qPCR results showed that overexpression of NNT-AS1 in OS cells significantly increased beta-catenin, RUNX2 and IGF-1R protein expression level, as well as those genes regulated by the two transcription factors, such as c-Myc, cyclin D1 and MMP-13, and phosphorylation level of Akt, while miR-320a expression was significantly inhibited in NNT-AS1 overexpressing cells; on the other hand, NNT-AS1 knockdown in OS cells showed opposite effect. Functionally, NNT-AS1 significantly increased cell proliferation, survival, migration and invasion of OS cells in vitro, which can be attenuated by miR-320a overexpression via miR-320a precursor transfection. NNT-AS1 knockdown significantly decreased these cell function parameters in OS cells, that can be partially rescued by miR-320a inhibition. Similarly, in vivo assay also suggested that NNT-AS1 is involved in OS development, possibly by increasing the protein expression of beta-catenin, RUNX2 and IGF-1R.
During the preparation of this manuscript, Ye et al also reported the OS promoting role of NNT-AS1 using similar methods as we employed in the present research; additionally, they demonstrated a significant correlation between increase in NNT-AS1 expression and OS development based on OS patients’ tissue specimens and clinical data.Citation33 Besides in MYC and cyclin D1, Ye et al also demonstrated that knockdown of NNT-AS1 decreased the protein expression of MMP-2/9 and Bcl-2 as well as the activation of Akt. Mutual upregulation and activation of AKT and RUNX2 has been suggested as a pivotal mechanism in OS progression.Citation34 In the present study, more than confirming the phenomena that NNT-AS1 is required for the cell growth and metastasis of OS cells in vitro and tumor formation in vivo, we also explored the molecular mechanism of these OS-promoting effect of NNT-AS1. Our data clearly suggested that NNT-AS1 can promote OS progression by inhibiting miR-320a and increasing the protein expression of beta-catenin, RUNX2 and IGF-1R, MYC, cyclin D1 and MMP-13 as well as the activation of PI3K/AKT signaling pathway.
The function of NNT-AS1 is still largely underexplored. As a lncRNA, NNT-AS1 may interact with multiple different miRNAs or mRNAs, influencing intracellular signal transductions and cell functions in different aspects. Besides miR-320a, other miRNAs belonging to the miR-320 family, including miR-320b, miR-320c and miR-320d, were also suggested to be interacting with NNT-AS1. Currently there’s only little reports regarding the function of miR-320 family miRNAs in OS. A recent report by Lv et al showed that lncRNA XIST promotes OS cell growth by inhibiting miR-320b and thus de-repressing the expression of Ras-related protein Rap-2b (RAP2B), suggesting the tumor-suppressing role of miR-320b in OS.Citation35 Besides, Wu et al also published their research on the OS-suppressing mechanism of “miR-320”, which we presume to be the previously used ID of miR-320a.Citation36 Interestingly, a microarray analysis performed by Braun et al yielded data implying that miR-320a might be a p53-responsive miRNA.Citation37 Their microarray analysis for miRNA expression profiling was performed using SJSA cells with or without Nutlin-3a treatment, a potent inhibitor of Mdm2-p53 interaction that liberates p53 and increases its transcriptional activity. Braun et al’s data showed that, upon Nutlin-3a treatment, miR-320a as well as miR-194 that was further investigated in their research were significantly upregulated, implying the OS-suppressing effect of this miRNA. Interestingly, Davaadelger et al showed that activation of IGF-1R/AKT axis significantly attenuated the Nutlin-3a-induced apoptosis in OS cells.Citation32 Except in OS, miR-320 family members were also found downregulated and functioning as tumor-suppressing miRNAs in cervical, colorectal and prostate cancer.Citation38–Citation40
OS has been considered as of low immunogenicity comparing to other solid tumors, and trials using immune checkpoint inhibitors for OS treatment showed disappointing results.Citation41 As a pilot study, we have examined the expression levels of several immune checkpoint molecules such as PD-L1, PD-L2 and CTLA-4 as well as the expression level of class I MHC molecules by western blot in our in vitro and in vivo models. However, we found no significant influence of NNT-AS1 overexpression or knockdown on the expression level of proteins mentioned above (data not shown). It seems that NNT-AS1 is not involved in the regulation of these immune checkpoint proteins or class I MHC molecules expression in osteosarcoma. Nevertheless, we propose that interaction of NNT-AS1 with these miR-320 family miRNAs and the role of these interactions should be further investigated in osteosarcoma.
Collectively, our data suggested that lncRNA NNT-AS1 promotes OS cell growth and tumor development by inhibiting miR-320a and hence increasing the protein expression of other OS-promoting genes, such as beta-catenin, RUNX2 and IGF-1R, which have been shown as direct targets of miR-320a, as well as the activation of PI3K/AKT signaling pathway. Targeting NNT-AS1/miR-320a axis might serve as novel therapeutic strategy for OS management.
Authors’ Contribution
This work was conceived and designed by Changhui Li and Shouyun Zhang. The experiments were carried out by Tongguo Qiu and Yuanji Wang. The manuscript was prepared by David M.Ricketts, Chao Qi and Changhui Li.
Competing interests
The authors have no conflicts of interests to declare.
Acknowledgments
This work was supported by 2018 Medicine and Health Scienceand Technology Development Planning Project of Shandong Province. Number 2018WSA11023.
References
- Lin YH, Jewell BE, Gingold J, Lu L, Zhao R, Wang LL, Lee DF. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol Med. 2017;23:737–755. doi:10.1016/j.molmed.2017.06.004.
- Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029–3035. doi:10.1200/jco.2014.59.4895.
- Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13:480–491. doi:10.1038/nrendo.2017.16.
- Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi:10.1038/nrc3838.
- Zanotti S, Canalis E. Notch signaling and the skeleton. Endocr Rev. 2016;37:223–253. doi:10.1210/er.2016-1002.
- Wagner ER, Luther G, Zhu G, Luo Q, Shi Q, Kim SH, Gao JL, Huang E, Gao Y, Yang K, et al. Defective osteogenic differentiation in the development of osteosarcoma. Sarcoma. 2011;2011:325238. doi:10.1155/2011/325238.
- Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016;7:e2389. doi:10.1038/cddis.2016.272.
- Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407–1419. doi:10.1159/000479205.
- Zhao J, Ma ST. Downregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-kappaB pathway. Mol Med Rep. 2018;17:7388–7394. doi:10.3892/mmr.2018.8746.
- Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi:10.1007/s13277-014-2631-4.
- Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8:15138–15142.
- Wang Q, Yang L, Hu X, Jiang Y, Hu Y, Liu Z, Liu J, Wen T, Ma Y, An G, et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8:3441–3453. doi:10.18632/oncotarget.13840.
- Lu YB, Jiang Q, Yang MY, Zhou JX, Zhang Q. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget. 2017;8:88804–88814. doi:10.18632/oncotarget.21321.
- Hua F, Liu S, Zhu L, Ma N, Jiang S, Yang J. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/beta-catenin signaling pathway in cervical cancer. Biomed Pharmacother. 2017;92:1128–1134. doi:10.1016/j.biopha.2017.03.057.
- Lv G, Wu M, Wang M, Jiang X, Du J, Zhang K, Li D, Ma N, Peng Y, Wang L, et al. miR-320a regulates high mobility group box 1 expression and inhibits invasion and metastasis in hepatocellular carcinoma. Liver Int. 2017;37:1354–1364. doi:10.1111/liv.13424.
- Lv Q, Jx H, Yj L, Xie N, Song DD, Zhao W, Yan YF, Bs L, Wang PY, Xie SY. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol Ther. 2017;18:142–151. doi:10.1080/15384047.2017.1281497.
- Yu J, Wang JG, Zhang L, Yang HP, Wang L, Ding D, Chen Q, Yang WL, Ren KH, Zhou DM, et al. MicroRNA-320a inhibits breast cancer metastasis by targeting metadherin. Oncotarget. 2016;7:38612–38625. doi:10.18632/oncotarget.9572.
- Wang W, Zhao L, Wei X, Wang L, Liu S, Yang Y, Wang F, Sun G, Zhang J, Ma Y, et al. MicroRNA-320a promotes 5-FU resistance in human pancreatic cancer cells. Sci Rep. 2016;6:27641. doi:10.1038/srep27641.
- Hamam D, Ali D, Vishnubalaji R, Hamam R, Al-Nbaheen M, Chen L, Kassem M, Aldahmash A, Alajez NM. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell Death Dis. 2014;5:e1499. doi:10.1038/cddis.2014.462.
- Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem Biophys Res Commun. 2012;420:787–792. doi:10.1016/j.bbrc.2012.03.075.
- Wang J, Shi C, Wang J, Cao L, Zhong L, Wang D. MicroRNA-320a is downregulated in non-small cell lung cancer and suppresses tumor cell growth and invasion by directly targeting insulin-like growth factor 1 receptor. Oncol Lett. 2017;13:3247–3252. doi:10.3892/ol.2017.5863.
- Guo T, Feng Y, Liu Q, Yang X, Jiang T, Chen Y, Zhang Q. MicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1R. Tumour Biol. 2014;35:11269–11275. doi:10.1007/s13277-014-2283-4.
- Wang W, Li X, Meng FB, Wang ZX, Zhao RT, Yang CY. Effects of the long non-coding RNA HOST2 on the proliferation, migration, invasion and apoptosis of human osteosarcoma cells. Cell Physiol Biochem. 2017;43:320–330. doi:10.1159/000480412.
- Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol. 2005;294:23–29.
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi:10.1158/2159-8290.cd-13-0202.
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–286. doi:10.1007/978-1-4939-3378-5_21.
- Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–8. doi:10.1093/nar/gkv1270.
- Zhang CL, Wang H, Yan CY, Gao XF, Ling XJ. Deregulation of RUNX2 by miR-320a deficiency impairs steroidogenesis in cumulus granulosa cells from polycystic ovary syndrome (PCOS) patients. Biochem Biophys Res Commun. 2017;482:1469–1476. doi:10.1016/j.bbrc.2016.12.059.
- Xie Y, Sun W, Deng Z, Zhu X, Hu C, Cai L. MiR-302b suppresses osteosarcoma cell migration and invasion by targeting Runx2. Sci Rep. 2017;7:13388. doi:10.1038/s41598-017-13353-9.
- He Y, Meng C, Shao Z, Wang H, Yang S. MiR-23a functions as a tumor suppressor in osteosarcoma. Cell Physiol Biochem. 2014;34:1485–1496. doi:10.1159/000366353.
- Chiu YJ, Hour MJ, Jin YA, Lu CC, Tsai FJ, Chen TL, Ma H, Juan YN, Yang JS. Disruption of IGF1R signaling by a novel quinazoline derivative, HMJ30, inhibits invasiveness and reverses epithelial-mesenchymal transition in osteosarcoma U2 OS cells. Int J Oncol. 2018. doi:10.3892/ijo.2018.4325.
- Davaadelger B, Perez RE, Zhou Y, Duan L, Gitelis S, Maki CG. The IGF-1R/AKT pathway has opposing effects on Nutlin-3a-induced apoptosis. Cancer Biol Ther. 2017;18:895–903. doi:10.1080/15384047.2017.1345397.
- Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of long non-coding RNA NNT-AS1 correlates with tumor progression and poor prognosis in osteosarcoma. Cell Physiol Biochem. 2018;45:1904–1914. doi:10.1159/000487966.
- Cohen-Solal KA, Boregowda RK, Lasfar A. RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol Cancer. 2015;14:137. doi:10.1186/s12943-015-0404-3.
- Lv GY, Miao J, Zhang XL. Long non-coding RNA XIST promotes osteosarcoma progression by targeting ras-related protein RAP2B via miR-320b. Oncol Res. 2017. doi:10.3727/096504017X14920318811721.
- Wu H, Li W, Zhang M, Zhu S, Zhang D, Wang X. Inhibitory roles of miR-320 in osteosarcoma via regulating E2F1. J Cancer Res Ther. 2016;12:68–71. doi:10.4103/0973-1482.191635.
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi:10.1158/0008-5472.CAN-08-1569.
- Vishnubalaji R, Hamam R, Yue S, Al-Obeed O, Kassem M, Liu FF, Aldahmash A, Alajez NM. MicroRNA-320 suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1. Oncotarget. 2016;7:35789–35802. doi:10.18632/oncotarget.8937.
- Zhang T, Zou P, Wang T, Xiang J, Cheng J, Chen D, Zhou J. Down-regulation of miR-320 associated with cancer progression and cell apoptosis via targeting Mcl-1 in cervical cancer. Tumour Biol. 2016;37:8931–8940. doi:10.1007/s13277-015-4771-6.
- Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, Yeh SD, Hong TM, Chen YL. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi:10.1093/carcin/bgs371.
- Saraf AJ, Fenger JM, Roberts RD. Osteosarcoma: accelerating progress makes for a hopeful future. Front Oncol. 2018;8:4. doi:10.3389/fonc.2018.00004.
