ABSTRACT
Accumulative reports have documented the critical functions of long non-coding RNAs (lncRNAs) in the progression of malignant tumors, including bladder cancer (BCa). LncRNA ARAP1-AS1 was chosen to be the object of this study due to it was significantly upregulated in the BCa samples of TCGA database. qRT-PCR further validated the dysregulation of ARAP1-AS1 in 88 pairs of BCa tissues and six BCa cells. Kaplan Meier analysis was utilized to analyze the prognostic value of ARAP1-AS1 for patients with BCa. To evaluate the oncogenic property of ARAP1-AS1 in bladder cancer, loss-of-function assays were conducted in two BCa cells in which ARAP1-AS1 was expressed highest. Mechanically, ARAP1-AS1 was enriched in the cytoplasm of BCa cells, suggesting that ARAP1-AS1 might act as a ceRNA to regulate gene expression and biological processes in bladder cancer. It was certified that ARAP1-AS1 can bind with miR-4735-3p in BCa cells. Moreover, functional assays supported the hypothesis that miR-4735-3p is a tumor suppressor in BCa. Additionally, NOTCH2 mRNA could be targeted by miR-4735-3p in BCa cells. The results of all mechanism experiments indicated that ARAP1-AS1 regulated miR-4735-3p/NOTCH2 axis in BCa by acting as a ceRNA. All our research findings may bring novel insights into the treatment of bladder cancer.
Introduction
Bladder cancer (BCa) is the ninth commonest malignant cancer throughout the world, and increasing new cases are diagnosed worldwide each year.Citation1–Citation4 In spite of the great progress has been made in the clinical treatment of BCa, the overall survival rate for BCa patients is still pessimistic.Citation5–Citation8 The prognosis of BCa patients is closely related to the clinical stage of BCa, but the specific early-stage symptoms for BCa patients are unclear.Citation9–Citation12 Thus, it is still urgent to differentiate novel biomarkers for the early diagnosis and efficient treatment for BCa patients.
On the basis of the genome sequencing technology, there are only about 2% of the mammalian genome is able to encode proteins, while almost the rest belong to non-protein-coding RNAs (ncRNAs).Citation13 As a main subgroup of the ncRNAs, long non-coding RNAs (lncRNAs) are longer than 200 nt and participate in various biological modifications including epigenetic modification, transcriptional or post-transcriptional regulation.Citation14,Citation15 According to the data of TCGA database, lncRNA ARAP1-AS1 was significantly upregulated in the BCa samples. The expression pattern of ARAP1-AS1 was further identified in BCa tissues and cell lines with qRT-PCR. Kaplan-Meier method was then utilized to evaluate the prognostic importance of ARAP1-AS1 for patients with BCa. Subsequently, the effects of ARAP1-AS1 on BCa cell processes were detected by performing functional assays. LncRNAs is well-known as endogenous competitively RNAs (ceRNA) in human cancers, thereby modulating their downstream gene expression.Citation16–Citation19 Here, we explored whether ARAP1-AS1 can act as a ceRNA to regulate cellular processes in BCa. Mechanism experiments were designed and applied to detect the ceRNA pathway which centered with lncRNA ARAP1-AS1. Intriguingly, ARAP1-AS1 upregulated the expression of NOTCH2 by sponging miR-4735-3p. Finally, rescue assays were utilized to demonstrate the function of ARAP1-AS1-miR-4735-3p-NOTCH2 pathway in BCa progression.
Materials and methods
Patient samples
88 pairs of bladder cancer (BCa) tissues and the corresponding non-tumorous tissues from patients were given by PLA Army General Hospital. Before the research, none of the patients were treated with any chemotherapy or radiotherapy. Each sample was immediately frozen with liquid nitrogen and saved at −80°C until use. Our study had obtained the recognition of the Ethics Committee of PLA Army General Hospital. All bladder cancer patients enrolled in this study signed written informed consent.
Cell culture
All cell lines used in this study including one immortalized human bladder epithelium cell line (SV-HUC) and six bladder cancer cell lines (T24, 5637, EJ, TCC, 253J and J82) were provided by the American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in RPMI-1640 medium (GiBCaao, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin and then incubated at 37°C in a humidified incubator with 5% CO2.
Cell transfection
The sequence of NOTCH2 was synthesized and sub-cloned into pcDNA 3.1 vector (pcDNA-NOTCH2; Invitrogen) by GenePharma Corporation (Shanghai, China). sh-ARAP1-AS1 and control shRNA (sh-NC), miR-4735-3p mimics/inhibitors and corresponding miR-NC were also synthesized by GenePharma Corporation (Shanghai, China). BCa cells (5 × 104 cells/ml) were plated into 6-well plates for 24 h. Lipofectamine 2000 reagents (Invitrogen) was used for transfection when the cell confluence reached more than 70%.
QRT-PCR
RNAs were isolated from BCa cells by Trizol reagent (Invitrogen, Carlsbad, USA). The RNA samples were then reversely transcribed into cDNA by the PrimeScript RT Reagent Kit (Takara, Japan). The SYBR Green PCR Kit (Takara, Japan) was used to measure gene expression on the ABI 7500 Fast Real-Time PCR system (Applied Biosystems, CA, USA). GAPDH acted as a normal control. All primers for qRT-PCR assay were synthesized by GenePharma (Shanghai, China). The relative expression levels of genes were evaluated using the 2−ΔΔCt method.
MTT assay
The 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay was used to detect the cell viability. Cells (1 × 105 cells/ml) were cultured in 96-well plates for 48 h. Then, cells in each well which was added into the 10 µl of MTT solution were incubated for 4 h. The remaining MTT solution was discarded and then 100 μl dimethyl sulfoxide (DMSO) was added to dissolve the formazan crystals in each well for 5 min. The cell viability was identified by detecting the absorbance at 490 nm using a microplate reader (Thermo Scientific).
Colony formation assay
After transfection, cells (500–800 cells/well) were seeded into 6-well plates with RPMI-1640 medium for 14 days and the medium was replaced every three days. Then, the cells were washed gently with PBS before they were fixed with methanol and stained with 0.1% crystal violet (Sigma). Colonies (more than 50 cells) were counted under a microscope and the rate of colony formation was calculated by using Image J software (NIH, Bethesda, MD).
Transwell assays
Transwell plate with a filter of 8 µm pores (BD Biosciences, San Diego, CA, USA) was used to test the migration of cells. 200 μl of serum-free medium with 5 × 105 cells was added into the upper chamber and 600 μl of 20% FBS-containing medium was added into the lower chamber. After 48 h of incubation, the migratory cells were fixed with methanol and dyed with 0.1% crystal violet. The number of migratory cells (five random fields per chamber) was identified by using a light microscope. Cell invasion assay was performed with a similar way except 50 µl Matrigel (BD Biosciences) was coated on the upper compartment.
Animal experiments
For in vivo experiments, 4 weeks old male BALB/c nude mice were grow at n normal environment free from pathogen (SPF). The nude mice were divided into two groups. These two groups were separately tail intravenously injected with J82 cells stably transfected with sh-ARAP1-AS1 or negative control (sh-NC) at a density of 1 × 106 cells per mouse. 28 days later, the mice were killed. Animal study was approved by the Animal Welfare and Research Ethics Committee at PLA Army General Hospital. The experiment was conducted in line with the Guide for the Care and Use of Laboratory Animals.
Western blot assay
BCa cells were lysed by RIPA buffer (Sigma-Aldrich) supplemented with protease inhibitors cocktail (Roche, USA). Proteins were determined by bicinchoninic acid method. Cellular proteins were isolated by 10% SDS-PAGE and then transferred onto PVDF membrane (Millipore, Billerica, MA, USA). After sealing with skim milk, membranes were incubated overnight at 4°C with primary antibodies against NOTCH2 (1: 1000; Proteintech, Proteintech Group, USA). Then, membranes were cultivated with second antibody (1: 5000) for 2 h at room temperature. Finally, the blots were measured by the EZ-ECL chemiluminescence Detection kit for HRP (Biological Industries, Beit-Haemek, Israel). GAPDH (1: 3000; Proteintech) acted as the loading control.
Subcellular fractionation assay
The extracts of cytoplasmic and nuclear were isolated from BCa cells by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA). RNAs isolated from cytoplasm or nucleus were examined by qRT–PCR. The distribution of cytoplasmic control (GAPDH) and nuclear control (U6) was also determined with qRT–PCR.
RIP assay
RIP assay was performed with a MagnaRIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). The BCaA cell lysates were incubated in RIP buffer containing magnetic beads which were coated with Ago2 antibodies. Input and normal IgG acted as controls. After digesting protein using proteinase K, the immunoprecipitated RNA was isolated. Purified RNAs were examined with quantitative real-time PCR (qRT-PCR).
Dual luciferase reporter assay
Dual luciferase reporter assay was performed in J82 or T24 cells to examine whether ARAP1-AS1 interacted with miR-4735-3p. Recognize the binding sites of miR-4735-3p and ARAP1-AS1 on DIANA (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php). The recombinant plasmids (ARAP1-AS1-WT and ARAP1-AS1-Mut) were respectively co-transfected with miR-4735-3p mimic or miR-NC into J82 or T24 cells. After 48 h of incubation, luciferase activity was detected by dual-luciferase assay system (Promega).
Statistical analysis
SPSS (vision, 19.0; SPSS, Inc., Chicago, IL, USA) was applied to analyze the statistical data. All data obtained from three independent experiments were shown as mean ± standard deviation (SD). Student’s t-test or one-way ANOVA was used to compare the differences among groups. Kaplan-Meier method and the log-rank test were utilized to determine the survival rate of patients with different ARAP1-AS1 level. p < 0.05 was acknowledged to be statistically significant.
Results
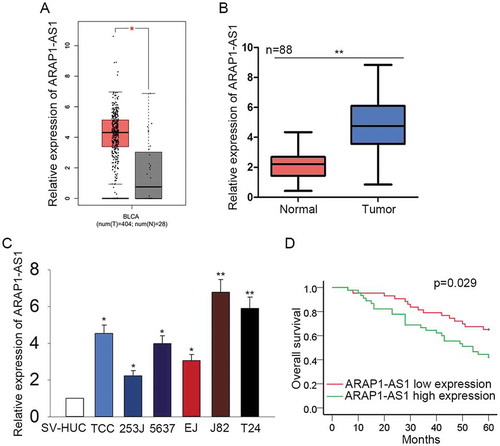
Highly expressed ARAP1-AS1 predicted the unfavorable prognosis for patients with BCa
ARAP1-AS1 was chosen to be the research object of this study due to it was highly expressed in the BCa samples of TCGA database (). qRT-PCR was performed to quantify the expression of ARAP1-AS1 in both BCa tissues and cell lines. It was uncovered that ARAP1-AS1 was up-regulated in bladder cancer tissues and cell lines (-). According to the mean expression value of ARAP1-AS1, 88 BCa specimens were divided into two groups: ARAP1-AS1 high expression (n = 45) and ARAP1-AS1 low expression (n = 43). According to the Kaplan-Meier analysis, the up-regulation of ARAP1-AS1 indicated the poor overall survival for patients with BCa ().
Figure 1. Highly expressed ARAP1-AS1 predicted the unfavorable prognosis for patients with BCa.
A. The expression condition of ARAP1-AS1 in the BCa samples and normal controls was detected in TCGA database. B-C. qRT-PCR was performed to quantify the expression of ARAP1-AS1 in both BCa tissues and cell lines. D. Kaplan-Meier analysis was used to reveal the correlation between the expression of ARAP1-AS1 and the overall survival of patients with BCa. *p < 0.05 and **p < 0.01 vs. control group.
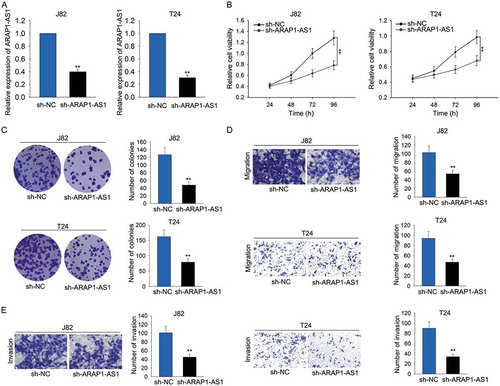
Knockdown of ARAP1-AS1 suppressed bladder cancer cell proliferation, migration and invasion
As illustrated in , ARAP1-AS1 was expressed highest in J82 and T24 cells. To elucidate the oncogenic effects of ARAP1-AS1 on BCa cell processes, high expression of ARAP1-AS1 was knocked down in J82 and T24 cells via transfecting specific shRNA (sh-ARAP1-AS1). Cells transfected with control shRNA (sh-NC) were used as the control group for subsequent experiments. Two days after transfection, qRT-PCR was applied to harvest the transfection results (). Subsequently, cell proliferative assays including MTT and colony formation assays were performed in indicated T24 and J82 cells. Unsurprisingly, the cell proliferative ability was markedly weakened by sh-ARAP1-AS1 (-C). In addition, transwell assays were utilized to examine the migratory and invasive ability of J82 and T24 cells transfected with sh-ARAP1-AS1. As expected, both migration and invasion of indicated cells were markedly repressed (-E).
Figure 2. Knockdown of ARAP1-AS1 suppressed bladder cancer cell proliferation, migration and invasion.
A. High expression of ARAP1-AS1 was knocked down in J82 and T24 cells via transfecting specific shRNA (sh-ARAP1-AS1). Cells transfected with control shRNA (sh-NC) were used as the control group for subsequent experiments. B-C. Cell proliferative assays (MTT and colony formation assays) were performed in indicated T24 and J82 cells. D-E. Transwell assays were utilized to examine the migratory and invasive ability of J82 and T24 cells transfected with sh-ARAP1-AS1. **p < 0.01 vs. control group.
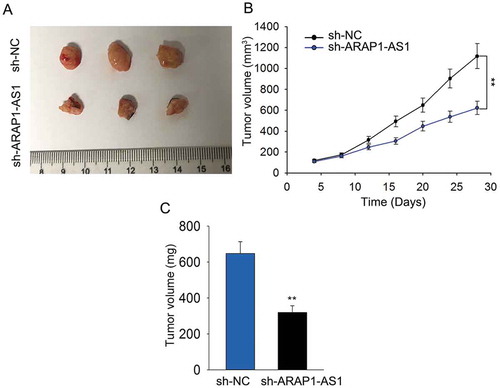
Knockdown of ARAP1-AS1 inhibited bca cell growth in vivo
In vivo experiment was conducted to validate the role of ARAP1-AS1. It was found that tumors derived from J82 cells stably transfected with sh-ARAP1-AS1 were smaller than tumors derived from J82 cells stably transfected with sh-NC (). Moreover, both tumor volume and tumor weight became smaller in response to ARAP1-AS1 knockdown (-).
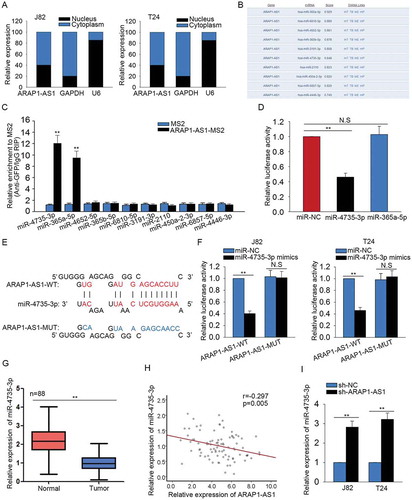
ARAP1-AS1 acted as a ceRNA to bind mir-4735-3p
Mechanically, lncRNAs can act as a ceRNA to modulate the gene expression in human cancers. Here, we investigated the mechanism of ARAP1-AS1 in BCa. The localization of ARAP1-AS1 was identified in J82 and T24 cells with subcellular fractionation assay. ARAP1-AS1 was obviously enriched in the cytoplasm of J82 and T24 cells (). Next, top ten miRNAs which can bind with ARAP1-AS1 were predicted and obtained from DIANA (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) (). To determine which one of those ten miRNAs can bind with ARAP1-AS1 in BCa cells. MS2-RIP assay was carried out. As presented in , two miRNAs (miR-4735-3p and miR-36a-5p) of ten miRNAs were enriched in ARAP1-AS1 immunoprecipitates which were treated with MS2 vector. Furthermore, the luciferase reporter vector containing ARAP1-AS1 was constructed to demonstrate the binding relation between ARAP1-AS1 and two miRNAs above. The results indicated that miR-4735-3p is the only one could decreased the luciferase activity of ARAP1-AS1 vector (). For further analysis, the binding sequence between the wild type ARAP1-AS1 (ARAP1-AS1-WT) or mutant type ARAP1-AS1 (ARAP1-AS1-MUT) and miR-4735-3p was obtained and illustrated (). Further luciferase reporter assay was performed to confirm the combination between ARAP1-AS1 and miR-4735-3p. As a result, miR-4735-3p mimics efficiently repressed the luciferase activity of ARAP1-AS1-WT but not that of ARAP1-AS1-MUT (). The low level of ARAP1-AS1 was observed in BCa tissues using qRT-PCR (). The negative expression relevance between ARAP1-AS1 and miR-4735-3p in BCa tissues was then analyzed (). At last, the expression of miR-4735-3p was increased in APAP1-AS1-dwonregulated BCa cells ().
Figure 4. ARAP1-AS1 acted as a ceRNA to bind miR-4735-3p.
A. The localization of ARAP1-AS1 was identified in J82 and T24 cells with subcellular fractionation assay. B. Top ten miRNAs which can bind with ARAP1-AS1 were predicted and obtained from DIANA. C. MS2-RIP assay was carried out to determine which one of those ten miRNAs can bind with ARAP1-AS1 in BCa cells. D. The luciferase activity analysis was utilized to demonstrate the binding relation between ARAP1-AS1 and two candidate miRNAs. E. The binding sequence between the wild type ARAP1-AS1 (ARAP1-AS1-WT) or mutant type ARAP1-AS1 (ARAP1-AS1-MUT) and miR-4735-3p was obtained and illustrated. F. Further luciferase reporter assay was performed to confirm the combination between ARAP1-AS1 and miR-4735-3p. G. The level of ARAP1-AS1 was observed in BCa tissues using qRT-PCR. H. The expression relevance between ARAP1-AS1 and miR-4735-3p in BCa tissues was analyzed. I. The expression change of miR-4735-3p was detected in APAP1-AS1-dwonregulated BCa cells. **p < 0.01 vs. control group. N.S: no significance.
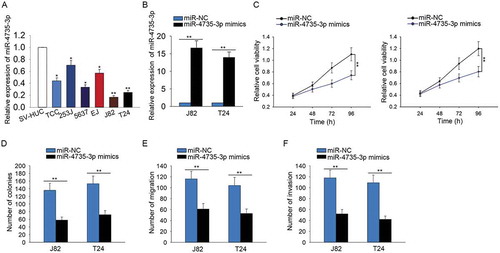
miR-4735-3p exerted anti-oncogenic function in BCa
The expression of miR-4735-3p was identified to be lower in BCa cells than that in SV-HUC cell (). To identify the effect of miR-4735-3p on BCa cell proliferation, migration and invasion, the gain-of-function assays were carried out. For gain-of-function assay, the level of miR-4735-3p was increased in J82 and T24 cells by transfecting with miR-4735-3p mimics (). According to the results of proliferation assays, the proliferative ability of cells transfected with miR-4735-3p mimics was significantly weakened compared with that of cells transfected with miR-NC (-D). Additionally, the inhibitory effects of miR-4735-3p on the migration and invasion of BCa cells were identified (-).
Figure 5. MiR-4735-3p exerted anti-oncogenic function in BCa.
A. The specific expression of miR-4735-3p in BCa cells and SV-HUC cell was identified. B. The level of miR-4735-3p was increased in J82 and T24 cells by transfecting with miR-4735-3p mimics. C-D. The proliferative ability of cells transfected with miR-4735-3p mimics was examined with MTT and colony forming assays. E-F. The influences of miR-4735-3p mimics on the migration and invasion of BCa cells were identified using transwell assays. *p < 0.05 and **p < 0.01 vs. control group.
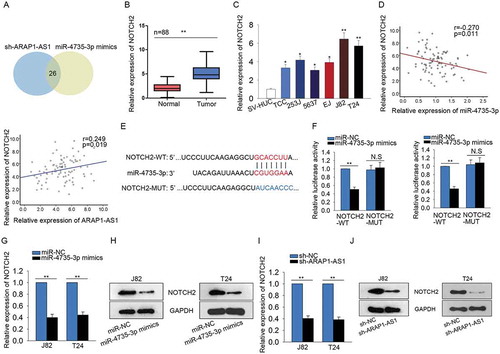
ARAP1-AS1 positively regulated NOTCH2 by sponging miR-4735-3p
457 putative target mRNAs of miR-4735-3p were found from targetScan (http://www.targetscan.org/vert_72/). Next, we examined the expression level of these 457 potential targets in response to ARAP1-AS1 knockdown or miR-4735-3p overexpression. At a result, we found 26 potential targets can be downregulated by both ARAP1-AS1 knockdown or miR-4735-3p overexpression (). Among these 26 mRNAs, NOTCH2 has been studied in bladder cancer as an oncogene. Thus, we chose NOTCH2 for further detection. Subsequently, the higher expression of NOTCH2 was tested in BCa tissues and cell lines (-). Combining with the data above, the negative relationship between NOTH2 and miR-4735-3p in BCa tissues was analyzed. Whereas, the expression correlation between ARAP1-AS1 and NOTCH2 in BCa tissues was found to be positive (). The putative binding sequence between NOTCH2 and miR-4735-3p was shown in . Dual luciferase reporter assay further validated the combination between miR-4735-3p and NOTCH2. The results manifested that miR-4735-3p mimics efficiently decreased the luciferase activity of wild type NOTCH2 (NOTCH2-WT) but not that of mutated NOTCH2 (NOTCH2-MUT) (). Both mRNA and protein levels of NOTCH2 were negatively regulated by miR-4735-3p (-), while was positively regulated by ARAP1-AS1 (-). Taken together, ARAP1-AS1 regulated miR-4735-3p/NOTCH2 axis by acting as a ceRNA.
Figure 6. ARAP1-AS1 positively regulated NOTCH2 by sponging miR-4735-3p.
A. 457 potential target mRNAs of miR-4735-3p were found from targetScan. The expression change of all these mRNAs was detected in response to both sh-ARAP1-AS1 and miR-4735-3p mimics. B-C. The expression level of NOTH2 was examined with qRT-PCR in both BCa tissues and cell lines. D. The expression correlation between NOTH2 and miR-4735-3p in BCa tissues was analyzed (left). The expression correlation between ARAP1-AS1 and NOTCH2 was assessed (right). E. The putative binding sequence between NOTCH2 and miR-4735-3p obtained from targetScan. F. Dual luciferase reporter assay further validated the combination between miR-4735-3p and NOTCH2. G-H. Both mRNA and protein levels of NOTCH2 were tested in J82 and T24 cells transfected with miR-4735-3p mimics. I-J. The expression of NOTCH2 was tested in cells transfected with sh-ARAP1-AS1. *p < 0.05 and **p < 0.01 vs. control group.
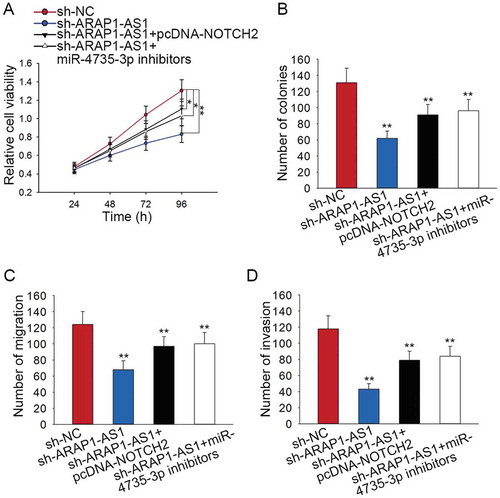
The effects of ARAP1-AS1-miR-4735-3p-NOTCH2 axis on the BCa progression
Rescue assays were designed and conducted in J82 cell to validate the function of ARAP1-AS1-miR-4735-3p-NOTCH2 axis in BCa progression. Results of MTT assay and colony formation assay suggested that cell proliferation suppressed by sh-ARAP1-AS1 was partially recovered by pcDNA-NOTCH2 or miR-4735-3p inhibitors (-). Moreover, the inhibitory effects of sh-ARAP1-AS1 on the migration and invasion of J82 cell were partly abolished by pcDNA-NOTCH2 or miR-4735-3p inhibitors (-).
Figure 7. The effects of ARAP1-AS1-miR-4735-3p-NOTCH2 axis on the BCa progression.
A-B. Cell proliferation ability of indicated J82 cell was examined by applying rescue assays. C-D. The migration and invasion of J82 cell were tested by performing rescue assays. *p < 0.05 and **p < 0.01 vs. control group.
Discussion
Increasing studies have disclosed that lncRNAs are critical participants in the tumorigenesis.Citation20–Citation22 Long non-coding RNAs (lncRNAs) are a group of RNA molecules with more than 200 nt in length and little protein-coding ability.Citation23 In present, the effects of lncRNAs on the biological processes have attracted great research attention. Moreover, the dysregulation of lncRNAs is crucial for the pathogenesis of cancers and provide the novel pathway for cancer treatment.Citation24,Citation25 This study focused on the underlying molecular mechanism and biological function of a certain lncRNAs in bladder cancer. Based on the analysis of TCGA data, lncRNA ARAP1-AS1 was markedly upregulated in BCa tissues than that in the normal controls. The ectopic expression of ARAP1-AS1 in BCa tissues and cell lines was further confirmed by applying qRT-PCR. The prognostic significance of ARAP1-AS1 expression for patients with BCa was analyzed and identified with Kalan-Meier method. The data showed that highly expressed ARAP1-AS1 was certified to be a poor prognostic element for patients with BCa. The biological function of dysregulated ARAP1-AS1 in BCa was then examined with loss-of-function assay. As expected, silenced ARAP1-AS1 inhibited the proliferation migration and invasion of BCa cells, indicating ARAP1-AS1 exerted oncogenic function in the progression of BCa. In vivo experiment further validated the oncogenic role of ARAP1-AS1 in BCa progression.
As reported previously, lncRNAs can modulate gene expression in human cancers by acting as ceRNAs.Citation26–Citation30 Here, we designed mechanism experiments to detect whether ARAP1-AS1 can act as a ceRNA in BCa. The localization of ARAP1-AS1 was detected in BCa cells. The expression of ARAP1-AS1 was enriched in the cytoplasm of BCa cells, suggesting the potential ceRNA role of ARAP1-AS1 in BCa cells. It is well-known that lncRNAs usually exert ceRNA function in human cancers via sponging miRNAs.Citation30–Citation32 Based on the bioinformatics analysis, ten candidate miRNAs were chosen for further analysis. Further mechanism experiments demonstrated that ARAP1-AS1 can bind with miR-4735-3p in BCa cells. Moreover, ARAP1-AS1 negatively regulated the expression of miR-4735-3p. Gain-of-function assays validated the anti-oncogenic role of miR-4735-3p in BCa cells. Bioinformatics analysis and mechanism investigation were utilized to find the target mRNA of miR-4735-3p in BCa cells. NOTCH2 was verified to be the target mRNA of miR-4735-3p in BCa cells. Additionally, NOTCH2 was negatively regulated by miR-4735-3p, while was positively regulated by ARAP1-AS1. Taken all together, ARAP1-AS1, miR-4735-3p and NOTCH2 contributed to a ceRNA pathway in BCa. Finally, rescue assays were applied to certify the effects of ARAP1-AS1-miR-4735-3p-NOTCH2 axis on the proliferation and migration of BCa cells. In conclusion, we confirmed that lncRNA ARAP1-AS1 contributes to the proliferation and migration of BCa cells by regulating miR-4735-3p-NOTCH2 axis. It was expected that the findings in this study could bring novel insights into the treatment of bladder cancer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors convey their sincere thanks to all members engaged in this study.
Additional information
Funding
References
- Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–241. doi:10.1016/j.eururo.2012.07.033.
- Comprehensive molecular characterization of urothelial bladder carcinoma Cancer Genome Atlas Research Network.. Nature. 2014;507:315–322. doi:10.1038/nature12965.
- Chamie K, Ballon-Landa E, Bassett JC, Daskivich TJ, Leventhal M, Deapen D, Litwin MS. Quality of diagnostic staging in patients with bladder cancer: a process-outcomes link. Cancer. 2015;121:379–385. doi:10.1002/cncr.29071.
- Chamie K, Litwin MS, Bassett JC, Daskivich TJ, Lai J, Hanley JM, Konety BR, Saigal CS. Recurrence of high-risk bladder cancer: a population-based analysis. Cancer. 2013;119:3219–3227. doi:10.1002/cncr.28147.
- Chou R, Selph SS, Buckley DI, Gustafson KS, Griffin JC, Grusing SE, Gore JL. Treatment of muscle-invasive bladder cancer: A systematic review. Cancer. 2016;122:842–851. doi:10.1002/cncr.29843.
- Galsky MD, Hendricks R, Svatek R, Bangs R, Hoffman-Censits J, Clement J, Dreicer R, Guancial E, Hahn N, Lerner SP, et al. Critical analysis of contemporary clinical research in muscle-invasive and metastatic urothelial cancer: a report from the bladder cancer advocacy network clinical trials working group. Cancer. 2013;119:1994–1998. doi:10.1002/cncr.27973.
- Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng S-L, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi:10.1038/nature13904.
- Zhan Y, Liu Y, Lin J, Fu X, Zhuang C, Liu L, Xu W, Li J, Chen M, Zhao G, et al. Synthetic Tet-inducible artificial microRNAs targeting beta-catenin or HIF-1alpha inhibit malignant phenotypes of bladder cancer cells T24 and 5637. Sci Rep. 2015;5:16177. doi:10.1038/srep16177.
- Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee I-L, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi:10.1016/j.ccr.2014.01.009.
- Christodouleas JP, Baumann BC, He J, Hwang WT, Tucker KN, Bekelman JE, Tangen CM, Lerner SP, Guzzo TJ, Malkowicz SB, et al. Optimizing bladder cancer locoregional failure risk stratification after radical cystectomy using SWOG 8710. Cancer. 2014;120:1272–1280. doi:10.1002/cncr.28544.
- Casadio V, Molinari C, Calistri D, Tebaldi M, Gunelli R, Serra L, Falcini F, Zingaretti C, Silvestrini R, Amadori D, et al. DNA Methylation profiles as predictors of recurrence in non muscle invasive bladder cancer: an MS-MLPA approach. J Exp Clin Cancer Res. 2013;32:94. doi:10.1186/1756-9966-32-94.
- Feng Y, Liu J, Kang Y, He Y, Liang B, Yang P, Yu Z. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J Exp Clin Cancer Res. 2014;33:67. doi:10.1186/s13046-014-0067-8.
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi:10.1093/hmg/ddl046.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Reviews Genet. 2009;10:155–159. doi:10.1038/nrg2521.
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Reviews Genet. 2014;15:7–21. doi:10.1038/nrg3606.
- Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, Chang C, Qi J, Yeh S. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018. doi:10.1038/s41388-018-0175-6.
- Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, Chen K, Zhang Y, Liu H, Gan L, et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37:3822–3838. doi:10.1038/s41388-018-0237-9.
- Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, Wu D, Liu L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112–1122. doi:10.1038/onc.2016.278.
- Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang S-M, He F-T, Yang S-M. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. doi:10.1038/onc.2015.413.
- Huang W, Cui X, Chen J, Feng Y, Song E, Li J, Liu Y. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget. 2016;7:62520–62532. doi:10.18632/oncotarget.11528.
- Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi:10.1038/onc.2015.223.
- Ma L, Tian X, Wang F, Zhang Z, Du C, Xie X, Kornmann M, Yang Y. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2016;17:1051–1061. doi:10.1080/15384047.2016.1219814.
- Frith MC, Bailey TL, Kasukawa T, Mignone F, Kummerfeld SK, Madera M, Sunkara S, Furuno M, Bult CJ, Quackenbush J, et al. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006;3:40–48.
- Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–6. doi:10.1093/nar/gks1099.
- Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu L-M, Chen L-M, Zheng -S-S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi:10.1007/s12032-011-0004-z.
- Zhang S, Ma H, Zhang D, Xie S, Wang W, Li Q, Lin Z, Wang Y. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742. doi:10.1038/s41419-018-0793-5.
- Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, Zhao Q, Wu D, Gong W, Du M, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17:87. doi:10.1186/s12943-018-0829-6.
- Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi:10.1186/s12943-018-0837-6.
- Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, Cheng D, Chen H, Deng X, Peng C, et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169. doi:10.1186/s12943-017-0738-0.
- Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, Dou C, Xu M, Liu Q, Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123. doi:10.1186/s12943-017-0702-z.
- Wang K, Jin W, Song Y, Fei X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol Cancer. 2017;16:166. doi:10.1186/s12943-017-0735-3.
- YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. doi:10.1186/s12943-017-0743-3.