ABSTRACT
Objective
DNA repair pathways are potential targets of molecular therapy in cancer patients. The FANCD2, BRIP1, BRCA1/2, and FANCF genes are involved in homologous recombination DNA repair, which implicates their possible role in cell response to DNA-damaging agents. We evaluated a clinical significance of pre-treatment expression of these genes at mRNA level in 99 primary, advanced-stage ovarian carcinomas from patients, who later received taxane-platinum (TP) or platinum-cyclophosphamide (PC) treatment.
Methods
Gene expression was determined with the use of Real-Time PCR. The BRCA2 and BRIP1 gene sequence was investigated with the use of SSCP, dHPLC, and PCR-sequencing.
Results
Increased FANCD2 expression occurred to be a negative prognostic factor for all patients (PC+TP:HR 3.85, p = 0.0003 for the risk of recurrence; HR 1.96, p = 0.02 for the risk of death), and this association was even stronger in the TP-treated group (HR 6.7, p = 0.0002 and HR 2.33, p = 0.01, respectively). Elevated BRIP1 expression was the only unfavorable molecular factor in the PC-treated patients (HR 8.37, p = 0.02 for the risk of recurrence). Additionally, an increased FANCD2 and BRCA1/2 expression levels were associated with poor ovarian cancer outcome in either TP53-positive or -negative subgroups of the TP-treated patients, however these groups were small. Sequence analysis identified one protein truncating variant (1/99) in BRCA2 and no mutations (0/56) in BRIP1.
Conclusions
Our study shows for the first time that FANCD2 overexpression is a strong negative prognostic factor in ovarian cancer, particularly in patients treated with TP regimen. Moreover, increased mRNA level of the BRIP1 is a negative prognostic factor in the PC-treated patients. Next, changes in the BRCA2 and BRIP1 genes are rare and together with other analyzed FA genes considered as homologous recombination deficiency may not affect the expression level of analyzed genes.
Introduction
Ovarian cancer ranks at the top of the list of the most lethal gynecological malignancies.Citation1 Therefore, it is of utmost importance to identify molecular biomarkers predicting prognosis and response to chemotherapy, and potential new targets for molecular inhibition.
Currently, taxanes combined with cisplatin or its analogs (the TP regimen) are the standard first-line treatment of ovarian cancer patients.Citation2,Citation3 It replaced platinum-cyclophosphamide (the PC regimen) and other protocols based on DNA damaging agents. Nevertheless, in patients with advanced disease, the overall survival rates are still poor.
Platinum compounds induce DNA damage by the formation of DNA adducts and interstrand crosslinks (ICL). These lesions inhibit DNA replication, transcription and induce cell cycle arrest and apoptosis.Citation4 Homologous recombination (HR) during the S phase of the cell cycle is one of the mechanisms removing DNA adducts.Citation5–Citation8 HR-mediated DNA repair requires activation of Fanconi anemia (FA) pathway. Abnormalities of genes involved in the FA pathway, resulting in the homologous recombination deficiency (HRD) have been described to be essential for cell sensitivity to DNA damaging agents (for reviews, see refs.Citation9–Citation11) and PARP inhibitors.Citation12-Citation14 On the other hand, taxanes impair the cell tubular system through polymerization and stabilization of β-tubulin in G2 and M phases of the cell cycle. This leads to activation of the spindle assembly checkpoint (SAC). A prolonged cell-cycle arrest may lead to apoptosis or to mitotic exit, by slippage into G1 state, in which cells develop resistance to antimitotic agents.Citation15,Citation16 Recently, studies on cell lines have shown that FA genes are involved in regulation of the SAC.Citation17
Fanconi anemia is a genetic disease characterized by chromosomal instability and a high risk of cancer development. The FA pathway involves proteins encoded by 19 genes, including FANCD2, BRIP1 (FANCJ, BACH1), BRCA1 (FANCS), BRCA2 (FANCD1) and FANCF (for a review, see ref.Citation18). The FANCD2 is a central component of the FA DNA repair pathway, which protects the stalled replication fork and localizes to centrosomes during mitosis. BRIP1 is RECQ-like helicase that participates in FANCD2 loading onto chromatin and in ATR-mediated DNA damage checkpoint activation. BRCA1 and BRCA2 participate in the RAD51 loading to DNA, stalled replication fork protection, and interact with FANCD2. FANCF is a component of FA core complex that is responsible for the mono-ubiquitination of FANCD2. The FANCD2 gene mutations have been found in breast cancer,Citation16 acute leukemiaCitation19,Citation20, and ovarian cancer.Citation21 Germline mutations in BRIP1 have been associated with an increased risk of epithelial ovarian and breast cancers,Citation22–Citation25 just as germline mutations in the BRCA1/2 genes [for review see ref.Citation26–Citation28]. The FANCF mutation studies in breast,Citation16,Citation29 cervicalCitation30 and ovarian cancerCitation21 have not revealed any mutations leading to the loss of protein function. Still, epigenetic silencing of FANCF by promoter hypermethylation has been reported in several tumors.Citation30–Citation33 To date, the prognostic and/or predictive significance of FA genes was analyzed in many cancers. However, there are few data available on the clinical importance of the expression of these genes at the mRNA level, especially in ovarian cancers.
TP53 is one of the most frequently mutated genes in ovarian carcinomas. TP53 dysfunction, as determined by TP53 protein accumulation in the nuclei of tumor cells, may influence the clinical importance of other molecular factors, particularly of those regulated by, or interfering with TP53.Citation34–Citation37
In the present study, the prognostic and predictive value of tumor FANCD2, BRIP1 (FANCJ, BACH1), BRCA1 (FANCS), BRCA2 (FANCD1) and FANCF expression at the mRNA level, was investigated in ovarian cancer patients treated with PC- or TP-regimen. Moreover, the significance of the expression level was analyzed in the context of the TP53 protein accumulation status and HR-deficiency status. We also evaluated mutation frequency in the BRCA2 and BRIP1 genes.
Results
FANCD2 expression
Increased FANCD2 mRNA level significantly enhanced the risk of recurrence (), ) and death (), ) in all patients (TP+PC, n = 99), in both univariate and multivariate analyses. A particularly unfavorable prognosis, considering both the risk of recurrence (), ) and the risk of death (), ), was observed in TP-treated patients (TP, n = 66) with an increased expression of FANCD2. A mean disease-free survival time of patients with high and low FANCD2 expression in this group was 507 days and 636 days, respectively, while the same values of overall survival time of patients with high and low FANCD2 expression were 991 days and 1263 days, respectively. Kaplan-Meier survival curves also showed a trend toward poorer prognosis for patients with high FANCD2 expression compared to those with low expression, in terms of both the risk of recurrence (,)) and death ()).
Table 1. Statistically significant associations of the BRCA1, BRIP1 and FANCD2 mRNA expression with disease-free survival (DFS) in ovarian cancer patients, assessed in multivariate Cox proportional hazards models. Univariate analyses showed similar but weaker associations.
Table 2. Statistically significant associations of the BRCA2 and FANCD2 mRNA expression with overall survival (OS) in ovarian cancer patients, assessed in multivariate Cox proportional hazard models. Univariate analyses showed similar but weaker associations.
Figure 1. Disease-free survival (DFS) according to the FANCD2 gene expression at the mRNA level in the (a, b) combined TP- and PC-treated groups of patients; (c, d) TP-treated group of patients (e, f) group of TP-treated patients with TP53-positive carcinomas; (a, c, e) univariate analysis of a continuous variable; (b, d, f) analysis of Kaplan–Meier curves, cut-off point at the median value of 0.4.
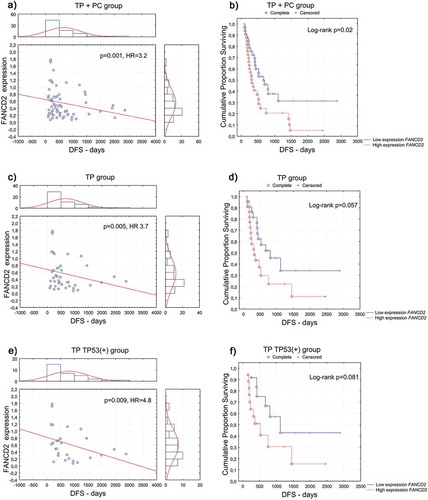
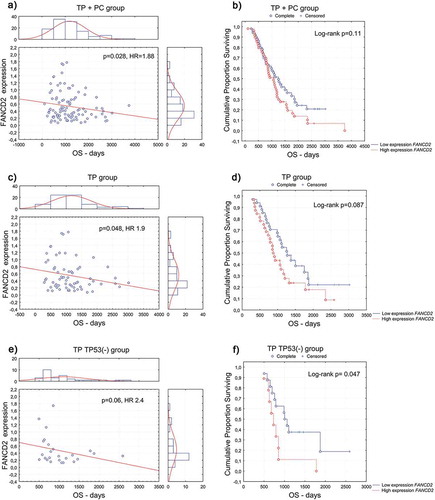
Figure 2. Overall survival (OS) according to the FANCD2 gene expression at the mRNA level in the (a, b) combined TP- and PC-treated groups of patients; (c, d) TP-treated group of patients; (e, f) group of TP-treated patients with TP53-negative carcinomas. (a, c, e) univariate analysis of a continuous variable; (b, d, f) analysis of Kaplan–Meier curves, cut-off point at the median value of 0.4.
Furthermore, we investigated whether mutations in FA genes: FANCD2, BRIP1, BRCA1, BRCA2, FANCF, and PALB2 which contribute to the homologous recombination deficiency status, may affect the relevance of FANCD2 expressions as an independent prognostic factor. Multivariate analysis revealed that HRD status was not significantly associated with the prediction of OS and DFS, and confirmed that patients with increased FANCD2 expression had a significantly greater risk of death and recurrence (,).
FANCD2 expression did not associate with complete remission and platinum sensitivity in any of the analyzed groups.
BRIP1, BRCA1, BRCA2, and FANCF expression
Elevated BRIP1 expression was the only molecular factor which enhanced the risk of recurrence in the PC-treated patients (n = 22, ), in both univariate and multivariate analyses. Additional multivariate analysis with HRD status also confirmed obtained association. Although this association was not significant in Kaplan-Meier analysis (Log-rank p = 0.27).
We did not find any significant association between the BRCA1, BRCA2 or FANCF gene expression at mRNA level and the analyzed clinical endpoints in the whole series of ovarian cancer patients, and separately, in the PC- and TP-treated groups.
Analysis of gene expression considering the TP53 protein accumulation status
In the TP-treated patients, the clinical importance of the FANCD2, BRCA1, and BRCA2 genes expression was observed in small subgroups related to TP53 accumulation status. Increased FANCD2 expression enhanced the risk of recurrence (), ) in patients with TP53-positive carcinomas (n = 29), and the risk of death (), ) in those with TP53-negative carcinomas (n = 25).
Increased expression of the BRCA1 gene was associated with a higher risk of recurrence in patients with TP53-positive carcinomas (, n = 29). On the other hand, increased expression of the BRCA2 gene negatively influenced the probability of complete remission (OR 0.063, p = 0.05) and increased the risk of death in patients with TP53-negative carcinomas (, n = 25). The latter association also proved significant in the Kaplan-Meier analysis (Log-rank p = 0.011).
BRCA2 and BRIP1 sequence analysis
Sequence analyses of the BRCA2 gene revealed a previously knownCitation38, four base-pair duplication c.3975_3978dupTGCT (exon 11, Figure S1) in one tumor sample (1%) of 99 investigated ovarian carcinomas. It resulted in p.(Ala1327Cysfs*4) and generated a premature stop codon at position 1330. Since no matched normal tissue was available, it was not possible to classify this alteration as somatic vs. germline in origin. In addition, eight previously known germline substitutions were revealed (Table S1). One of the identified substitutions was missense variant of unknown significance (VUS) and conflicting interpretations of pathogenicity (c.9371A>T, p.(Asn3124Ile)), revealed in three (3%) of 99 investigated tumor samples (Table S1).
The BRIP1 gene sequence analyzes, performed for 56 out of 99 tumors indicated the presence of nine previously known single nucleotide changes (Table S1).
Analysis of FANCD2, BRIP1, BRCA1, BRCA2, and FANCF expression considering the HR-deficiency status
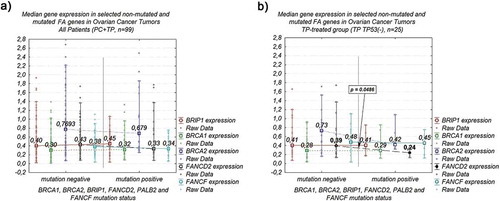
A relationship between the expression of the studied genes at the mRNA level and the HR-deficiency (HRD) status based on mutations in the FA genes was assessed. To this end we found no differences in the expression levels of the studied genes between tumors without mutation in FANCD2, BRIP1, BRCA1, BRCA2, FANCF, and PALB2 (n = 71) and tumors harboring mutations in BRCA1, BRCA2, and PALB2 (n = 28), either in the whole series of ovarian cancer patients ()) or in the PC- and TP-treated groups (Table S2). In the TP-treated patients with TP53-negative carcinomas, the median FANCD2 mRNA level was significantly higher in tumors with no FA mutations (0.39) than in mutation-positive tumors (0.24; p = 0.048; n = 25; Table S2; )). We also found that HRD status did not affect the disease-free survival DFS and OS overall survival (Table S3, Figure S2). This relationship was observed for the DFS even if we extended the group of tumors harboring mutations for the p.(Asn3124Ile) variant, which deleteriousness is not well determined and the studies on the clinical importance of this variant are limited (data not shown). However, the revised HRD status had the slight impact on OS in the TP TP53(-) subgroup, where the risk of death was significantly lower for the mutation carries (log-rank p = 0.025).
Figure 3. Association of analyzed FA genes median expression level and a mutation status of selected FA genes in ovarian cancer tumors: (a) combined TP- and PC-treated groups of patients – no significant relationship; (b) group of TP-treated patients with TP53-negative carcinomas – significant difference in median FANCD2 expression level.
Discussion
This study demonstrated that increased FANCD2 mRNA expression is an independent negative prognostic marker for the risk of recurrence and death in ovarian cancer patients. To date, the FANCD2 mRNA level in relation to prognosis of ovarian cancer patients was analyzed only in one study, and no association with patient outcomes has been found.Citation39 This discrepancy might partly be influenced by the statistical approach. While Ganzinelli et al. (2011)Citation39 analyzed FANCD2 expression as a categorical variable with three values, our analysis comprised a continuous or dichotomous variable. Our results are in line with those obtained in other types of cancer, including breast, cervical and colorectal carcinomas, multiple myeloma, alveolar rhabdomyosarcoma, and hepatocellular carcinoma, where high FANCD2 mRNA level has been shown to relate to poor prognosis.Citation40–Citation45 With regard to response to treatment, RNAi silencing of FANCD2 in NSCLC cell lines enhanced cisplatin and oxaliplatin sensitivityCitation46 and in taxol-treated HeLa cells led to inactivation of spindle assembly checkpoint (SAC), accumulation of extra centrosomes and multinucleation.Citation17 Recently, it was also shown that BRCA1/2 mutated breast cell lines are hypersensitive to the loss of FANCD2, and that FANCD2 gene overexpression was critical for the resistance of BRCA1/2 deficient cells to PARP inhibitors, by stabilizing the replication fork.Citation47 Other literature data also indicate that mutations of BRCA1/2 or other FA genes in ovarian cancers result in homologous recombination deficiency that promotes DNA repair through the FANCD2-dependent, mutagenic alternative end-joining (alt-EJ) pathway.Citation48 Taken together, these data suggest that inhibition of FANCD2 expression may have a potential of sensitizing cancer cells to chemotherapeutic agents and PARP inhibitors, and may provide a tool to improve survival of cancer patients.
Further investigations of the FANCD2 expression significance in relation to HR-deficiency showed that the prognostic significance of FANCD2 expression was not affected by the HRD status. Moreover, the FANCD2 gene mRNA levels did not relate to the HRD status. Contrary to this, Kais et al. (2016)Citation47, in a study based on the TCGA data set for which gene expression and the whole-exome DNA sequencing were available, presented increased FANCD2 expression in tumors with BRCA1/2 mutation-related HR-deficiency. This discrepancy may result from several reasons. Firstly, our analysis did not comprise the whole sequence of the analyzed genes; thus, some mutations might have been missed. Second, apart from germline and somatic mutations in FA genes, HRD may result from BRCA1/2 silencing by promoter methylation and due to interactions with other proteins involved in DNA repairCitation13, the aspect that was not addressed in our study. Third, it has been hypothesized that not all BRCA1 mutations are equal, and some may not induce HRD.Citation49 Moreover, a recent study based on the Next Generation Sequencing (NGS) allowed for a more specific description of HR-deficiency as an HRD score based on an analysis of Genome-wide LOH combined with HR gene mutation profiling, telomeric allelic imbalance, and large-scale state transitions.Citation50 Future studies with the use of NGS approach would be very helpful to assess the homologous recombination deficiency status more precisely.
Herein, we have shown that increased mRNA level of the BRIP1 gene was associated with shorter disease-free survival (DFS) of the PC-treated ovarian cancer patients. Although this observation needs to be confirmed in a larger series of patients, this is consistent with the results in other cancers. Elevated level of BRIP1 mRNA has been associated with a significantly shorter DFS in five-FU-treated patients with metastatic colorectal cancerCitation51 and with poor prognosis in breast cancer patients.Citation52 BRIP1 plays an important role in DNA repair mechanism. Available data indicate that BRIP1 and BRCA1 proteins co-localize before and after exposure to ionizing radiation in ovarian and breast cancer cell lines.Citation53 BRIP1 silencing led to dissociation of BRCA1 protein from chromatin, which resulted in an inhibition of RB1- and TP53-dependent DNA repair-activation of pro-apoptotic pathways, while restoration of BRIP1 expression reversed this effect.Citation53,Citation54 It has also been shown that overexpression of BRIP1 protein correlated with an increased cell proliferation rate,Citation54 which may contribute to earlier tumor recurrence.Citation55,Citation56 Thus, both our analysis and published data indicate that high level of BRIP1 in tumor cells is an unfavorable factor, especially in patients treated with DNA damaging compounds.
Our study did not reveal the association between BRCA1/2 mRNA levels and clinical endpoints in PC- and TP-treated patients. To date, there is no consensus on the impact of BRCA1 expression on ovarian cancer patients’ outcome, and there are few studies on the clinical importance of BRCA2 expression. Several studies have indicated the lack of relationship between the BRCA1/2 expression and the response to TP-treatment,Citation56,Citation57 while others have reported that low level of the BRCA1 mRNA positively influenced prognosis (OS) of the PC-treated patientsCitation58, and the TP-treated patients with the residual tumor less than 2 cm.Citation59 In some other cancers, high BRCA1 gene expression has been considered to be a negative prognostic and/or predictive factor,Citation60–Citation62 while high expression of BRCA2 at mRNA level in breast cancer patients had a negative impact on docetaxel response.Citation63
In the current study, there was no significant association between FANCF gene expression at the mRNA level and clinical endpoints, which is consistent with previous reports.Citation39,Citation64
Finally, analyses considering the TP53 protein accumulation status were performed. Due to the relatively small size of both TP53 subgroups, the results obtained should be interpreted with caution. Nevertheless, the clinical importance of increased FANCD2 expression was observed in the TP-treated patients with TP53-positive (shorter DFS) and TP53-negative carcinomas (shorter OS). Moreover, gene expression analysis considering the HR-deficiency status revealed that the FANCD2 mRNA level was higher in tumors with no TP53 protein accumulation and no mutations in any of the analyzed genes, as compared to tumors carrying those mutations. Although the obtained results appear ambiguous, there is an evidence in the literature, that TP53 status may influence the biological and clinical importance of FANCD2. Studies on wild-type and TP53-mutant mice embryonic fibroblasts revealed that decreased expression of FANCD2 mRNA and reduced capacity to repair the DNA interstrand crosslinks may depend on TP53 role in promoting the recruitment of the E2F4 repressor of the FANCD2 promoter.Citation65 Interestingly, the authors reported that the analysis of the transcriptome data from the Australian Ovarian Cancer Study confirmed that the loss of TP53 function leads to an increased expression of the FANCD2 gene in high-grade ovarian tumors. Moreover, Wysham et al. (2012)Citation66 observed that ovarian cancer patients with the co-expression of FANCD2, PARP, and TP53 proteins had unfavorable prognosis.
We also found a significant relationship between an increased BRCA1 expression and shorter DFS in the TP-treated patients with TP53-positive carcinomas. The clinical significance of BRCA1 gene expression in relation to the TP53 status was examined in ovarian cancer cell lines with wild-type TP53, mutant TP53 and without the TP53 protein.Citation67 The authors have shown that the reduced BRCA1 mRNA expression resulted in approximately five-fold increase of platinum, but not taxane sensitivity of TP53 wild-type cells, but not of those with mutated TP53. Although our patient group was small, the obtained result is in line with our other study, where we have shown that TP53 accumulation status may determine the prognosis of patients who carry BRCA1 mutations.Citation37
The present study demonstrates the clinical importance of BRCA2 mRNA level in ovarian cancer patients. A negative impact of increased BRCA2 expression on complete remission (CR) and overall survival (OS) was observed in the TP-treated patients with TP53-negative carcinomas. The relationship between BRCA2 and TP53 investigated in breast cancer cell lines pointed to the importance of TP53 status for the regulation of BRCA2 gene promoter, as normal TP53 has been considered as a repressor of the BRCA2 gene.Citation68 These observations together with our findings suggest that the prognostic and predictive value of BRCA1/2 expression in the context of TP53 accumulation deserves further investigations.
To better characterize the molecular background of the analyzed tumor samples, sequence analyses of the studied genes were performed. One, protein-truncating mutation in the BRCA2 gene (n = 1/99), and no mutations in the BRIP1 gene (n = 0/56) have been identified. The BRCA2 mutation was located in one of the high ovarian cancer risk-associated cluster regions (OCCR) of the BRCA2 gene.Citation69,Citation70 We also detected missense, germline substitution (c.9371A>T, p.(Asn3124Ile), ), classified as a variant of uncertain significance and conflicting interpretations of pathogenicity (ClinVar). Previous studies have indicated that this BRCA2 gene variant was frequently identified in patients with breast and ovarian cancers, especially from the Polish population.Citation71–Citation73 Interestingly, in order to investigate significance of the HRD status in the context of DFS and OS prediction with c.9371A>T variant classified as mutation the present study demonstrated that revised HRD status had no impact on the risk of recurrence, however, it might have the slight impact on risk of death in the TP53(-) subgroup (data not shown). Because of the low number of patients with the c.9371A>T variant in our analysis and the lack of the literature data about the impact of this variant on patients outcome, the further studies are necessary in order to determine its clinical significance.
Our previous studies have revealed that the analyzed tumor samples harbored BRCA1 (n = 26/99; 26.3%) and PALB2 (n = 1/99; 1%) mutationsCitation74,Citation75 and no FANCD2 and FANCF (n = 0/99) mutations.Citation21 Taken together, it may be concluded that besides the most common mutations of the BRCA1 gene that frequently result in a loss of protein function (which are mostly point changes or small deletions and insertions, located across the entire coding gene sequence and at splice sites),Citation25,Citation76 other deleterious variants in FA genes are relatively rare. This is in line with literature data which show germline and somatic mutations of BRCA1/2 genes in about 20% of cases, and much less frequent mutations in other FA pathway genes, BRIP1 (0,9–1,72%), PALB2 (0,2–0,5%), RAD51C (0,41–2,9%).Citation77
In summary, in the present study, we provided the evidence that the increased tumor FANCD2 mRNA expression level is an unfavorable prognostic factor in ovarian cancer patients treated with a taxane-platinum regimen. For platinum-cyclophosphamide treated patients, only BRIP1 expression turned out to be clinically significant. Our results also demonstrate that overexpression of the FANCD2, BRCA1, and BRCA2 genes, depending on TP53 accumulation status, has a value of an adverse prognostic factor in TP-treated ovarian cancer patients. Additionally, we found no significant association between significance of analyzed genes expression and HR-deficiency based on BRCA1/2 and other FA genes mutation status. Taken together, we showed that increased expression of HR DNA repair pathway genes may negatively influence prognosis in ovarian cancer patients.
Materials and methods
Patients and tumors
The study was performed on 99 fresh frozen samples of ovarian carcinomas from patients, who were subsequently treated with a taxane-platinum chemotherapy (TP, n = 66) or a platinum-cyclophosphamide chemotherapy (PC, n = 33). Tumors obtained during the surgical procedure as well as the relevant blood samples anticoagulated with EDTA were snap-frozen in liquid nitrogen and stored at −70°C.
The material was carefully selected, as previously described.Citation78 The study included only tumors containing less than 15% stromal cell contamination (scc) and meeting the following criteria: no chemotherapy before staging laparotomy; adequate staging procedure; International Federation of Gynecologists and Obstetricians (FIGO) stage IIB to IV diseaseCitation79; tumor tissue from the first laparotomy available; moderate (G2) or poor tumor differentiation (G3); availability of clinical data, including residual tumor size and follow-up. All tumors were uniformly reviewed histopathologically, classified according to the criteria of the World Health Organization (WHO) and graded in a three-grade scale.Citation80 Clinicopathological characteristics are presented in Table S4. Previous sequence analysis of these tumor samples revealed mutations in the BRCA1 gene (26/99; 26%),Citation74 the PALB2 gene (1/70; 1%)Citation75 and no mutations in the FANCD2 and FANCF genes (0/99)Citation21.
Response to chemotherapy was evaluated retrospectively, according to the WHO response evaluation criteria.Citation81 The evaluation was based on data retrieved from medical records referring to the patients’ clinical condition and CA125 levels assessed in 3 to 4-week intervals. Complete remission (CR) was defined as the disappearance of all clinical and biochemical symptoms of ovarian cancer, evaluated after completion of first-line chemotherapy and confirmed 4 weeks later. Within the CR group, a platinum-sensitive group (PS), with disease-free survival (DFS) longer than six months was identified. Other tumors were described as platinum-resistant.Citation82
The study was approved by the bioethics committee of Maria Sklodowska-Curie Institute – Oncology Center (ref. no. 39/2007).
DNA, mRNA extraction, cDNA synthesis
Genomic DNA was extracted from frozen tissues and relevant blood samples with the use of the QIAmp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. Total mRNA was extracted from frozen tissues with the NucleoSpin RNA Kit (Macherey-Nagel), according to the manufacturer’s protocol. mRNA quantity was measured with the use of UV spectrophotometer, and mRNA quality was assessed by the 260/280 ratio and in a 1% agarose gel. One microgram of total mRNA was transcribed to cDNA using the Super Script III First Strand kit (Invitrogen).
Gene expression
Expression of FANCD2, BRIP1, and BRCA1/2 at the mRNA level
Quantitative RT-PCR (Q-PCR) was run on the 7500 Fast Real-Time PCR System (Applied Biosystems), with the use of the FAM-labeled, TaqMan Gene Expression Assays (Applied Biosystems): FANCD2 (assay ID: Hs00276992_m1), BRIP1 (assay ID: Hs00230743_m1), BRCA1 (assay ID: Hs00173233_m1), BRCA2 (assay ID: Hs00609060_m1). All Real-Time PCR reactions were carried out in triplicates, in the final volume of 10 μl, with TaqMan Universal PCR Master Mix, no AmpErase™ UNG (Applied Biosystems) and about of 10 ng of cDNA, for 40 cycles, according to the following protocol: each cycle at 95°C for the initial 10 min, then at 95°C for 15 s and 60°C for 1 min. The obtained results were averaged, and gene expression levels were normalized to the HGPRTgene expression (VIC-labeled, Applied Biosystems, assay ID: 4326321E). A standard curve, used in all experiments, was prepared from serial dilutions of cDNA from one of the analyzed tumors.
Expression of FANCF at the mRNA level
FANCF gene consists of one exon. FANCF TaqMan probe (FAM-labeled, Applied Biosystems, assay ID: Hs00256030_s1) detects both the cDNA and genomic DNA (gDNA), which remains after the isolation of RNA. Purification of RNA from gDNA with the use of enzymatic method, i.e. digestion with DNase using RNase inhibitor has failed. Therefore, two separate Q-PCR experiments were performed. In the first experiment, gene expression was obtained based on both cDNA and gDNA, as described in the previous section. The second experiment was carried out for measurement of the gene expression based on gDNA, with the use of total mRNA instated of cDNA. Final values of FANCF expression were obtained by subtracting the value of gene expression evaluated in the second experiment from the value of gene expression obtained in the first experiment and were normalized to the reference gene expression (HGPRT). A standard curve was prepared as described in the previous section.
Sequence analysis of the BRCA2 and BRIP1 genes
DNA sequence analysis was carried out for the BRCA2 and BRIP1 genes in 99 and 56 ovarian carcinomas, respectively. Germline origin of the detected changes was confirmed in the corresponding DNA from blood samples (if available). The selected regions of BRCA2 – exon 2,3, part of exon 11 – including the ovarian cancer cluster region (OCCRCitation70, nucleotides 3035 to 6629) and exon 25 [GenBank: NG_012772.3; NM_000059.3] were investigated with the use of the dHPLC method. The full coding sequence of the BRIP1 gene (22 exons with the intron bounders [GenBank: NG_007409.2; NM_032043.2]) was analyzed with the use of the PCR-SSCP method.
Polymerase chain reaction (PCR)
DNA fragments were amplified with the use of primers designed by Wagner et al. (1999)Citation83 for BRCA2 and by Lewis et al. (2005)Citation84 for BRIP1, or with the use of Primer3 software (Table S5). PCR mixtures were prepared according to the standard procedure (Applied Biosystems PCR Kit). PCR reactions were carried out for 36 cycles in a programmable thermal cyclers (Biometra, Eppendorff) with denaturation at 95°C, annealing at 54–64°C (depending on the exon) and extension at 72°C for 30 s each.
Denaturing high-performance liquid chromatography
Amplified DNA fragments of the BRCA2 gene were screened by the dHPLC method with the use of automated dHPLC instrumentation (Transgenomic Inc). PCR products were eluted with linear acetonitryle gradient. The gradient and the temperature required for a successful resolution of heteroduplex molecules was determined with the use of the dHPLC melting algorithm (Transgenomic Inc).
Single strand conformational polymorphism analysis (SSCP)
All amplified DNA fragments of the BRIP1 gene were analyzed with the use of the SSCP method. PCR products were denatured with 0.1 M NaOH and 2 mM EDTA at 55°C for 15 min. Subsequently, after 95% formamide, 0.05% xylene cyanol and 0.05% bromphenol blue were added, the samples were loaded to polyacrylamide gels (1:39 N,N’-methylenebisacrylamide to acrylamide in 0.5 x TBE with 10% glycerol). Electrophoresis was performed at 100 V, for 16–24 hours at room temperature. DNA bands were visualized with the silver-staining method compiled from several protocols. In our experience, this method detects 90% of all alterations, and 100% of deletions and insertions.Citation85
Sequencing
All variants detected using SSCP and dHPLC were further sequenced with the Sanger method and BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) on automated ABI PRISM 3100 Sequencer (Life Technologies) according to the manufacturer’s recommendations. Prior to sequencing, the PCR products were purified enzymatically with exonuclease I and alkaline phosphatase (Illustra ExoProStar, GE Healthcare Life Sciences).
TP53 protein accumulation status
Analysis of TP53 accumulation in the nuclei of tumor cells population was described previously by our team.Citation78,Citation85 Briefly, TP53 accumulation was visualized by an immunohistochemical method, with the use of PAb1801 monoclonal antibody (1/3000, Sigma-Genosys) on paraffin-embedded material, after heat-induced epitope retrieval (HIER). It was described as present (>10% of positive cells; TP53(+)) or absent (TP53(-)).
Statistical analysis
Associations between FANCD2, BRIP1, BRCA1, BRCA2, FANCF expression, and clinical endpoints were analyzed with the use of the Kaplan–Meier method, log-rank test, univariate and multivariate Cox’s proportional hazards models (OS, DFS) and logistic regression models (probability of CR, PS). Multivariate statistical analyses included the following independent variables: age of the patients (median: 53 years), the FIGO stage, histopathological type, grade, residual tumor size, and the TP53 accumulation status. The genes expression was analyzed as a continuous variable, and for Kaplan–Meier analysis – as a categorical variable (the median value of expression for the entire group was used as a cut-off point). Important factors were selected using a backward selection technique, where factors not significant at 0.1 (for OS, DFS), and 0.2 (for CR, PS) were removed stepwise from the model. To estimate the association between the analyzed genes’ expression and the homologous recombination deficiency status (HRD, based on mutation analysis of six FA genes: FANCD2, BRIP1, BRCA1, BRCA2, FANCF, PALB2), the Kruskal–Wallis test was used. The analyses were performed in 1) the entire group of patients, 2) in the PC- and 3) TP-treated groups, and 4) in the TP-treated patients, subgrouped with respect to TP53 accumulation status. Additionally, associations between HRD status and clinical endpoints (OS, DFS) were analyzed with the use of the Kaplan–Meier method, log-rank test, and Cox’s proportional hazard model. Furthermore, the clinical significance of the FANCD2 and BRIP1 gene expression (OS, DFS) was analyzed also with the HRD variable included in the multivariate Cox’s proportional hazard model.
A p-value <0.05 was considered significant. All calculations were performed using SAS or Statistica softwares.
Abbreviations
| CR | = | complete remission |
| DFS | = | disease-free survival |
| HR | = | hazard ratio |
| OR | = | odds ratio |
| OS | = | overall survival |
| PC | = | platinum-cyclophosphamide chemotherapy |
| PS | = | platinum sensitivity |
| TP | = | taxane-platinum chemotherapy |
| HRD | = | homologous recombination deficiency |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ contributions
JM-S designed the study, performed laboratory research, analyzed the data and wrote the manuscript. IKR analyzed the data and drafted the manuscript. JC carried out a statistical analysis. AD-M performed laboratory research and critically reviewed the manuscript. AB, LMS, RL performed laboratory research. PS, JK collected and described the clinical material. JK made a histopathological evaluation of the tumors, analyzed the data and drafted the manuscript.
Supplemental Material
Download Zip (1.4 MB)Acknowledgments
The authors would like to thank Magdalena Chechlinska, PhD for the critical reading of the manuscript, all valuable comments and for the English editing of the manuscript. We would like to thank also Dr Renata Zub for DNA sequencing.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61(2):69–90. doi:10.3322/caac.20107.
- McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med. 1996;334(1):1–6. doi:10.1056/NEJM199601043340101.
- Piccart MJ, Du Bois A, Gore ME, Neijt JP, Pecorelli S, Pujade-Lauraine E. A new standard of care for treatment of ovarian cancer. Eur J Cancer. 2000;36(1):10–12. doi:10.1016/S0959-8049(99)00210-5.
- Boulikas T, Pantos A, Bellis E, Christofis P. Designing platinum compounds in cancer: structures and mechanisms. Cancer Ther. 2007;5:537–583. doi:10.1016/j.jinorgbio.2014.07.011.
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7(2):249–262. doi:10.1016/S1097-2765(01)00173-3.
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15(4):607–620. doi:10.1016/j.molcel.2004.08.009.
- Cummings M, Higginbottom K, McGurk CJ, Wong OG, Köberle B, Oliver RT, Masters JR. XPA versus ERCC1 as chemosensitising agents to cisplatin and mitomycin C in prostate cancer cells: role of ERCC1 in homologous recombination repair. Biochem Pharmacol. 2006;72(2):166–175. doi:10.1016/j.bcp.2006.04.025.
- Natarajan AT, Palitti F. DNA repair and chromosomal alterations. Mutat Res. 2008;657(1):3–7. doi:10.1016/j.mrgentox.2008.08.017.
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11(7):467–480. doi:10.1038/nrc3088.
- Furgason JM, Bahassi El M. Targeting DNA repair mechanisms in cancer. Pharmacol Ther. 2013;137(3):298–308. doi:10.1016/j.pharmthera.2012.10.009.
- Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137–1154. doi:10.1158/2159-8290.CD-15-0714.
- Telli ML, Hellyer J, Audeh W, Jensen KC, Bose S, Timms KM, Gutin A, Abkevich V, Peterson RN, Neff C, et al. Homologous recombination deficiency (HRD) status predicts response to standard neoadjuvant chemotherapy in patients with triple-negative or BRCA1/2 mutation-associated breast cancer. Breast Cancer Res Treat. 2018;168(3):625–630. DOI:10.1007/s10549-017-4624-7
- Frey MK, Pothuri B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: a review of the literature. Gynecol Oncol Res Pract. 2017;4:4. doi:10.1186/s40661-017-0039-8.
- De Picciotto N, Cacheux W, Roth A, Chappuis PO, Labidi-Galy SI. Ovarian cancer: status of homologous recombination pathway as a predictor of drug response. Crit Rev Oncol Hematol. 2016;101:50–59. doi:10.1016/j.critrevonc.2016.02.014.
- Rossio V, Galati E, Piatti S. Adapt or die: how eukaryotic cells respond to prolonged activation of the spindle assembly checkpoint. Biochem Soc Trans. 2010;38(6):1645–1649. doi:10.1042/BST0381645.
- Seal S, Barfoot R, Jayatilake H, Smith P, Renwick A, Bascombe L, McGuffog L, Evans DG, Eccles D, Easton DF, et al. Evaluation of Fanconi Anemia genes in familial breast cancer predisposition. Cancer Res. 2003;63(24):8596–8599.
- Nalepa G, Enzor R, Sun Z, Marchal C, Park SJ, Yang Y, Tedeschi L, Kelich S, Hanenberg H, Clapp DW. Fanconi anemia signaling network regulates the spindle assembly checkpoint. J Clin Invest. 2013;123(9):3839–3847. doi:10.1172/JCI67364.
- Michl J, Zimmer J, Buffa FM, McDermott U, Tarsounas M. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat Struct Mol Biol. 2016;23(8):755–757. doi:10.1038/nsmb.3252.
- Offman J, Gascoigne K, Bristow F, Macpherson P, Bignami M, Casorelli I, Leone G, Pagano L, Sica S, Halil O, et al. Repeated sequences in CASPASE-5 and FANCD2 but not NF1 are targets for mutation in microsatellite-unstable acute leukemia/myelodysplastic syndrome. Mol Cancer Res. 2005;3(5):251–260. doi:10.1158/1541-7786.MCR-04-0182.
- Borriello A, Locasciulli A, Bianco AM, Criscuolo M, Conti V, Grammatico P, Cappellacci S, Zatterale A, Morgese F, Cucciolla V, et al. A novel Leu153Ser mutation of the Fanconi anemia FANCD2 gene is associated with severe chemotherapy toxicity in a pediatric T-cell acute lymphoblastic leukemia. Leukemia. 2007;21(1):72–78. doi:10.1038/sj.leu.2404468
- Moes-Sosnowska J, Budzilowska A, Kupryjanczyk J. Mutation analysis of the FANCD2 gene in ovarian cancer patients from the Polish population. J Oncol. 2015;65(1):7–13. doi:10.5603/NJO.2015.0002.
- Rafnar T, Gudbjartsson DF, Sulem P, Jonasdottir A, Sigurdsson A, Jonasdottir A, Besenbacher S, Lundin P, Stacey SN, Gudmundsson J, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43(11):1104–1107. doi:10.1038/ng.955
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8(3):255–265. doi:10.1016/j.ccr.2005.08.004.
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, et al. mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38(11):1239–1241. doi:10.1038/ng1902.
- Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, Fraser L, Gentry-Maharaj A, Hayward J, Philpott S, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107(11):pii: djv214. DOI:10.1093/jnci/djv214
- Petrucelli N, Daly MB, Feldman GL. 2010. Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet Med. 12(5):245–259. doi:10.1097/GIM.0b013e3181d38f2f.
- Stoppa-Lyonnet D. The biological effects and clinical implications of BRCA mutations: where do we go from here? Eur J Hum Genet. 2016;24(Suppl 1):S3–9. doi:10.1038/ejhg.2016.93.
- Godet I, Gilkes DM. BRCA1 and BRCA2 mutations and treatment strategies for breast cancer. Integr Cancer Sci Ther. 2017;4:1. doi:10.15761/ICST.1000228.
- Barroso E, Pita G, Arias JI, Menendez P, Zamora P, Blanco M, Benitez J, Ribas G. The Fanconi anemia family of genes and its correlation with breast cancer susceptibility and breast cancer features. Breast Cancer Res Treat. 2009;118(3):655–660. doi:10.1007/s10549-009-0439-5.
- Narayan G, Arias-Pulido H, Nandula SV, Basso K, Sugirtharaj DD, Vargas H, Mansukhani M, Villella J, Meyer L, Schneider A, et al. Promoter hypermethylation of FANCF: disruption of Fanconi Anemia-BRCA pathway in cervical cancer. Cancer Res. 2004 May 1;64(9):2994–2997. doi:10.1158/0008-5472.CAN-04-0245.
- Wang Z, Li M, Lu S, Zhang Y, Wang H. Promoter hypermethylation of FANCF plays an important role in the occurrence of ovarian cancer through disrupting Fanconi anemia-BRCA pathway. Cancer Biol Ther. 2006;5(3):256–260. doi:10.4161/cbt.5.3.2380.
- Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23(4):1000–1004. doi:10.1038/sj.onc.1207256.
- Tokunaga E, Okada S, Kitao H, Shiotani S, Saeki H, Endo K, Morita M, Kakeji Y, Maehara Y. Low incidence of methylation of the promoter region of the FANCF gene in Japanese primary breast cancer. Breast Cancer. 2011;18(2):120–123. doi:10.1007/s12282-009-0175-z.
- Ziółkowska-Seta I, Madry R, Kraszewska E, Szymańska T, Timorek A, Rembiszewska A, Kupryjańczyk J. TP53, BCL-2 and BAX analysis in 199 ovarian cancer patients treated with taxane-platinum regimens. Gynecol Oncol. 2009;112(1):179–184. doi:10.1016/j.ygyno.2008.09.008.
- Felisiak-Golabek A, Rembiszewska A, Rzepecka IK, Szafron L, Madry R, Murawska M, Napiorkowski T, Sobiczewski P, Osuch B, Kupryjanczyk J. Nuclear survivin expression is a positive prognostic factor in taxane-platinum-treated ovarian cancer patients. J Ovarian Res. 2011 Nov 10;4(1):20. doi:10.1186/1757-2215-4-20.
- Szafron LM, Balcerak A, Grzybowska EA, Pienkowska-Grela B, Podgorska A, Zub R, Olbryt M, Pamula-Pilat J, Lisowska KM, Grzybowska E, et al. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget. 2015;6(41):43897–43910. DOI:10.18632/oncotarget.6016
- Rzepecka IK, Szafron LM, Stys A, Felisiak-Golabek A, Podgorska A, Timorek A, Sobiczewski P, Pienkowska-Grela B, El-Bahrawy M, Kupryjanczyk J. Prognosis of patients with BRCA1-associated ovarian carcinomas depends on TP53 accumulation status in tumor cells. Gynecol Oncol. 2017;144(2):369–376. doi:10.1016/j.ygyno.2016.11.028.
- Ratajska M, Krygier M, Stukan M, Kuźniacka A, Koczkowska M, Dudziak M, Sniadecki M, Debniak J, Wydra D, Brozek I, et al. Mutational analysis of BRCA1/2 in a group of 134 consecutive ovarian cancer patients. Novel and recurrent BRCA1/2 alterations detected by next generation sequencing. J Appl Genet. 2015 May;56(2):193–198. DOI:10.1007/s13353-014-0254-5.
- Ganzinelli M, Mariani P, Cattaneo D, Fossati R, Fruscio R, Corso S, Ricci F, Broggini M, Damia G. Expression of DNA repair genes in ovarian cancer samples: biological and clinical considerations. Eur J Cancer. 2011;47(7):1086–1094. doi:10.1016/j.ejca.2010.11.029.
- van der Groep P, Hoelzel M, Buerger H, Joenje H, de Winter JP, van Diest PJ. Loss of expression of FANCD2 protein in sporadic and hereditary breast cancer. Breast Cancer Res Treat. 2008;107(1):41–47. doi:10.1007/s10549-007-9534-7.
- Balacescu O, Balacescu L, Tudoran O, Todor N, Rus M, Buiga R, Susman S, Fetica B, Pop L, Maja L, et al. Gene expression profiling reveals activation of the FA/BRCA pathway in advanced squamous cervical cancer with intrinsic resistance and therapy failure. BMC Cancer. 2014;14:246. doi:10.1186/1471-2407-14-246.
- Ozawa H, Iwatsuki M, Mimori K, Sato T, Johansson F, Toh H, Watanabe M, Mori M. FANCD2 mRNA overexpression is a bona fide indicator of lymph node metastasis in human colorectal cancer. Ann Surg Oncol. 2010;17(9):2341–2348. doi:10.1245/s10434-010-1002-7.
- Han SS, Tompkins VS, Son DJ, Han S, Yun H, Kamberos NL, Dehoedt CL, Gu C, Holman C, Tricot G, et al. CDKN1A and FANCD2 are potential oncotargets in Burkitt lymphoma and multiple myeloma. Exp Hematol Oncol. 2015;4:9. doi:10.1186/s40164-015-0005-2.
- Singh M, Leasure JM, Chronowski C, Geier B, Bondra K, Duan W, Hensley LA, Villalona-Calero M, Li N, Vergis AM, et al. FANCD2 is a potential therapeutic target and biomarker in alveolar rhabdomyosarcoma harboring the PAX3-FOXO1 fusion gene. Clin Cancer Res. 2014;20(14):3884–3895. DOI:10.1158/1078-0432.CCR-13-0556
- Komatsu H, Masuda T, Iguchi T, Nambara S, Sato K, Hu Q, Hirata H, Ito S, Eguchi H, Sugimachi K, et al. Clinical significance of FANCD2 gene expression and its association with tumor progression in hepatocellular carcinoma. Anticancer Res. 2017;37(3):1083–1090. DOI:10.21873/anticanres.11420
- Kachnic LA, Li L, Fournier L, Willers H. Fanconi anemia pathway heterogeneity revealed by cisplatin and oxaliplatin treatments. Cancer Lett. 2010;292(1):73–79. doi:10.1016/j.canlet.2009.11.009.
- Kais Z, Rondinelli B, Holmes A, O’Leary C, Kozono D, D’Andrea AD, Ceccaldi R. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 2016;15(11):2488–2499. doi:10.1016/j.celrep.2016.05.031.
- Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26(1):52–64. doi:10.1016/j.tcb.2015.07.009.
- Hollis RL, Churchman M, Gourley C. 2017. Distinct implications of different BRCA mutations: efficacy of cytotoxic chemotherapy, PARP inhibition and clinical outcome in ovarian cancer. Onco Targets Ther. 10:2539–2551. doi:10.2147/OTT.S102569.
- Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, Szallasi Z, Barry WT, Winer EP, Tung NM, et al. Homologous Recombination Deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016 Aug 1;22(15):3764–3773. doi:10.1158/1078-0432.CCR-15-2477.
- Nakanishi R, Kitao H, Fujinaka Y, Yamashita N, Iimori M, Tokunaga E, Yamashita N, Morita M, Kakeji Y, Maehara Y. FANCJ expression predicts the response to 5-fluorouracil-based chemotherapy in MLH1-proficient colorectal cancer. Ann Surg Oncol. 2012;19(11):3627–3635. doi:10.1245/s10434-012-2349-8.
- Eelen G, Vanden Bempt I, Verlinden L, Drijkoningen M, Smeets A, Neven P, Christiaens MR, Marchal K, Bouillon R, Verstuyf A. Expression of the BRCA1-interacting protein Brip1/BACH1/FANCJ is driven by E2F and correlates with human breast cancer malignancy. Oncogene. 2008 Jul;27(30):4233–4241. doi:10.1038/onc.2008.51.
- Peng M, Litman R, Jin Z, Fong G, Cantor SB. 2006. BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene. 25(15):2245–2253. doi:10.1038/sj.onc.1209257.
- Tu Z, Aird KM, Bitler BG, Nicodemus JP, Beeharry N, Xia B, Yen TJ, Zhang R. Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev Cell. 2011;21(6):1077–1091. doi:10.1016/j.devcel.2011.10.010.
- Frontini M, Vijayakumar M, Garvin A, Clarke N. A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res. 2009;37(4):1073–1085. doi:10.1093/nar/gkn1051.
- Pontikakis S, Papadaki C, Tzardi M, Trypaki M, Sfakianaki M, Koinis F, Lagoudaki E, Giannikaki L, Kalykaki A, Kontopodis E, et al. Predictive value of ATP7b, BRCA1, BRCA2, PARP1, UIMC1 (RAP80), HOXA9, DAXX, TXN (TRX1), THBS1 (TSP1) and PRR13 (TXR1) genes in patients with epithelial ovarian cancer who received platinum-taxane first-line therapy. Pharmacogenomics J. 2016. doi:10.1038/tpj.2016.63.
- Dann RB, DeLoia JA, Timms KM, Zorn KK, Potter J, Flake DD2, Lanchbury JS, Krivak TC. BRCA1/2 mutations and expression: response to platinum chemotherapy in patients with advanced stage epithelial ovarian cancer. Gynecol Oncol. 2012;125(3):677–682. doi:10.1016/j.ygyno.2012.03.006.
- Quinn JE, James CR, Stewart GE, Mulligan JM, White P, Chang GK, Mullan PB, Johnston PG, Wilson RH, Harkin DP. BRCA1 mRNA expression levels predict for overall survival in ovarian cancer after chemotherapy. Clin Cancer Res. 2007;13(24):7413–7420. doi:10.1158/1078-0432.CCR-07-1083.
- Weberpals J, Garbuio K, O’Brien A, Clark-Knowles K, Doucette S, Antoniouk O, Goss G, Dimitroulakos J. The DNA repair proteins BRCA1 and ERCC1 as predictive markers in sporadic ovarian cancer. Int J Cancer. 2009;124(4):806–815. doi:10.1002/ijc.23987.
- Taron M, Rosell R, Felip E, Mendez P, Souglakos J, Ronco MS, Queralt C, Majo J, Sanchez JM, Sanchez JJ, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet. 2004;13(20):2443–2449. DOI:10.1093/hmg/ddh260
- Rosell R, Skrzypski M, Jassem E, Taron M, Bartolucci R, Sanchez JJ, Mendez P, Chaib I, Perez-Roca L, Szymanowska A, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One. 2007;2(11):e1129. DOI:10.1371/journal.pone.0001129
- Gao Y, Zhu J, Zhang X, Wu Q, Jiang S, Liu Y, Hu Z, Liu B, Chen XBRCA1 mRNA expression as a predictive and prognostic marker in advanced esophageal squamous cell carcinoma treated with cisplatin- or docetaxel-based chemotherapy/chemoradiotherapy. PLoS One. 2013;8(1):e52589. doi:10.1371/journal.pone.0052589.
- Egawa C, Miyoshi Y, Takamura Y, Taguchi T, Tamaki Y, Noguchi S. Decreased expression of BRCA2 mRNA predicts favorable response to docetaxel in breast cancer. Int J Cancer. 2001;95(4):255–259.
- Lim SL, Smith P, Syed N, Coens C, Wong H, van der Burg M, Szlosarek P, Crook T, Green JA. Promoter hypermethylation of FANCF and outcome in advanced ovarian cancer. Br J Cancer. 2008;98(8):1452–1456. doi:10.1038/sj.bjc.6604325.
- Jaber S, Toufektchan E, Lejour V, Bardot B, Toledo F. p53 downregulates the Fanconi anaemia DNA repair pathway. Nat Commun. 2016;7:11091. doi:10.1038/ncomms11091.
- Wysham WZ, Mhawech-Fauceglia P, Li H, Hays L, Syriac S, Skrepnik T, Wright J, Pande N, Hoatlin M, Pejovic T. BRCAness profile of sporadic ovarian cancer predicts disease recurrence. PLoS One. 2012;7(1):e30042. doi:10.1371/journal.pone.0030042.
- Horiuchi A, Wang C, Kikuchi N, Osada R, Nikaido T, Konishi I. BRCA1 expression is an important biomarker for chemosensitivity: suppression of BRCA1 increases the apoptosis via up-regulation of p53 and p21 during cisplatin treatment in ovarian cancer cells. Biomark Insights. 2007;1:49–59.
- Wu K, Jiang SW, Couch FJ. p53 mediates repression of the BRCA2 promoter and down-regulation of BRCA2 mRNA and protein levels in response to DNA damage. J Biol Chem. 2003;278(18):15652–15660. doi:10.1074/jbc.M211297200.
- Gayther SA, Mangion J, Russell P, Seal S, Barfoot R, Ponder BA, Stratton MR, Easton D. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet. 1997;15(1):103–105. doi:10.1038/ng0197-103.
- Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, Mazoyer S, Chenevix-Trench G, Easton DF, Antoniou AC, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313(13):1347–1361. DOI:10.1001/jama.2014.5985
- Kluska A, Balabas A, Paziewska A, Kulecka M, Nowakowska D, Mikula M, Ostrowski J. New recurrent BRCA1/2 mutations in Polish patients with familial breast/ovarian cancer detected by next generation sequencing. BMC Med Genomics. 2015;8:19. doi:10.1186/s12920-015-0092-2.
- Wojcik P, Jasiowka M, Strycharz E, Sobol M, Hodorowicz-Zaniewska D, Skotnicki P, Byrski T, Blecharz P, Marczyk E, Cedrych I, et al. Recurrent mutations of BRCA1, BRCA2 and PALB2 in the population of breast and ovarian cancer patients in Southern Poland. Hered Cancer Clin Pract. 2016;14:5. doi:10.1186/s13053-016-0046-5.
- Kowalik A, Siołek M, Kopczyński J, Krawiec K, Kalisz J, Zięba S, Kozak-Klonowska B, Wypiórkiewicz E, Furmańczyk J, Nowak-Ozimek E, et al. BRCA1 founder mutations and beyond in the polish population: a single-institution BRCA1/2 next-generation sequencing study. PLoS One. 2018;13(7):e0201086. DOI:10.1371/journal.pone.0201086
- Rzepecka IK, Szafron L, Stys A, Bujko M, Plisiecka-Halasa J, Madry R, Osuch B, Markowska J, Bidzinski M, Kupryjanczyk J. High frequency of allelic loss at the BRCA1 locus in ovarian cancers: clinicopathologic and molecular associations. Cancer Genet. 2012;205(3):94–100. doi:10.1016/j.cancergen.2011.12.005.
- Dansonka-Mieszkowska A, Kluska A, Moes J, Dabrowska M, Nowakowska D, Niwinska A, Derlatka P, Cendrowski K, Kupryjanczyk J. A novel germline PALB2 deletion in polish breast and ovarian cancer patients. BMC Med Genet. 2010;11:20. doi:10.1186/1471-2350-11-20.
- Karakasis K, Burnier JV, Bowering V, Oza AM, Lheureux S. Ovarian cancer and BRCA1/2 testing: opportunities to improve clinical care and diseaseprevention. Front Oncol. 2016;6:119. doi:10.3389/fonc.2016.00119.
- Ledermann JA, Drew Y, Kristeleit RS. Homologous recombination deficiency and ovarian cancer. Eur J Cancer. 2016;60:49–58. doi:10.1016/j.ejca.2016.03.005.
- Kupryjanczyk J, Kraszewska E, Ziolkowska-Seta I, Madry R, Timorek A, Markowska J, Stelmachow J, Bidzinski M. TP53 status and taxane-platinum versus platinum-based therapy in ovarian cancer patients: a non-randomized retrospective study. BMC Cancer. 2008;8:27. doi:10.1186/1471-2407-8-27.
- Peterson F, Kolstad P, Ludwig H, Ulfelder H. Annual report on the results of treatment in gynecological cancer Vol. 20. Stockholm: International Federation of Gynecology and Obstetrics; 1988.
- Tavassoli FA, Devilee P. WHO classification of tumors. Pathology and genetics of tumors of breast and female genital organs. Lyon (France): IARC Press; 2003.
- Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47(1):207–214.
- Christian MC, Trimble EL. Salvage chemotherapy for epithelial ovarian carcinoma. Gynecol Oncol. 1994;55(3 Pt 2):S143–50. doi:10.1006/gyno.1994.1354.
- Wagner T, Stoppa-Lyonnet D, Fleischmann E, Muhr D, Pagès S, Sandberg T, Caux V, Moeslinger R, Langbauer G, Borg A, et al. Denaturing high-performance liquid chromatography detects reliably BRCA1 and BRCA2 mutations. Genomics. 1999;62(3):369–376. doi:10.1006/geno.1999.6026.
- Lewis AG, Flanagan J, Marsh A, Pupo GM, Mann G, Spurdle AB, Lindeman GJ, Visvader JE, Brown MA, Chenevix-Trench G, et al. Mutation analysis of FANCD2, BRIP1/BACH1, LMO4 and SFN in familial breast cancer. Breast Cancer Res. 2005;7(6):R1005–R16. Epub 2005 Oct 21. doi:10.1186/bcr1336.
- Dansonka-Mieszkowska A, Ludwig AH, Kraszewska E, Kupryjańczyk J. Geographical variations in TP53 mutational spectrum in ovarian carcinomas. Ann Hum Genet. 2006 Sep;70(Pt 5):594–604. doi:10.1111/j.1469-1809.2006.00257.x.
