ABSTRACT
Long non-coding RNAs (lncRNAs) are regarded as a group of biomarkers in the initiation and development of various cancers, including hepatocellular carcinoma (HCC). LncRNA FOXD2-AS1 has been studied in human colorectal cancer and glioma as an oncogene. However, the function and mechanism of lncRNA FOXD2-AS1 in hepatocellular carcinoma are marked. In this study, we found that high expression of FOXD2-AS1 predicted poor prognosis of HCC patients in the TCGA database. The dysregulation of FOXD2-AS1 was determined in HCC tissues and cell lines by qRT-PCR. Functionally, silenced FOXD2-AS1 efficiently suppressed HCC progression by regulating cell proliferation, apoptosis, migration and epithelial–mesenchymal transition (EMT). Mechanistically, FOXD2-AS1 was found to be activated by the transcription factor EGR1. Furthermore, FOXD2-AS1 could activate the Wnt/β-catenin signaling pathway. The mechanism contributed to the interaction between FOXD2-AS1 and Wnt/β-catenin signaling pathway was analyzed. It was uncovered that FOXD2-AS1 enhanced the activity of Wnt/β-catenin signaling pathway by epigenetically silencing the inhibitor of Wnt/β-catenin signaling pathway (DKK1). Rescue assays demonstrated that DKK1 and Wnt/β-catenin signaling pathway involved in FOXD2-AS1-mediated HCC progression. In conclusion, our study demonstrated that EGR1-induced upregulation of lncRNA FOXD2-AS1 promotes the progression of hepatocellular carcinoma via epigenetically silencing DKK1 and activating Wnt/β-catenin signaling pathway.
Introduction
Hepatocellular carcinoma (HCC) has become one of the commonest malignant tumors in the past years. It has gradually become the second leading cause of cancer-related deaths all over the world.Citation1,Citation2 Although the methods for diagnosis and therapy of HCC are continually upgrading, the overall survival rates of HCC patients are still low.Citation3 Additionally, the potential molecular mechanism and biological processes correlated with HCC are still marked. As a result, one of the most challenging thing for the treatment of HCC is to find novel therapeutic targets.Citation4
Non-coding RNAs (ncRNAs) are a group of molecular regulators which can affect diverse cellular functions.Citation5–Citation7 Based on length, ncRNAs are divided into three groups: lncRNAs (>200 nt), small ncRNAs (<200 nt) and miRNAs (18–22 nt). As a typical kind of ncRNAs, lncRNAs have been demonstrated to participate in various biological and pathological processes in cancers.Citation8–Citation10 A lot of lncRNAs have been reported to be regulators in hepatocellular carcinoma.Citation11–Citation17 All the above reports indicated the importance of lncRNAs in human cancers. As a member of lncRNA family, FOXD2-AS1 has been proved to be an oncogene in human colorectal cancer,Citation18 nasopharyngeal carcinoma,Citation19 and lung cancer.Citation20 Moreover, it has been proved to be a prognostic factor for patients with hepatocellular carcinoma.Citation21 Nevertheless, the biological function and mechanism of FOXD2-AS1 in HCC progression are still marked. The aim of this study is to explore the role of FOXD2-AS1 in HCC progression. Based on The Cancer Genome Atlas (TCGA) dataset, we analyzed the effect of FOXD2-AS1 dysregulation on the overall survival rate of HCC patients. Moreover, the high expression of FOXD2-AS1 was examined and identified in HCC tissues and cell lines. To explore and identify the function of FOXD2-AS1 in HCC progression and development, loss-of-function assays were conducted. As a result, the cell proliferation, migration, and EMT progress were inhibited, while the cell apoptosis was promoted. Therefore, we confirmed the oncogenic effects of FOXD2-AS1 on HCC progression. Mechanistically, FOXD2-AS1 was found to be activated by the transcription activator EGR1. And high expression of FOXD2-AS1 led to the activation of β-catenin signaling pathway. Further mechanism investigation revealed that lncRNA FOXD2-AS1 activated Wnt/β-catenin signaling pathway via epigenetically silencing DKK1. In conclusion, this study elucidated that lncRNA FOXD2-AS1 activated by EGR1 contributed to the progression of HCC via activating Wnt/β-catenin signaling pathway.
Materials and methods
Tissue samples
Eighty-eight pairs of fresh HCC tissues and the adjacent normal tissues were all collected from patients who were diagnosed with HCC at Luoyang Central Hospital, affiliated with Zhengzhou University. All patients had signed the informed consent before they enrolled in this study. This study had acquired the approval from the ethics committee of Zhengzhou University.
Cell culture
All cell lines used in this study were commercially obtained from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China). The cell lines were shown as follows: one normal cell (L-02) and four HCC cell lines (HepG2, Hep3B, SMMC-7721, LM3). All cell lines were preserved in a moist incubator with 5% CO2 at 37°C and maintained in DMEM (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS, Gibco).
Plasmid construction and transfection
The specific shRNAs against FOXD2-AS1 or DKK1 and their control shRNAs (sh-NC) were commercially obtained from Genepharm Co. (Shanghai, China). Transfections were conducted and finished by using Lipofectamine 2000 (Invitrogen, NY, USA). In order to construct pcDNA3.1-YY1 vector or pcDNA3.1-EGR1 vector, the whole sequence of them was synthesized and sub-cloned into a pcDNA3.1 (+) vector (GenePharma, Shanghai, China). The transfection efficiency was evaluated with qRT-PCR.
qRT-PCR
Total RNA was isolated and collected from HCC cells with TRIzol Reagent (Takara, Tokyo, Japan) in accordance with the users’ instruction. A total of 1 μg RNA was reversely transcribed with HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China). Next, AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) was utilized for real-time PCR. The conductions for PCR reaction were shown as follows: 95°C for 15 s, 40 cycles of 95°C for 5 s and 60°C for 30 s. The expression levels of genes were normalized to the internal control GAPDH and were calculated by using the equation: 2−ΔΔCt.
MTT assay
Cell viability was evaluated by using MTT Kit (Roche Applied Science, Basel, Switzerland). HepG2 and LM3 cells which had been transfected with sh-FOXD2-AS1 were grown in 96-well plates at a density of 3000 cells per well. According to the instruction, cell viability was assessed at different time points.
Cell viability assay
To assess cell viability, cells were seeded in a six-well plate at a density of 5 × 105 cells per well. Then, cells were transfected with sh-FOXD2-AS1 or sh-NC. Forty-eight hours later, cells were trypsinized and diluted (1:1) in trypan blue in a 96-well plate. Cell viability was evaluated by trypan exclusion. Twenty fields were calculated per condition with an automated cell counter (TC20, BioRad).
Colony formation assay
Cell proliferation was further determined by performing colony formation assay. The indicated HCC cells were placed on six-well plates (500 cells/well) and preserved in DMEM containing 10% FBS for about two weeks. In this process, the medium was replaced every 4 days. Two weeks later, methanol was used to fix colonies, 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) was used to stain the colonies and made them visible. Finally, the visible colonies were manually calculated.
Flow cytometry analysis
HepG2 and LM3 cells were harvested at two days’ post-transfection. To analyze cell apoptosis, FITC Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA) was used for double staining (FITC Annexin V and PI). The apoptotic cells were analyzed with Cell Quest software 0.9.3.1 (Biosciences, Franklin Lakes, NJ, USA).
Immunofluorescence assay
Cells were grown on glass slides and fixed in 4% formaldehyde for about 10 min. 0.3% Triton X-100 was used to permeate cell, while the goat serum was utilized to block slices for 15 min at 37°C. And the slices were incubated at 4°C overnight with anti-E-cadherin (1:80, Bioworld, MN, USA) and anti-N-cadherin (1:80, Bioworld) and with goat TRITC labeled secondary antibody (1:70, Bioworld) at 37°C for about 1 h. DAPI (Genview Inc, Shanghai, China) was used for staining. At length, fluorescence was visualized under a microscope (×400). The quantification was made by using the Image-Pro Plus 7.0 version software (Media Cybernetics, Inc., Rockville, MD, USA) as previously described.Citation22
Cell migration assay
The cell migration ability was assessed by performing the transwell migration assay. At 48-h post-transfection, HCC cells (5 × 104) in serum-free medium were put into the upper chamber of an insert (8 μm pore size; Millipore). Medium with 10% FBS was added into the lower chamber. After incubation for about one day, the cells remain stayed on the upper chamber were removed with cotton wool. Meanwhile, methanol and 0.1% crystal violet was used to stain cells that migrated through the membrane. Next, cells were stained and calculated on an IX71 inverted microscope (Olympus, Tokyo, Japan).
Western blot assay
Cells were lysed in RIPA buffer (Beyotime Biotechnolog, Shanghai, China) which contains a protease inhibitor. The BCA protein assay kit (Thermo Fisher Scientific, MA, USA) was utilized to measure the protein concentration. Subsequently, the protein extracts were subjected to 10% SDS-PAGE electrophoresis (40 mg/lane) and were transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were then blocked with 5% defatted milk. Next, they were incubated with the primary antibodies for all night at 4°C and with anti-Mouse IgG (ab193651) conjugated to horseradish peroxidase for 2 h at room temperature. All antibodies were commercially obtained from Abcam (Cambridge, Mass, USA). The blots were detected by using the Enhanced chemiluminescence detection reagent (Thermo Fisher Scientific). The levels of protein were normalized to GAPDH.
Luciferase reporter assay
To analyze the binding of EGR1 in the site 2 of FOXD2-AS1 promoter, HepG2 and LM3 cells were co-transfected with the wide type or mutant type of site 2 along with EGR1 and NC. The relative luciferase activity was assessed via the Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA) in accordance with the supplier’s protocols.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using the SimpleChIP® Enzymatic Chromatin IP Kit (CST, USA) in line with the recommendation of manufacturers. HepG2 and LM3 cell lines were subjected to 4% formaldehyde at room temperature for 15 min in order to cross-link DNA to protein. The DNA-protein complex was sonicated to generate chromatin fragments of 200–500 base pairs and immunoprecipitated with EGR1-specific antibody (Millipore, Bedford, MA, USA) at 4°C overnight. Normal rabbit IgG antibody (Santa Cruz Biotechnology, CA, USA) acted as a control. The next day, the immune-complexes were incubated with Dynabeads Protein G (Life Technologies, CA, USA) for 2 h at 4°C and DNA was retrieved using magnets. Recovered DNA was analyzed by qRT-PCR using SYBR Green Mixture.
RNA immunoprecipitation (RIP) assay
Based on the supplier’s introduction, RIP assay was performed to confirm the interaction between EZH2 and FOXD2-AS1 using the EZMagna RIP Kit (Millipore, Bellerica, MA, USA). In brief, HepG2 and LM3 cells were lysed with the RIP lysis buffer. Subsequently, cell lysates were cultured with the RIP buffer containing magnetic beads at 4°C for 4 h. The magnetic beads were coated with anti-IgG or anti-EZH2. Normal rabbit IgG (Proteintech) was used as the negative control. Finally, qRT‐PCR was performed to detect the enrichment of EZH2 and FOXD2-AS1.
Statistical analysis
SPASS 20.0 software (IBM) was utilized for all statistical analyses in this study. Differences between groups were compared and estimated by using Student’s t-test and one-way ANOVA. The overall survival rate of HCC patients was assessed by the Kaplan–Meier method and compared with the log-rank test. Relevance between and FOXD2-AS1 expression and EGR1 expression in HCC tissues was analyzed with Pearson correlation analysis. Data were considered statistically significant when P value less than 0.05.
Results
Upregulation of FOXD2-AS1 predicted poor prognosis for HCC patients
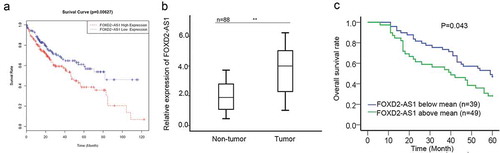
Based on the TCGA data, high expression of FOXD2-AS1 was closely correlated with the poor prognosis of HCC patients (). To identify the potential role of FOXD2-AS1 in HCC progression, we examined the expression of FOXD2-AS1 in 88 pairs of HCC tissues and adjacent non-malignant tissues. According to the result of qRT-PCR, we found that FOXD2-AS1 was higher in HCC tissues than that in normal tissues (). Next, HCC tissues were divided into two groups (FOXD2-AS1 above mean or blow mean) in accordance with the mean value of FOXD2-AS1 expression in 88 HCC tissues. Using the Kaplan Meier method, we analyzed the negative relevance between FOXD2-AS1 expression and overall survival rate of HCC patients ().
Figure 1. Upregulation of FOXD2-AS1 predicted poor prognosis for HCC patients. A. Based on the TCGA data, high expression of FOXD2-AS1 was closely correlated with the poor prognosis of HCC patients. B. The expression of FOXD2-AS1 was examined in 88 pairs of HCC tissues and adjacent non-malignant tissues. C. Kaplan Meier method was used to analyze the relevance between FOXD2-AS1 expression and overall survival rate of HCC patients. *P < 0.05, **P < 0.01 vs control group.
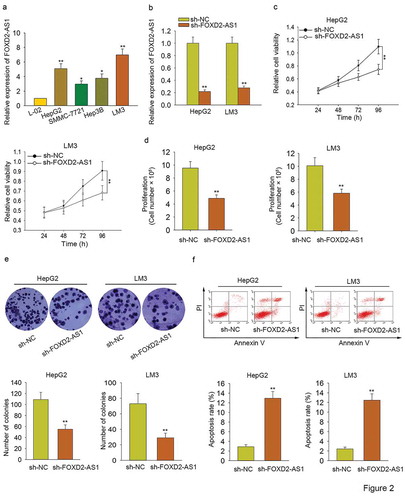
Knockdown of FOXD2-AS1 inhibited HCC cell proliferation and induced HCC cell apoptosis
To further analyze the biological role of FOXD2-AS1 in HCC progression, functional assays were carried out. To prepare for functional assays, we tested the expression level of FOXD2-AS1 in one normal cell line and four HCC cell lines. Obviously, it was highly expressed in HCC cells, especially in HepG2 and LM3 (). For loss-of-function assays, FOXD2-AS1 was silenced in HepG2 and LM3 cells by specific shRNA (termed sh-FOXD2-AS1). The transfection efficiency was examined with qRT-PCR (). Subsequently, MTT and trypan blue exclusion were applied to assess the cell viability. It was found that cell viability was efficiently suppressed by the knockdown of FOXD2-AS1 (–). Colony formation assay was performed to test the cell proliferation. As displayed in , silenced FOXD2-AS1 markedly suppressed cell proliferation. Flow cytometry analysis was then utilized to assess cell apoptosis. As a consequence, cell apoptosis was accelerated by the knockdown of FOXD2-AS1 (). Based on all the above findings, we confirmed that FOXD2-AS1 acted as an oncogene in HCC progression through affecting cell proliferation and cell apoptosis.
Figure 2. Knockdown of FOXD2-AS1 inhibited HCC cell proliferation and induced HCC cell apoptosis. A. The expression level of FOXD2-AS1 in one normal cell line and four HCC cell lines was tested. B. FOXD2-AS1 was silenced in HepG2 and LM3 cells by specific shRNA (termed sh-FOXD2-AS1). The transfection efficiency was examined with the qRT-PCR examination. C-D. MTT and trypan blue exclusion were carried out to determine the cell viability in HCC cells transfected with sh-FOXD2-AS1 or sh-NC. E. Colony formation assay was performed to test the proliferative ability of HepG2 and LM3 cells transfected with sh-FOXD2-AS1 or sh-NC. F. Flow cytometry analysis was then utilized to assess cell apoptosis. *P < 0.05, **P < 0.01 vs control group.
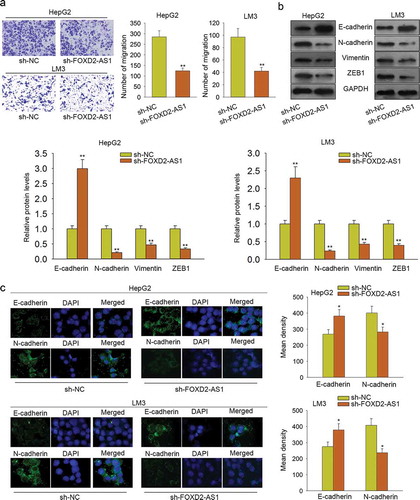
Knockdown of FOXD2-AS1 suppressed cell migration and EMT progress
Furthermore, we conducted a transwell assay to detect the cell migration ability. We found that cell migration was significantly inhibited by FOXD2-AS1 knockdown (). Western blot and immunofluorescence were applied to analyze the effect of sh-FOXD2-AS1 on EMT progress. The experimental results suggested that EMT progress was negatively affected by transfecting sh-FOXD2-AS1 (–).
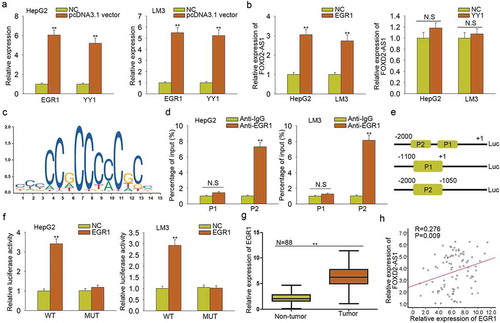
FOXD2-AS1 was activated by the transcription factor EGR1
Furthermore, we conducted mechanism experiments to analyze the molecular mechanism which led to the upregulation of FOXD2-AS1. According to the ChIP-seq result of UCSC (http://genome.ucsc.edu), we found some potential transcription factors of FOXD2-AS1. Among which, EGR1 and YY1 have been reported as the transcription activators of lncRNAs. Next, we overexpressed EGR1 and YY1 in both HepG2 and LM3 cells (). It was found that EGR1 efficiently enhanced the expression level of FOXD2-AS1 (). The binding motif of EGR1 obtained from JASPAR (http://jaspar.genereg.net/) was illustrated (). We chose the top two binding sites between EGR1 and FOXD2-AS1 promoter to do further study. These two binding sequenced were defined as part 1 (P1) and part 2 (P2) of FOXD2-AS1 promoter. ChIP assay revealed the strong affinity of EGR1 in the P2 of FOXD2-AS1 promoter (). Luciferase reporter assay demonstrated the binding of EGR1 in part 2 of FOXD2-AS1 promoter (–). Furthermore, we found that EGR1 was highly expressed in HCC tissues (), which is consistent with the expression of FOXD2-AS1 (). All these experimental results suggested that EGR1 is the transcription activator of FOXD2-AS1 in HCC.
Figure 4. FOXD2-AS1 was activated by the transcription factor EGR1. A. EGR1 and YY1 were overexpressed in both HepG2 and LM3 cells. B. The expression level of FOXD2-AS1 was measured in response to the overexpression of EGR1 and YY1. C. The binding motif of EGR1 obtained from JASPAR was illustrated. D. ChIP assay revealed the strong affinity of EGR1 in the P2 of FOXD2-AS1 promoter. E-F. Luciferase reporter assay demonstrated the binding of EGR1 in the site 2 of FOXD2-AS1 promoter. G. EGR1 was highly expressed in HCC tissues. H. The expression association between FOXD2-AS1 and EGR1 in HCC tissues was analyzed. **P < 0.01 vs control group. N.S: no significance.
FOXD2-AS1 enhanced the activity of Wnt/β-catenin signaling pathway
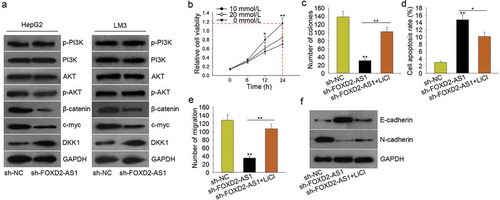
PI3K/AKT signaling pathway and Wnt/β-catenin signaling pathway are involved in lncRNAs-mediated tumor progression. In this study, we hypothesized that FOXD2-AS1 might exert oncogenic function in HCC progression by regulating PI3K/AKT signaling pathway or Wnt/β-catenin signaling pathway. At first, we examined the protein levels of the core factors of these two signaling pathways. The results showed that silenced FOXD2-AS1 significantly decreased the protein levels of β-catenin, c-myc but increased the level of DKK1 (the inhibitor of Wnt/β-catenin signaling pathway) (). Therefore, we speculated that FOXD2-AS1 promoted HCC progression by activating Wnt/β-catenin signaling pathway. To validate our viewpoint, we designed and conducted rescue assays. LiCl is the activator of Wnt/β-catenin signaling pathway. We examined the viability of HepG2 cell which was treated with a different dose of LiCl. The result of the MTT assay showed that the cell viability was strongest when HepG2 cell was treated with 10 mmol/L for 24 h (). Therefore, we conducted rescue assays in HepG2 cell treated with 10 mmol/L LiCl. The colony formation and transwell migration assays demonstrated that LiCl rescued the decreased cell proliferation and migration induced by silenced FOXD2-AS1 (, ), whereas the cell apoptosis increased by FOXD2-AS1 knockdown was partly decreased by LiCl (). Moreover, the reversal effect of silenced FOXD2-AS1 on the EMT progress was attenuated by LiCl (). Based on all these data, we confirmed that FOXD2-AS1 promoted HCC progression by activating Wnt/β-catenin signaling pathway.
Figure 5. FOXD2-AS1 enhanced the activity of Wnt/β-catenin signaling pathway. A. The protein levels of the core factors of PI3K/AKT signaling pathway and Wnt/β-catenin signaling pathway were examined in HepG2 and LM3 cells transfected with sh-FOXD2-AS1. B. The viability of HepG2 cell treated with a different dose of LiCl was evaluated with MTT assay. C. The proliferative ability of cell treated with sh-FOXD2-AS1 and LiCl was examined with colony formation assay. D. The cell apoptosis increased by FOXD2-AS1 knockdown was partly decreased by LiCl. E. The migratory ability of cell treated with sh-FOXD2-AS1 and LiCl was detected with transwell migration assay. F. The reversal effect of silenced FOXD2-AS1 on the EMT progress was attenuated by LiCl. *P < 0.05, **P < 0.01 vs control group.
FOXD2-AS1 activated Wnt/β-catenin signaling pathway by epigenetically silencing DKK1
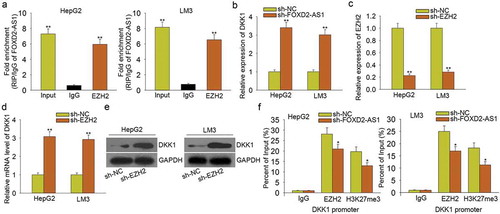
According to the above data, FOXD2-AS1 negatively modulated the expression of DKK1. Previous reports suggested that lncRNAs can epigenetically silence their downstream target genes by binding with EZH2. Similarly, FOXD2-AS1 has been certified to bind with EZH2 to epigenetically silence its downstream target gene in gastric cancer.Citation23 Moreover, EZH2 can repress the expression of DKK1.Citation24 Here, we hypothesized that FOXD2-AS1 activated Wnt/β-catenin signaling pathway by epigenetically silencing DKK1. The results of the RIP assay showed that FOXD2-AS1 could bind with EZH2 in HCC cells (). Moreover, we found that knockdown of FOXD2-AS1 led to the increased mRNA level of DKK1 (), further indicating the negative regulatory effect of FOXD2-AS1 on DKK1. Then, we found that knockdown of EZH2 () led to the increased mRNA and protein levels of DKK1 (–). Finally, ChIP assay revealed that EZH2 could directly bind to the DKK1 promoter and reconcile H3K4me2 demethylation. Nevertheless, FOXD2-AS1 knockdown partially attenuated the above phenomenon (). Therefore, we confirmed that FOXD2-AS1 activated Wnt/β-catenin signaling pathway by epigenetically silencing DKK1.
Figure 6. FOXD2-AS1 activated Wnt/β-catenin signaling pathway by epigenetically silencing DKK1. A. RIP assay was conducted to demonstrate that FOXD2-AS1 could bind with EZH2 in HCC cells. B. The mRNA level of DKK1 was measured in HCC cells transfected with sh-FOXD2-AS1. C. EZH2 was knocked down in HCC cells by sh-EZH2. D-E. The mRNA and protein level of DKK1 were tested in cells transfected with sh-EZH2. F. ChIP assay revealed that EZH2 could directly bind to the DKK1 promoter and reconcile H3K4me2 demethylation. Nevertheless, FOXD2-AS1 knockdown partially attenuated the above phenomenon. *P < 0.05, **P < 0.01 vs control group.
DKK1 involved in FOXD2-AS1-mediated HCC progression
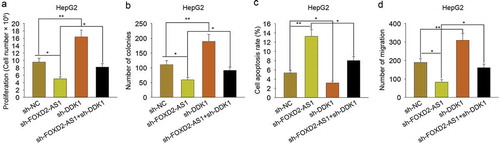
To evaluate the role of DKK1 in FOXD2-AS1-mediated HCC progression, rescue assays were carried out in HepG2 cells. As shown in , knockdown of FOXD2-AS1 suppressed cell viability, while knockdown of DKK1 increased cell viability. More importantly, knockdown of DKK1 rescued the decreased cell viability induced by FOXD2-AS1 knockdown. The colony formation assay indicated the inhibitory effect of sh-FOXD2-AS1 on cell proliferation and the positive effect of sh-DKK1 on cell proliferation (). Moreover, DKK1 knockdown rescued the decreased cell proliferation induced by silenced FOXD2-AS1, whereas cell apoptosis was increased by silenced FOXD2-AS1, while was decreased by DKK1 knockdown. Cell apoptosis increased by FOXD2-AS1 knockdown was partly decreased by silenced DKK1 (). Transwell assay revealed the inhibitory effect of FOXD2-AS1 knockdown on cell migration and the positive effect of DKK1 knockdown on cell migration (). Moreover, DKK1 knockdown rescued the decreased cell migration induced by silenced FOXD2-AS1. Thus, we confirmed that DKK1 involved in FOXD2-AS1-mediated HCC progression.
Figure 7. DKK1 involved in FOXD2-AS1-mediated HCC progression. A. The cell viability was examined in HepG2 cell transfected with sh-NC, sh-FOXD2-AS1, sh-DKK1 or co-transfected with sh-FOXD2-AS1 and sh-DKK1. B. Colony formation assay was carried out to detect the proliferative ability of HepG2 cell transfected with sh-NC, sh-FOXD2-AS1, sh-DKK1 or co-transfected with sh-FOXD2-AS1 and sh-DKK1. C. Apoptosis rate of HepG2 cell transfected with sh-NC, sh-FOXD2-AS1, sh-DKK1 or co-transfected with sh-FOXD2-AS1 and sh-DKK1 was measured with flow cytometry analysis. D. Transwell assay was applied to detect cell migration in HepG2 cell transfected with sh-NC, sh-FOXD2-AS1, sh-DKK1 or co-transfected with sh-FOXD2-AS1 and sh-DKK1. *P < 0.05, **P < 0.01 vs control group.
Discussion
Over the past years, lncRNAs have been verified to be potential therapeutic biomarkers for HCC.Citation25–Citation28 This study focused on investigating the biological effects of FOXD2-AS1 on HCC progression. It has been reported that FOXD2-AS1 can act as a prognostic factor for cancer patients.Citation29–Citation31 Based on the data of the TCGA database, we analyzed the prognostic influence of FOXD2-AS1 on the patients with HCC. High expression of FOXD2-AS1 was found to predict unfavorable prognosis, indicating the research value of FOXD2-AS1 in HCC progression. Furthermore, we found that FOXD2-AS1 was expressed stronger in tumor tissues collected from 88 HCC patients. Kaplan–Meier method was used to analyze the prognostic value of FOXD2-AS1 for these 88 HCC patients. All these data proved the importance of FOXD2-AS1 for researching HCC.
Dysregulation of lncRNAs is closely associated with tumor progression and development.Citation32–Citation34 Moreover, ectopic expression of lncRNAs can contribute to proliferation, migration and EMT progress.Citation35–Citation38 To identify the role of FOXD2-AS1 in HCC progression, we designed and conducted functional assays. Since FOXD2-AS1 was overexpressed in HCC tissues and cell lines, we silenced FOXD2-AS1 to examine cell proliferation, migration, and EMT progress. According to the experimental results, we found that knockdown of FOXD2-AS1 negatively regulated cell proliferation, migration, and EMT progress while promoted cell apoptosis, indicating that FOXD2-AS1 served as a tumor facilitator in HCC progression through modulating cell proliferation, apoptosis, migration and EMT progress.
Dysregulation of lncRNAs may be caused by various molecular mechanisms. It has been reported that transcription factors can contribute to the dysregulation of lncRNAs by activating the transcription of lncRNAs.Citation39–Citation42 In this study, we found the potential transcription factors of FOXD2-AS1 from UCSC. Among which, YYA and EGR1 have been certified to activate the transcription of lncRNAs.Citation43–Citation45 Therefore, we chose them for subsequent mechanism experiments. However, the qRT-PCR analysis revealed that only EGR1 overexpression can lead to the upregulation of FOXD2-AS1 in HCC cells. Further mechanism investigation proved that EGR1 enhanced the transcription activity of FOXD2-AS1 by binding to the promoter of FOXD2-AS1.
It has been widely reported that lncRNAs can act as tumor suppressors or tumor promoters through modulating some signaling pathway.Citation46–Citation53 In this study, we tried to analyze whether FOXD2-AS1 exerts functions by regulating a certain signaling pathway. According to western blot analysis, we found that FOXD2-AS1 might activate Wnt/β-catenin signaling pathway. Rescue assays demonstrated that FOXD2-AS1 promoted HCC progression via activation of Wnt/β-catenin signaling pathway. Furthermore, we continued to detect the mechanism which contributed to the interaction between FOXD2-AS1 and Wnt/β-catenin signaling pathway. FOXD2-AS1 has been certified to bind with EZH2 to epigenetically silence its downstream target. Moreover, EZH2 can repress the expression of DKK1. Therefore, we designed and conducted mechanism experiments to demonstrate that whether FOXD2-AS1 activated Wnt/β-catenin signaling pathway by epigenetically silencing DKK1. Based on the results, we confirmed that FOXD2-AS1 epigenetically silencing DKK1 by binding with EZH2. Rescue assays indicated that DKK1 involved in FOXD2-AS1-medicated HCC progression. In conclusion, our research findings revealed that lncRNA FOXD2-AS1 activated by EGR1 contributed to HCC progression by activating Wnt/β-catenin signaling pathway. The findings of this study may help to find the novel therapeutic target in the treatment of HCC.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank all lab members.
References
- Baker E, Jacobs C, Martinie J, Iannitti DA, Vrochides D, Swan RZ. Mixed hepatocellular carcinoma, neuroendocrine carcinoma of the liver. Am Surg. 2016;82:1121–1125.
- Zhao L, Mao Y, Zhou J, Zhao Y, Cao Y, Chen X. Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways. Am J Cancer Res. 2016;6:387–402.
- Yang PC, Ho CM, Hu RH, Ho MC, Wu YM, Lee PH. Prophylactic liver transplantation for high-risk recurrent hepatocellular carcinoma. World J Hepatol. 2016;8:1309–1317. doi:10.4254/wjh.v8.i31.1309.
- Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: an update. World J Gastroenterol. 2016;22:9069–9095. doi:10.3748/wjg.v22.i41.9069.
- He X, Sun F, Guo F, Wang K, Gao Y, Feng Y, Li, Y. Knockdown of long noncoding RNA FTX inhibits proliferation, migration, and invasion in renal cell carcinoma cells. Oncol Res. 2017;25:157–166. doi:10.3727/096504016X14719078133203.
- Li Q, Li C, Chen J, Liu P, Cui Y, Zhou X, Zu, X. High expression of long noncoding RNA NORAD indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Urol Oncol. 2018;36:310.e15–310.e22. doi:10.1016/j.urolonc.2018.02.019.
- Jiang H, Li T, Qu Y, Wang X, Li B, Song J, Zhan, J. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018;425:78–87. doi:10.1016/j.canlet.2018.03.038.
- Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng SH, Zhang DH. Long non-coding RNA NEAT1 promotes malignant progression of thyroid carcinoma by regulating miRNA-214. Int J Oncol. 2017;50:708–716. doi:10.3892/ijo.2016.3803.
- Zeng J, Du T, Song Y, Gao Y, Li F, Wu R, Pei, Z. Knockdown of long noncoding RNA CCAT2 inhibits cellular proliferation, invasion, and epithelial-mesenchymal transition in glioma cells. Oncol Res. 2017;25:913–921. doi:10.3727/096504016X14792098307036.
- Durzynska J, Lesniewicz K, Poreba E. Human papillomaviruses in epigenetic regulations. Mutat Res Rev Mutat Res. 2017;772:36–50. doi:10.1016/j.mrrev.2016.09.006.
- Yang J, Li J, Liu B, Zhang R, Gu F, Zhao J, Cheng, S. Long noncoding RNA AK021443 promotes cell proliferation and migration by regulating epithelial-mesenchymal transition in hepatocellular carcinoma cells. DNA Cell Biol. 2018;37:481–490. doi:10.1089/dna.2017.4030.
- Chen H, Yang F, Li X, Gong ZJ, Wang LW. Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2018;499:702–710. doi:10.1016/j.bbrc.2018.03.215.
- Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J. LncRNA NR2F1-AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR-363. J Cell Mol Med. 2018;22:3238–3245. doi:10.1111/jcmm.13605.
- Zhang K, Zhao Z, Yu J, Chen W, Xu Q, Chen L. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem. 2018;119:6045–6056. doi:10.1002/jcb.26802.
- Dong Y, Yan W, Zhang SL, Zhang MZ, Zhou YP, Ling HH, Zhang, P. Prognostic values of long non-coding RNA MIR22HG for patients with hepatocellular carcinoma after hepatectomy. Oncotarget. 2017;8:114041–114049. doi:10.18632/oncotarget.23110.
- Xie CR, Wang F, Zhang S, Wang FQ, Zheng S, Li Z, Yin, ZY. Long noncoding RNA HCAL facilitates the growth and metastasis of hepatocellular carcinoma by acting as a ceRNA of LAPTM4B. Mol Ther Nucleic Acids. 2017;9:440–451. doi:10.1016/j.omtn.2017.10.018.
- Jang SY, Kim G, Park SY, Lee YR, Kwon SH, Kim HS, Chun, JM. Clinical significance of lncRNA-ATB expression in human hepatocellular carcinoma. Oncotarget. 2017;8:78588–78597. doi:10.18632/oncotarget.21094.
- Zhu Y, Qiao L, Zhou Y, Ma N, Wang C, Zhou J. Long non-coding RNA FOXD2-AS1 contributes to colorectal cancer proliferation through its interaction with microRNA-185-5p. Cancer Sci. 2018;109:2235–2242. doi:10.1111/cas.13632.
- Chen G, Sun W, Hua X, Zeng W, Yang L. Long non-coding RNA FOXD2-AS1 aggravates nasopharyngeal carcinoma carcinogenesis by modulating miR-363-5p/S100A1 pathway. Gene. 2018;645:76–84. doi:10.1016/j.gene.2017.12.026.
- Rong L, Zhao R, Lu J. Highly expressed long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer progression via Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2017;484:586–591. doi:10.1016/j.bbrc.2017.01.141.
- Zhao QJ, Zhang J, Xu L, Liu FF. Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma. World J Gastroenterol. 2018;24:3426–3439. doi:10.3748/wjg.v24.i30.3426.
- Zhong WQ, Chen G, Zhang W, Xiong XP, Ren JG, Zhao Y, Zhao, YF. Down-regulation of connexin43 and connexin32 in keratocystic odontogenic tumours: potential association with clinical features. Histopathology. 2015;66:798–807. doi:10.1111/his.12569.
- Xu TP, Wang WY, Ma P, Shuai Y, Zhao K, Wang YF, Shu, YQ. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene. 2018;37:5020–5036. doi:10.1038/s41388-018-0308-y.
- Yang Y, Chen XX, Li WX, Wu XQ, Huang C, Xie J, Li, J. EZH2-mediated repression of Dkk1 promotes hepatic stellate cell activation and hepatic fibrosis. J Cell Mol Med. 2017;21:2317–2328. doi:10.1111/jcmm.13153.
- Zeisel MB, Baumert TF. Translation and protein expression of lncRNAs: impact for liver disease and hepatocellular carcinoma. Baltimore (Md): Hepatology; 2016.
- Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. doi:10.1016/j.pharmthera.2016.03.004.
- Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Fan, J. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma via de-repression of CTNNB1. Hepatology: Baltimore (Md); 2015.
- Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, Sun, SH. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi:10.1002/hep.27239.
- Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Andersen, JB. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi:10.1002/hep.26740.
- Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Li, H. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi:10.1038/onc.2015.223.
- Yu G, Lin J, Liu C, Hou K, Liang M, Shi B. Long non-coding RNA SPRY4-IT1 promotes development of hepatic cellular carcinoma by interacting with ERRα and predicts poor prognosis. Sci Rep. 2017;7:17176. doi:10.1038/s41598-017-16781-9.
- Shih JW, Chiang WF, Wu ATH, Wu MH, Wang LY, Yu YL, Changou, CA. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer progression. Nat Commun. 2017;8:15874. doi:10.1038/ncomms15874.
- Herriges MJ, Tischfield DJ, Cui Z, Morley MP, Han Y, Babu A, Anderson, SA. The NANCI-Nkx2.1 gene duplex buffers Nkx2.1 expression to maintain lung development and homeostasis. Genes Dev. 2017. doi:10.1101/gad.298018.117.
- Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Wang, H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2015;65:1494–504. doi:10.1136/gutjnl-2014-308392.
- Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol Cell. 2015;59:698–711. doi:10.1016/j.molcel.2015.06.023.
- Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Khashab, M. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi:10.1136/gutjnl-2013-305266.
- Zhang E, Han L, Yin D, He X, Hong L, Si X, Shu, Y. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2016;45:3086–3101. doi:10.1093/nar/gkw1247.
- Rodríguez-Mateo C, Torres B, Gutiérrez G, Pintor-Toro JA. Downregulation of Lnc-Spry1 mediates TGF-β-induced epithelial-mesenchymal transition by transcriptional and posttranscriptional regulatory mechanisms. Cell Death Differ. 2017;24:785–797. doi:10.1038/cdd.2017.9.
- Chen X, Zeng K, Xu M, Hu X, Liu X, Xu T, Wang, S. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9:982. doi:10.1038/s41419-018-0962-6.
- Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, Ponnusamy, M. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29. doi:10.1038/s41467-017-02280-y.
- Jiang Y, Jiang YY, Xie JJ, Mayakonda A, Hazawa M, Chen L, Sun, QY. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun. 2018;9:3619. doi:10.1038/s41467-018-06081-9.
- Chen Z, Gao Y, Yao L, Liu Y, Huang L, Yan Z, Weng, H. LncFZD6 initiates Wnt/β-catenin and liver TIC self-renewal through BRG1-mediated FZD6 transcriptional activation. Oncogene. 2018;37:3098–3112. doi:10.1038/s41388-018-0203-6.
- Zhang Y, Jin F, Li XC, Shen FJ, Ma XL, Wu F, Lin, Y. The YY1-HOTAIR-MMP2 signaling axis controls trophoblast invasion at the maternal-fetal interface. Mol Ther. 2017;25:2394–2403. doi:10.1016/j.ymthe.2017.06.028.
- Knauss JL, Miao N, Kim SN, Nie Y, Shi Y, Wu T, Sun, T. Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis. 2018;9:799. doi:10.1038/s41419-018-0840-2.
- Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell cycle progression in gastric cancer. Cancer Res. 2018. doi:10.1158/0008-5472.CAN-18-1011.
- Lu Z, Chen Z, Li Y, Wang J, Zhang Z, Che Y, Gao, Y. TGF-beta-induced NKILA inhibits ESCC cell migration and invasion through NF-kappaB/MMP14 signaling. J Mol Med (Berl). 2018;96:301–313. doi:10.1007/s00109-018-1621-1.
- Wen X, Han XR, Wang YJ, Wang S, Shen M, Zhang ZF, Hu, B. Down-regulated long non-coding RNA ANRIL restores the learning and memory abilities and rescues hippocampal pyramidal neurons from apoptosis in streptozotocin-induced diabetic rats via the NF-kappaB signaling pathway. J Cell Biochem. 2018;119:5821–5833. doi:10.1002/jcb.v119.7.
- Zhang L, Shi SB, Zhu Y, Qian TT, Wang HL. Long non-coding RNA ASAP1-IT1 promotes cell proliferation, invasion and metastasis through the PTEN/AKT signaling axis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22:142–149. doi:10.26355/eurrev_201801_14111.
- Li WD, Zhou DM, Sun LL, Xiao L, Liu Z, Zhou M, Li, XQ. LncRNA WTAPP1 promotes migration and angiogenesis of endothelial progenitor cells via MMP1 through microRNA 3,120 and Akt/PI3K/autophagy pathways. Stem cells: Dayton (Ohio); 2018.
- Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B, Jia, L. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. 2018;11:89. doi:10.1186/s13045-018-0632-2.
- Li C, Liang G, Yang S, Sui J, Yao W, Shen X, Wu, W. Dysregulated lncRNA-UCA1 contributes to the progression of gastric cancer through regulation of the PI3K-Akt-mTOR signaling pathway. Oncotarget. 2017;8:93476–93491. doi:10.18632/oncotarget.19281.
- Fernández-Barrena MG, Perugorria MJ, Banales JM. Novel lncRNA T-UCR as a potential downstream driver of the Wnt/β-catenin pathway in hepatobiliary carcinogenesis. Gut. 2016,66:1177–1178. doi:10.1136/gutjnl-2016-312899.
- Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, Du, X. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11:113. doi:10.1186/s13045-018-0656-7.