ABSTRACT
Purpose: It remains unclear that long noncoding RNAs’ role in cancer initiation and progression, including osteosarcoma. Long noncoding RNA LINC00963 was found to be participated in carcinogenesis and progression of osteosarcoma. However, the molecular mechanisms of LINC00963 engaged in osteosarcoma (OS) still needs to be explored.
Methods: LINC00963 and miR-204-3p RNA expression levels were quantified by PCR in OS tissues and cells. CCK 8 assay, wound healing assay and transwell migration and invasion assay were chosen to assess cell growth, viability, migration, and invasion. Luciferase reporter assays were performed to verify direct interaction between LINC00963 and miR-204-3p and miR-204-3p and Fibronectin-1. Western blot was conducted to evaluate Fibronectin-1 expression in OS cells.
Results: LINC00963 was verified to be highly expressed in OS samples and cells. Specifically, elevated expression of LINC00963 was correlated with poor prognosis in patients. Furthermore, LINC00963 overexpression was found to promote proliferation, migration, and invasion in vitro. The luciferase reporter assay showed that LINC00963 can suppress miR-204-3p by directly binding miR-204-3p. Rescue experiment results indicated that function of LINC00963 in osteosarcoma was miR-204-3p dependant. Besides, we initially explored Fibronectin-1 (FN1) as the target of LINC00963/miR-204-3p axis in osteosarcoma.
Conclusions: Our findings implied that LINC00963/miR-204-3p/FN1 can play an important role in proliferation and progression in osteosarcoma. LINC00963 has the potential to be a therapeutic target for osteosarcoma treatment.
KEYWORDS:
Introduction
Osteosarcoma is the most common primary malignant bone tumor, which occurs most often in adolescence.Citation1 Because of aggressive biological behaviors including early metastasis and micro-metastasis, amputation was regarded as the main surgical treatment.Citation2 As with deep understanding of osteosarcoma development and huge advancement of surgical techniques, most patients are treated by surgical limb-salvage resection in combination with neoadjuvant chemotherapy when possible.Citation2–Citation4 Correspondingly, the 5-year survival rate has been improved from no more than 30% to over 70%.Citation2 However, the unclear and sophisticated molecular mechanisms in osteosarcoma development is still a key barrier to further prognosis.Citation3,Citation5 Consequently, identification of therapeutic targets is a good path to improve osteosarcoma outcomes.
LncRNAs, also called long noncoding RNAs, is a non-protein-coding RNA transcript whose length is greater than 200 nt.Citation6 Compared to those protein-coding genes, the number of LncRNA genes is far larger.Citation7,Citation8 Once considered as junk or useless RNA, LncRNAs is now being realized playing a vital role in biological functions.Citation6 It is increasingly evident that aberrant LncRNAs expression is involved with many different cancers including osteosarcoma.Citation6,Citation9–Citation12 Many studies revealed that LncRNAs can influence various aspects of cancer biology, like proliferation, migration, metastasis, drug resistanceCitation13, and cancer stem-like cells.Citation14 Some researchers revealed that a lot of lncRNAs were statistically differentially expressed in squamous cell lung cancer compared with corresponding normal tissues.Citation15 HOTAIR was verified to assess the therapeutic effect for advanced gastric adenocarcinoma patients receiving chemotherapy.Citation16 A long noncoding RNA RBM5-AS1 was reported to play a key role in self-renewal of colon cancer-initiating cell.Citation14 Likewise, several studies have reported that aberrantly expressed lncRNAs played a vital role in osteosarcoma development.Citation17,Citation18 However, the exact mechanisms of lncRNAs regulation in osteosarcoma are still unclear. LINC00963 (also named MetaLnc9) was found to promote lung cancer metastasis,Citation19 but it has not been reported in osteosarcoma. Consequently, we decided to explore the potential function of LINC00963 in osteosarcoma.
In this study, we found that LINC00963 was highly expressed in osteosarcoma tissues and cell lines. Poor prognosis was found in patients with relatively high LINC00963 expression. LINC00963 was identified to promote osteosarcoma cell proliferation, migration, and invasion in vitro. On the basis of LncBase v2’s prediction, miR-204-3p was verified to be one target miRNA of LINC00963. Furthermore, we found that miR-204-3p was significantly underexpressed in osteosarcoma tissues and cell lines. By performing the rescue experiment, the results showed that effects of LINC00963 overexpression or underexpression on osteosarcoma will be attenuated by knocking down or upregulating miR-204-3p expression. Via western blot assay, we verified that LINC00963 can increase Fibronectin-1 expression by suppressing miR-204-3p. All our findings demonstrate that LINC00963 can promote osteosarcoma proliferation, migration, and invasion by suppressing miR-204-3p/FN1 axis. LINC00963 can be an underlying biomarker for OS poor prognosis and a potential therapeutic target for OS treatment.
Results
LINC00963 is upregulated in osteosarcoma and indicates a poor prognosis
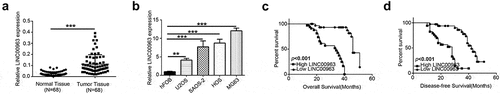
Initially, real-time PCR was performed to compare LINC00963 expression in 68 osteosarcoma and adjacent normal tissues. LINC00963 expression was predominantly increased in all osteosarcoma tissues by comparison with matched normal tissues (). Besides, we confirmed significantly higher LINC00963 expression in different OS cell lines (MG63, U2OS, HOS, Saos-2) compared to hFOB cell (). Next, all the osteosarcoma patients were divided into two groups according to those real-time PCR results using a median LINC00963 expression value. Then, we further investigated the relationship between LINC00963 expression and some vital clinical parameters of those 68 osteosarcoma patients. As shown in Supplementary Table 1, the expression of LINC00963 was significantly correlated with Enneking Classification, recurrence and lung metastasis in OS patients. Furthermore, to examine the relationship between the prognosis and LINC00963 expression, Kaplan-Meier survival analysis was conducted. Both overall survival (OS) and disease-free survival (DFS) were shorter in high LINC00963 expression patients than low expression patients (&).
Figure 1. LINC00963 is highly expressed in OS and associated with poor outcomes in OS patients. (a) LINC00963 RNA expression levels were assessed quantitatively in 68 pairs of osteosarcoma tissues and corresponding normal tissues utilizing qRT-PCR. (b) LINC00963 RNA expression levels were examined quantitatively in human osteoblast (hFOB) and osteosarcoma (U2OS, SAOS-2, HOS and MG63) cell lines. (c) OS patients with higher LINC00963 had a poor overall survival (N = 68, median survival:34 months vs. 46 months, Long Rank p< 0.001). (d) OS patients with higher LINC00963 had poor disease-free survival (N = 68, median survival:28 months vs. 42 months, Long Rank p < 0.001). Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).
LINC00963 overexpression enhances proliferation and migration of OS cells
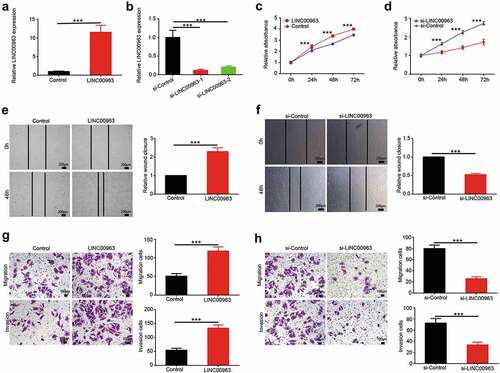
To investigate underlying tumorigenic function of LINC00963, stably overexpressed LINC00963 U2OS cell line was constructed. As is shown in , LINC00963 expression level was verified in LINC00963 overexpressed U2OS cells compared to control cells. In CCK8 assays, the growth curve showed that overexpression of LINC00963 promoted U2OS cells growth over time compared to the control group (). Trypan blue assays were also performed to prove that LINC00963 can enhance U2OS cell viability (Supplementary Figure 3A). In wound healing assays, the results revealed that LINC00963-overexpression U2OS cells can migrate more distance than the control cells (). Consistently, in transwell migration assays, we found that more cells migrated in LINC00963-overexpression group than the control group (). Moreover, transwell invasion assays were performed to determine the effects of LINC00963 on cell invasion (). The results showed that LINC00963 overexpression in U2OS cells resulted in increasing the invasion capacity.
Figure 2. LINC00963 enhances proliferation and migration of OS cells. (a) The transfection effect of LINC00963 overexpression was determined by qRT-PCR. (b) The transfection effect of LINC00963 underexpression was determined by qRT-PCR. (c) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after LINC00963 overexpression. (d) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after LINC00963 underexpression. (e) Wound healing assays were performed to assess cell mobility after LINC00963 overexpression. (f) Wound healing assays were performed to assess cell mobility after LINC00963 underexpression. (g) Transwell migration and invasion assays were conducted to examine cell migration and invasion after LINC00963 overexpression. (h) Transwell migration and invasion assays were conducted to examine cell migration and invasion after LINC00963 underexpression. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).
LINC00963 knockdown reduces proliferation and migration of OS cells
To further examine the biological role of LINC00963, LINC00963 was stably underexpressed in osteosarcoma cell line MG63. As is shown in , LINC00963 expression level was verified in LINC00963 underexpressed MG63 cells compared to control cells. si-LINC00963-1 was chosen to perform the following function experiment. In CCK8 assays, we found that knockdown of LINC00963 impeded MG63 cells growth over time compared to the control group (). Trypan blue assays were also performed to prove that LINC00963 knockdown can reduce MG63 cell viability (Supplementary Figure 3B). In wound healing assays, the results indicated that knockdown of Lcn00963 also decreased the migrated distance of MG63 cells (). In transwell migration assays, the data significantly manifested that less cells migrated in LINC00963-underexpression group than the control group (). To investigate the effects of LINC00963-knockdown on cell invasion, transwell invasion assays were performed in LINC00963 underexpressed MG63 cells (). The results suggested that MG63 cells underexpressing LINC00963 had markedly less invasion capacity than control cells.
LINC00963 downregulates mir-204-3p expression
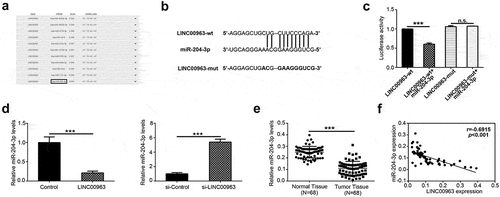
One of LncRNAs’ main function is as competing endogenous RNA. To explore the potential “sponge” mechanism of LINC00963 in osteosarcoma, a bioinformatic prediction was performed to seek targeted miRNAs of LINC00963 in LncBase Predicted v.2.Citation20 As shown in , miR-204-3p was chosen to be the candidate targeted miRNA of LINC00963. We, therefore, examined whether LINC00963 regulate the expression of miR-204-3p. To verify how LINC00963 modulate miR-204-3p, luciferase reporter assays were conducted (). The results showed that luciferase activity of LINC00963 wild-type 3ʹ-UTR was reduced while that of its mutant 3ʹ-UTR was not affected (). To explore the interaction relationship between LINC00963 and miR-204-3p, both stably overexpressed LINC00963 U2OS and underexpressed MG63 cell line were established. Real-time PCR was performed to verify the miR-204-3p expression. The results indicated that LINC00963 overexpression notably increased the expression level of miR-204-3p in U2OS cells (). Inversely, RNA levels of miR-204-3p were prominently reduced in the LINC00963 underexpressed MG63 cells (). However, LINC00963 RNA expression level was not affected after miR-204-3p mimics or miR-204-3p inhibitors or corresponding control was transfected in OS cells (Supplementary Figure 1). Taken altogether, we can conclude that LINC00963 can suppress miR-204-3p by directly binding with miR-204-3p.
Figure 3. LINC00963 downregulates miR-204-3p expression and negative correlation between LINC00963 RNA and miR-204-3p RNA expression. (a) Potential binding targets were predicted on LncBase Predicted v.2. (b) Predicted sequence of LINC00963 wild-type 3ʹ-UTR was predicted to bind with miR-204-3p and its corresponding LINC00963 mutant 3ʹ-UTR. (c) Luciferase activity assay showed that LINC00963 can bind directly with miR-204-3p. (d) miR-204-3p RNA expression levels were measured after LINC00963 overexpression or knockdown in OS cells. (e)miR-204-3p RNA expression levels were assessed quantitatively in 68 pairs of osteosarcoma tissues and corresponding normal tissues utilizing qRT-PCR. (f) LINC00963 RNA expression levels were inversely correlated with miR-204-3p RNA expression levels in 68 osteosarcoma tissues. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).
Negative correlation between LINC00963 and mir-204-3p RNA expression
miR-204-3p mRNA expression levels were also measured by real-time PCR in 68 osteosarcoma tissues compared to match normal tissues. The mRNA expression of miR-204-3p was predominantly lower in tumor tissues than normal tissues (). Furthermore, the relationship between LINC00963 and miR-204-3p expression in 68 osteosarcoma patients was investigated. We found that miR-204-3p expression was negatively correlated with LINC00963 expression ().
MiR-204-3p overexpression attenuates the effects of LINC00963 overexpression on osteosarcoma cells
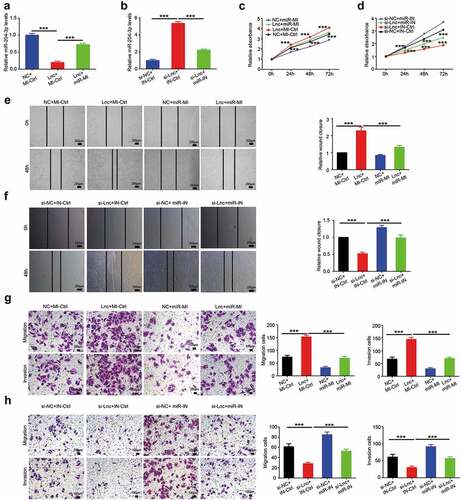
As miR-204-3p has been confirmed to be regulated by LINC00963, we next verified if miR-204-3p is essential for LINC00963-induced biological function in osteosarcoma. miR-204-3p expression was overexpressed in LINC00963 stably overexpressed U2OS cells by transfecting miR-204-3p specific overexpression plasmids. As shown in , the expression level of miR-204-3p was increased by miR-204-3p specific overexpression plasmids in LINC00963 stably overexpressed U2OS cells. CCK8 growth curve data indicated that the elevated expression of miR-204-3p reversed the proliferation induced by LINC00963 overexpression (). Trypan blue assays were also performed to prove that the overexpression of miR-204-3p can attenuate the cell viability induced by LINC00963 overexpression (Supplementary Figure 3C). Next, in wound healing assays and transwell migration assays, the results revealed that the elevated expression of miR-204-3p attenuated the increased migration capacity caused by LINC00963 overexpression (&). Furthermore, the elevated invasion capacity induced by LINC00963 overexpression was abolished by miR-204-3p overexpression (). All the results suggest that overexpression of miR-204-3p partly reverse the growth and progression effects induced by LINC00963 overexpression in OS cells.
Figure 4. miR-204-3p attenuates the effects of LINC00963 on osteosarcoma cells. (a) miR-204-3p RNA expression level was examined after LINC00963 or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (b) miR-204-3p RNA expression level was examined after si-LINC00963 or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. (c) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after LINC00963 or miR-204-3p mimics or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (d) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after si-LINC00963 or miR-204-3p inhibitors or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. (e) Wound healing assays were performed to assess cell mobility after LINC00963 or miR-204-3p mimics or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (f) Wound healing assays were performed to assess cell mobility after si-LINC00963 or miR-204-3p inhibitors or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. (g) Transwell migration and invasion assays were conducted to examine cell migration and invasion after LINC00963 or miR-204-3p mimics or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (h) Transwell migration and invasion assays were conducted to examine cell migration and invasion after si-LINC00963 or miR-204-3p inhibitors or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance). NC:LINC00963 control, si-NC:si-LINC00963 control, Lnc: LINC00963, si-Lnc: si-LINC00963, miR-MI: miR-204-3p mimics, miR-IN: miR-204-3p inhibitors, MI-Ctrl: miR-204-3p mimics control, IN-Ctrl: miR-204-3p inhibitors control.
MiR-204-3p underexpression reverses the effects of LINC00963 underexpression on osteosarcoma cells
To further assess the roles of miR-204-3p in osteosarcoma, we underexpressed miR-204-3p in LINC00963 stably knockdown MG63 cells. As shown in , the expression level of miR-204-3p was decreased by transfecting specific siRNAs in LINC00963 stably underexpressed MG63 cells. Next, we analyzed whether miR-204-3p underexpression can influence the proliferation, migration and invasion capacity decreased by LINC00963 underexpression. In CCK8 assays, we found that decreased miR-204-3p can promote the growth impeded by LINC00963 knockdown (). Trypan blue assays were also performed to prove that the knockdown of miR-204-3p can attenuate the cell viability decreased by LINC00963 underexpression (Supplementary Figure 3D). In wound healing assays, the results showed that the migrated distance increased in miR-204-3p underexpression +LINC00963 knockdown group compared to simply LINC00963 knockdown group (). In transwell migration assays, we observed that more MG63 cells migrated in miR-204-3p underexpression +LINC00963 knockdown group compared to simply LINC00963 knockdown group (). In transwell invasion assays, the results manifested that the underexpression of miR-204-3p reversed the hampered invasion capacity resulting from LINC00963 knockdown (). The results presented above imply that inhibition of miR-204-3p attenuates the growth and progression effects hindered by LINC00963 underexpression in OS cells.
LINC00963 can affect fibronectin-1 expression by suppressing miR-204-3p in OS
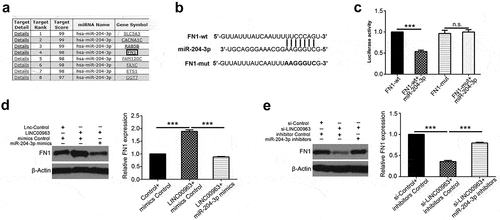
Previous study has demonstrated that miR-204-3p can suppress invasion by directly inhibiting Fibronectin-1 (FN1) in clear cell renal cell carcinoma.Citation21 Besides, FN1 was reported to promote proliferation and drug resistance in osteosarcoma.Citation22 Combined with predicted targets in miRDB (), we hypothesized that LINC00963 may function in OS by altering miR-204-3p/FN1 axis. To verify how miR-204-3p modulate FN1, luciferase reporter assays were conducted. In , predicted sequence of FN1 wild-type 3ʹ-UTR was predicted to bind with miR-204-3p and its corresponding FN1 mutant 3ʹ-UTR. The results showed that luciferase activity of FN1 wild-type 3ʹ-UTR was reduced while that of its mutant 3ʹ-UTR was not affected (). We can conclude that miR-204-3p can bind directly with FN1 3ʹ-UTR, in favor of results from the previous study. Also, we measured the FN1 RNA and protein expression after miR-204-3p mimics or inhibitors or corresponding control in OS cells. The data showed that miR-204-3p can suppress FN1 mRNA and protein expression (Supplementary Figure 2A&B). Moreover, western blot assays were to carry out to explore whether LINC00963 can affect the expression of FN1 (). The results showed that overexpression of LINC00963 elevated the expression of FN1 and inhibition of LINC00963 decreased the expression of FN1. However, FN1 elevation or reduction resulting from LINC00963 can be attenuated by miR-204-3p overexpression or underexpression. All results imply that LINC00963/miR-204-3p/FN1 axis may contribute to proliferation and progression of osteosarcoma.
Figure 5. LINC00963 increases FN1 expression by suppressing miR-204-3p in OS. (a) Potential binding targets were predicted on miRDB. (b) Predicted sequence of FN1 wild-type 3ʹ-UTR was predicted to bind with miR-204-3p and its corresponding FN1 mutant 3ʹ-UTR. (c) Luciferase activity assay showed that FN1 can bind directly with miR-204-3p. (d) FN1 protein level was examined after LINC00963 or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells and after si-LINC00963 or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).
Discussion
Once LncRNAs were regarded as junk RNA or transcriptional noise, increasing evidence has shown that LncRNAs contributed statistically to a great many biological activities.Citation6,Citation8 In cancer, recently many studies have revealed that an important role can be played by LncRNAs in tumor initiation and development including osteosarcoma.Citation6 Some LncRNA studies on osteosarcoma scratched the tip of icberg.Citation23,Citation24 Although thousands of long non-coding RNA have been identified and verified, the knowledge of its complicated functions is still limited.
In our present study, we verified that LINC00963 was aberrantly highly expressed in human osteosarcoma tissues in a cohort of patients in comparison with normal tissues. Besides, the relationship between LINC00963 expression level and multiple clinicopathological characteristics was also investigated. The results showed that the high expression level of LINC00963 was significantly related to Enneking Classification, recurrence and lung metastasis. The Kaplan-Meier survival analysis was performed to show that high LINC00963 expression was correlated with shorter OS and DFS. Consequently, LINC00963 can be a candidate prognostic marker for osteosarcoma. LINC00963 was investigated to be significantly upregulated in osteosarcoma cell lines compared to the normal cell line. Gain-of and loss-of-function approaches were chosen to study how LINC00963 can affect OS cells’ proliferation and progression. The data revealed that LINC00963 promoted OS cell proliferation, migration, and invasion.
Although the mechanism of LncRNAs remains unclear, many studies have revealed some potential biological mechanism. Competing endogenous RNAs (CeRNAs) can bind miRNAs directly and influence miRNAs and its target RNA expression, and lead to the change of cell biological process and activity.Citation25–Citation30 CeRNA is one of those mechanisms, which has been reported frequently in osteosarcoma study.Citation17,Citation24 Given the fact that LncRNAs can interact with miRNAs as CeRNAs, we sought for potential miRNAs interacting with LINC00963. Therefore, miR-204-3p was predicted as a binding target of LINC00963 by bioinformatics tools. We verified that miR-204-3p directly binds LINC00963 through Luciferase-reporter assay. qPCR results indicated that miR-204-3p expression level increased or decreased upon LINC00963 underexpression or overexpression. Furthermore, we found that LINC00963 transcript level was inversely related to miR-204-3p in OS patients. By applying genetic rescue experiment, we confirmed that overexpression or underexpression of miR-204-3p can reverse the effects of LINC00963 upregulation or downregulation in OS cells.
Fibronectin-1 (FN1) has been reported to be as a key role in cancer development.Citation31–Citation35 Fibronectin-1 has been studied and verified to bind miR-204-3p directly in a previous study,Citation21 the same result as we predicted on miRDB. Our data suggested that miR-204-3p can suppress FN1 by directly binding FN1. Consequently, western blot was performed to explore whether LINC00963 can influence FN1 expression by binding miR-204-3p. The data showed that overexpression or underexpression of LINC00963 can increase or decrease FN1 expression through binding miR-204-3p. LINC00963 may exert its influences by regulating miR-204-3p/FN1 axis in OS. However, the mechanism of LINC00963 downregulation in osteosarcoma still needs to be further elucidated.
In conclusion, our research revealed that LINC00963/miR-204-3p/FN1 can play an important role in proliferation and progression in osteosarcoma. All findings implied that LINC00963 could be an underlying therapeutic target for osteosarcoma treatment.
Materials and method
Tissue samples
Osteosarcoma tissues and their corresponding normal tissues were collected from 68 OS cases from 2009 to 2016 at the Second Xiangya Hospital, Central South University (Hunan, China). All samples were obtained by biopsy before patients received chemotherapy. All procedures were approved by the Ethics Committee of the Second Xiangya Hospital, Central South University (Hunan, China), and informed consent was acquired from every involved patient.
Cell lines and cell culture
MG63, U2OS, HOS, Saos-2, and hFOB cell lines were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). were cultured in the growth medium 1640 with 10% fetal calf serum and 1% penicillin-streptomycin under 5% CO2 humidified atmosphere at 37°C.
RNA extraction and qRT-PCR
RNA was isolated and acquired from osteosarcoma tissues and cells via the TRIzol reagent (Invitrogen, Carlsbad, CA) method. Total RNA was reverse-transcribed with All-in-oneTM First-Strand c-DNA Synthesis Kit or All-in-oneTM miRNA First-Strand c-DNA Synthesis Kit (GeneCopoeia, MD, USA). The quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed by the SYBR green method with All-in-oneTM qPCR Mix and All-in-oneTM miRNA Qpcr Kit (GeneCopoeia, MD, USA). The target RNA relative expression level was quantified by LightCycler 96 (Roche, IN, USA) thermocycler. β-actin was chosen as the internal control. All primer sequences and sequences for siRNA are listed in Supplementary Table 2.
Western blot
A Western blot (WB) assay was performed as our previous article described. Total protein was obtained using RIPA reagent method. Proteins were denatured with loading buffer by boiling at 100°C for 10 min. Then, proteins were separated by gel electrophoresis and transferred to nitrocellulose membrane. After blocking non-specific interaction, primary antibody and secondary antibody were incubated step by step. The blots were detected and filmed by the ChemiDoc™ XRS + System (BIO RAD, Hercules, CA). The results were quantified by normalizing to the corresponding internal control β-actin. Antibodies to FN1 were purchased from CST (Danvers, MA, USA).
Cell counting kit-8 proliferation assays
Cell Counting Kit-8 (CCK8) assays were performed to investigate the cell growth ability. OS cells were seeded in a 96-well plate and cultured under the conditions described above. Approximately 5 × 103 cells were seeded in each well. After incubation, CCK8 kit was added to each well in accordance with protocol at 24, 48, and 72 h. Accordingly, the relative optical density (OD) values were determined at a wavelength of 450 nm.
Trypan blue cell viability assays
Trypan blue assays were carried out to determine the cell viability of osteosarcoma cells. OS cells were seeded in a 24-well plate and cultured under the conditions described above. Around 5 × 104 cells were seeded in each well. After incubation in accordance with protocol at 24, 48, and 72 h, OS cells were trypsinized, centrifuged and resuspended. 0.4% trypan blue was mixed with an equivalent volume of cell suspensions. Cells are counted in the hemacytometer under a microscope.
Wound-healing assays
Wound-healing assay was conducted to investigate the cell mobility of osteosarcoma cells. OS cells were placed into six-well plates and incubated until 80%-90% coverage density. Sterile 200 μl pipette tip was used to produce the wound across the monolayer OS cells. Representative pictures were also obtained under the microscope by a 100× magnification at the various time point. Area of the wound was measured at a different time.
Transwell migration and invasion assays
For migration assays, 5 × 10s cells were placed into the upper chamber of each transwell chamber and maintained in serum-free medium. For invasion assays, the polycarbonate filter of transwell chamber was covered by a layer of reconstructed extracellular matrix. Then, the OS cells were incubated under the conditions described above for 24 h. All the cells that stuck to the lower surface of the membrane were dyed with crystal violet solution. Representative pictures were photographed, and we counted cells in different microscope fields.
Luciferase reporter assays
The wild-type and mutant 3ʹ-UTR sequences of LINC00963 and FN1 were amplified and separately subcloned into the pmiRRB-Report (RiboBio, Guangzhou, China). MG63 cells were seeded in 12-well plates. In addition, miR-204-3p mimics or mimics NC were cotransfected with wild-type or mutant 3ʹ-UTR reporter vectors into the cells above with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA). Then, luciferase activity was detected in accordance with the protocol of the dual‐luciferase reporter assay. All assays were conducted three times.
Statistics analysis
The results are shown as the mean ± SD of three separate experiments. The main statistical analyses were Student t test and one‐way ANOVA analysis of variance. P less than 0.05 was considered the standard for significance. All analyses were carried out in SPSS 19.0 (SPSS Inc, Chicago, IL) or GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA).
Compliance with ethical standards
Ethical approval The study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University, Hunan, China.
Informed consent All the study participants gave their written informed consent to participate in the study.
Acknowledgment and funding This work was supported by National Natural Science Foundation of China (No. 81302339 to Lihong Liu, 81502332 to Hui Li and 81372871 to Tao Xiao).
Conflict of interest The authors declare that they have no conflicts of interest.
Supplemental Material
Download MS Word (16.8 KB)Supplemental Material
Download MS Power Point (677.6 KB)Supplemental Material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi:10.1002/cncr.24121.
- Meyers PA, Schwartz CL, Krailo M, Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W, Gebhardt M, Goorin AM, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol. 2005;23:2004–2011. doi:10.1200/jco.2005.06.031.
- Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28:26–33. doi:10.1097/mop.0000000000000298.
- Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthop Belg. 2016;82:690–698.
- Ottaviani G, Jaffe N. The etiology of osteosarcoma. Cancer Treat Res. 2009;152:15–32. doi:10.1007/978-1-4419-0284-9_2.
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi:10.1016/j.ccell.2016.03.010.
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi:10.1016/j.cell.2013.06.009.
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi:10.1146/annurev-biochem-051410-092902.
- Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR, Liu YQ, Zhong H, Li H, Xiao T. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun. 2013;433:200–206. doi:10.1016/j.bbrc.2013.02.083.
- Liu K, Huang J, Ni J, Song D, Ding M, Wang J, Huang X, Li W. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle. 2017;16:578–587. doi:10.1080/15384101.2017.1288324.
- Cai W, Wu B, Li Z, He P, Wang B, Cai A, Zhang X. LncRNA NBR2 inhibits epithelial-mesenchymal transition by regulating Notch1 signaling in osteosarcoma cells. J Cell Biochem. 2018. doi:10.1002/jcb.27508.
- Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B, Zhou M, Li X, Xiong W, Li G, et al. The long noncoding RNA MALAT-1 is a novel biomarker in various cancers: a meta-analysis based on the GEO Database and Literature. J Cancer. 2016;7:991–1001. doi:10.7150/jca.14663.
- Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi:10.1016/j.ccell.2016.03.004.
- Di Cecilia S, Zhang F, Sancho A, Li S, Aguilo F, Sun Y, Rengasamy M, Zhang W, Del Vecchio L, Salvatore F, et al. RBM5-AS1 Is critical for self-renewal of colon cancer stem-like cells. Cancer Res. 2016;76:5615–5627. doi:10.1158/0008-5472.can-15-1824.
- Wang Y, Qian CY, Li XP, Zhang Y, He H, Wang J, Chen J, Cui JJ, Liu R, Zhou H, et al. Genome-scale long noncoding RNA expression pattern in squamous cell lung cancer. Sci Rep. 2015;5:11671. doi:10.1038/srep11671.
- Zhao W, Dong S, Duan B, Chen P, Shi L, Gao H, Qi H. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am J Transl Res. 2015;7:1295–1302.
- Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi:10.1186/s12943-018-0837-6.
- Li Z, Dou P, Liu T, He S. Application of Long Noncoding RNAs in Osteosarcoma: biomarkers and Therapeutic Targets. Cell Physiol Biochem. 2017;42:1407–1419. doi:10.1159/000479205.
- Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, Li Z, Chen B, Zhang X, Pan H, et al. MetaLnc9 Facilitates Lung Cancer Metastasis via a PGK1-Activated AKT/mTOR Pathway. Cancer Res. 2017;77:5782–5794. doi:10.1158/0008-5472.can-17-0671.
- Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–8. doi:10.1093/nar/gkv1270.
- Han Z, Zhang Y, Sun Y, Chen J, Chang C, Wang X, Yeh S. ERbeta-mediated alteration of circATP2B1 and miR-204-3p signaling promotes invasion of clear cell renal cell carcinoma. Cancer Res. 2018;78:2550–2563. doi:10.1158/0008-5472.can-17-1575.
- Kun-Peng Z, Chun-Lin Z, Xiao-Long M, Lei Z. Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J Cell Physiol. 2018. DOI:10.1002/jcp.27435
- Wang Y, Zhang R, Cheng G, Xu R, Han X. Long non-coding RNA HOXA-AS2 promotes migration and invasion by acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle. 2018;17:1637–1648. doi:10.1080/15384101.2018.1489174.
- Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33:4857–4866. doi:10.1038/onc.2013.433.
- Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5:317–333. doi:10.1002/wrna.1213.
- Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi:10.4161/21541272.2014.944014.
- Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi:10.3389/fgene.2014.00008.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi:10.1038/nature12986.
- Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi:10.1038/srep06088.
- Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479–13490. doi:10.18632/oncotarget.7266.
- Glasner A, Levi A, Enk J, Isaacson B, Viukov S, Orlanski S, Scope A, Neuman T, Enk CD, Hanna JH, et al. NKp46 Receptor-Mediated Interferon-gamma Production by Natural Killer Cells Increases Fibronectin 1 to Alter Tumor Architecture and Control Metastasis. Immunity. 2018;48(107–19.e4). doi:10.1016/j.immuni.2017.12.007.
- Lou X, Han X, Jin C, Tian W, Yu W, Ding D, et al. SOX2 targets fibronectin 1 to promote cell migration and invasion in ovarian cancer: new molecular leads for therapeutic intervention. OMICS. 2013;17:510–518. doi:10.1089/omi.2013.0058.
- Wang J, Deng L, Huang J, Cai R, Zhu X, Liu F, Wang Q, Zhang J, Zheng Y. High expression of Fibronectin 1 suppresses apoptosis through the NF-kappaB pathway and is associated with migration in nasopharyngeal carcinoma. Am J Transl Res. 2017;9:4502–4511.
- Ye Y, Zhuang J, Wang G, He S, Ni J, Xia W. MicroRNA-139 targets fibronectin 1 to inhibit papillary thyroid carcinoma progression. Oncol Lett. 2017;14:7799–7806. doi:10.3892/ol.2017.7201.
- Zhang H, Sun Z, Li Y, Fan D, Jiang H. MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomed Pharmacother. 2017;88:285–292. doi:10.1016/j.biopha.2017.01.023.
