ABSTRACT
Prostate cancer (PCa) is the second frequently newly diagnosed cancer in men. Androgen deprivation therapy has been widely used to inhibit PCa growth but eventually fails in many patients. Androgen receptor and its downstream molecules like microRNAs could be promising therapeutic targets. We aimed to investigate the involvement of miR-21 in PCa tumorigenesis. We found that miR-21 was an unfavorable factor and correlated positively with tumor grade in PCa patients from TCGA database. MiR-21 was more highly expressed in androgen-independent PCa cells than in androgen-dependent PCa cells. Overexpression of miR-21 promoted androgen-dependent and -independent PCa cell proliferation, migration, invasion, and resistance to apoptosis. Furthermore, increased miR-21 expression promoted mouse xenograft growth. We identified nine genes differentially expressed in PCa tumors and normal tissue which could be potential targets of miR-21 by bioinformatic analyses. We demonstrate that miR-21 directly targeted KLF5 and inhibited KLF5 mRNA and protein levels in PCa. STRING and functional enrichment analysis results suggest that GSK3B might be regulated by KLF5. Our findings demonstrate that miR-21 promotes the tumorigenesis of PCa cells by directly targeting KLF5. These biological effects are mediated through upregulation of GSK3B and activation of the AKT signaling pathway.
Introduction
Prostate cancer (PCa) is the second frequently newly diagnosed cancer in men around the world. Estimates predicted that as many as 1,276,106 men around the world would die because of PCa in 2018.Citation1 PCa has substantial heterogeneity, leading to variable clinical course and outcome.Citation2 It is known that androgen and androgen receptor (AR) drive PCa cell proliferation in the early stages. Thus, androgen deprivation therapy has been widely used in clinical practice to inhibit PCa growth.Citation3 Nevertheless, this therapy eventually fails in many patients, driving the transformation of androgen-dependent PCa into androgen-independent PCa, which is more aggressive, easily metastasized, and lacks effective therapy.Citation4 In androgen-independent PCa, AR signaling is aberrantly active without androgen. Therefore, AR and its downstream molecules may be promising therapeutic targets for both androgen-dependent and androgen-independent PCa.Citation5
Many molecules are involved in AR signaling, including AR co-regulators and microRNAs (miRNAs).Citation3,Citation6,Citation7 MiRNAs represent a class of small non-coding RNAs (18–25 nucleotides in length) binding to the 3′-untranslated region of the target mRNA and destabilizing mRNA or inhibiting its translation.Citation8 MiRNAs regulate multiple biological processes, especially in the pathogenesis of cancers.Citation9,Citation10 Some miRNAs may be promoted or inhibited by AR, such as miR-125b, miR-101, miR-21, and miR-34a.Citation5,Citation11,Citation12 Among these, miR-21 has been reported as an oncogene in PCa, regulating various molecular targets such as programmed cell death 4 (PDCD4), reversion-inducing-cysteine-rich protein with kazal motifs (RECK), and phosphatase and tensin homolog deleted on chromosome 10 (PTEN).Citation13 Recent studies suggested that miR-21 could be a potential biomarker or therapeutic target for PCa.Citation14,Citation15
A number of targets of miR-21 have been predicted by the microRNA database. Only a few of them were validated by experiments. Whether miR-21 can regulate the biological function of androgen-dependent and androgen-independent PCa cells through certain molecular targets remains unknown. Therefore, we aimed to investigate the biological role of miR-21 and its underlying mechanism in PCa tumorigenesis.
Results
MiR-21 promotes PCa cell proliferation and colony formation
As shown in and ), miR-21 expression was significantly greater in PCa tissue than in normal tissue (p < 0.001). The miR-21 levels increased gradually with Gleason score. High levels of miR-21 were associated with lower survival probability, suggesting that miR-21 is an unfavorable factor in PCa patients (). A similar result was obtained in PCa cells. MiR-21 was expressed to a greater extent in androgen-independent PCa cells (PC-3 and DU145) than in androgen-dependent PCa cells (VCaP and LNCaP) ().
Figure 1. MiR-21 promotes PCa cell proliferation and colony formation.
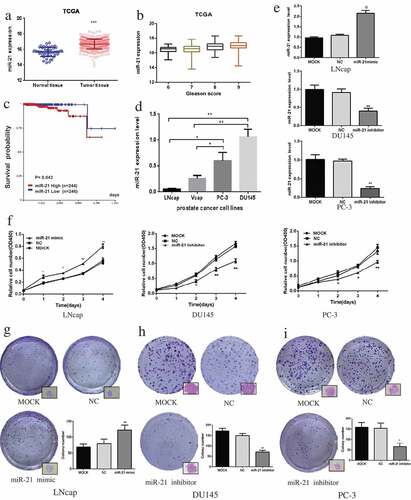
The miR-21 levels were increased in androgen-dependent LNCaP cells after transfection of miR-21 mimic (). As demonstrated in –), upregulation of miR-21 significantly promoted LNCaP cell viability and colony formation compared with the MOCK group, while downregulation of miR-21 in DU145 and PC-3 cells suppressed both cell proliferation and colony formation compared with the MOCK group (DU145, p < 0.01; PC-3, p < 0.05) (, , ).
MiR-21 inhibits PCa cell apoptosis
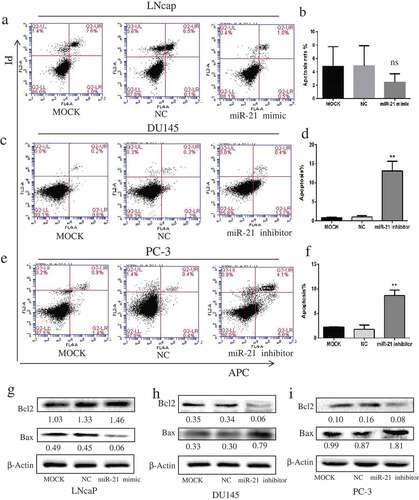
Upregulation of miR-21 via transfection with miR-21 mimic LNCaP moderately decreased the apoptosis rate of LNCaP cells (–), whereas downregulation of miR-21 markedly induced apoptosis of the androgen-independent cells (DU145, from 0.8% to 13.0%; PC3, from 2.2% to 8.7%) compared with the corresponding MOCK group (–), p < 0.01). The changes in proapoptotic (Bax) and anti-apoptotic (Bcl-2) protein levels induced by miR-21 were in line with the apoptosis results (–)). When the apoptotic rate was reduced in LNCaP cells, Bax protein was distinctly downregulated and Bcl-2 protein was slightly upregulated compared with the MOCK group (). However, when the apoptotic rate was increased through downregulation of miR-21, the Bax protein level was increased and the Bcl-2 protein level was decreased compared with the corresponding MOCK group (–)).
Figure 2. MiR-21 inhibits PCa cell apoptosis.
MiR-21 promotes PCa cell migration and invasion
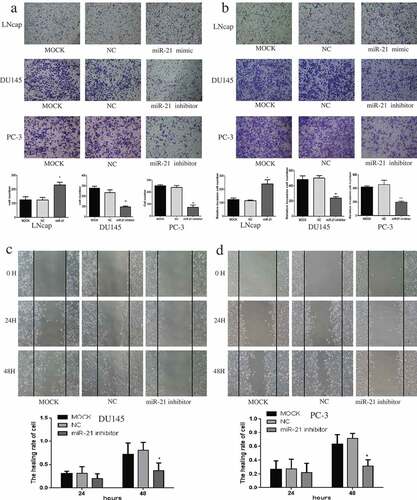
Overexpression of miR-21 in LNCaP cells, resulted in greater migration and invasion than in the MOCK group (p < 0.05), while inhibition of miR-21 in DU145 and PC-3 cells led to less migration and invasion (–); DU145 p < 0.05; PC-3 migration p < 0.01, invasion p < 0.001). Similar findings were observed in the wound healing assay, indicating that the healing ability of PCa cells was reduced by miR-21 downregulation at 48 h (–)).
Figure 3. MiR-21 promotes PCa cell migration and invasion.
MiR-21 directly targets KLF5 in PCa
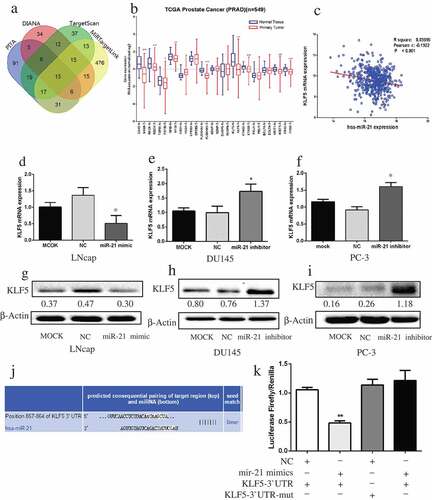
The targets of miR-21 were predicted by four prediction databases (PITA, DIANA, TargetScan, and MiRTargetLink), which together identified 15 common target genes (). Among the 15 genes, only nine were significantly downregulated in PCa compared with normal tissue (). Analysis of TCGA PCa data revealed a reverse correlation between KLF5 mRNA and miR-21 expression (). The expression of KLF5 mRNA and protein was lower in miR-21–overexpressing LNCaP cells than in MOCK or untreated controls (, )). In contrast, downregulation of miR-21 in DU145 and PC-3 cells increased KLF5 mRNA and protein expression (, , , ). The TargetScan database predicted the pairing target region of KLF5 and miR-21 (). Luciferase reporter assay confirmed the prediction that miR-21 could directly bind to the 3′-untranslated region of KLF5 ().
Figure 4. MiR-21 directly targets KLF5 in PCa.
MiR-21 promotes GSK3B and the AKT signaling pathway
Using STRING, we identified 26 KLF5-related proteins (). Functional enrichment analysis suggested that GSK3B and TP53 are co-involved in multiple pathways (KEGG Pathways: Pathways in cancer, prostate cancer) and biological processes (negative regulation of apoptotic process, regulation of growth). These two proteins, along with KLF5, are mainly located in the nucleoplasm (, Figure S1). Furthermore, our analysis of TCGA datasets revealed that GSK3B mRNA expression was noticeably increased in PCa patients with a high Gleason score, while KLF5 mRNA declined with a higher Gleason score, while TP53 mRNA showed no significant correlation with Gleason score (). KLF5 mRNA was negatively correlated with GSK3B mRNA in those patients (, p < 0.001). GSK3B was an unfavorable factor in PCa patients, in that patients with a high level of GSK3B showed a tendency for lower survival probability, and the survival rate of the high GSK3B and low GSK3B groups diverged and was fairly different after 5 years ().
Figure 5. MiR-21 promotes GSK3B and the AKT signaling pathway.
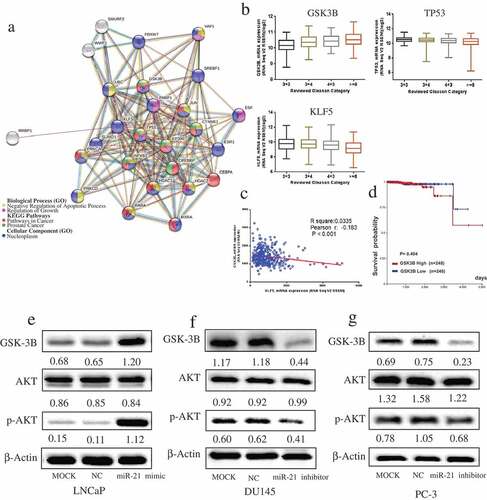
As illustrated in –), overexpression of miR-21 increased GSK3B expression and phosphorylation of AKT (Ser473), whereas downregulation of miR-21 decreased the expression of these proteins. Nevertheless, AKT phosphorylation on Thr308 was not affected by miR-21 (Figure S2).
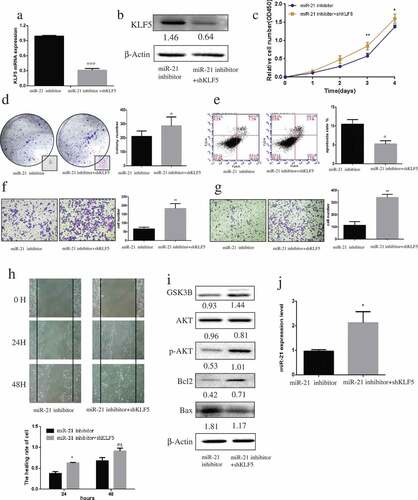
Downregulation of KLF5 promotes PCa tumorigenesis
KLF5 knockdown with shRNA suppressed KLF5 mRNA and protein expression levels in miR-21–downregulated DU145 cells (–)). Downregulation of KLF5 expression promoted DU145 cell proliferation (), colony formation (), migration (, ), and invasion (), and inhibited their apoptosis (p < 0.05; ) compared with cells treated with miR-21 inhibitor. After transfection of KLF5 shRNA, the levels of GSK3B, pS473-AKT, and Bcl-2 proteins were significantly increased, while Bax protein level was decreased (). Moreover, inhibition of KLF5 expression notably upregulated miR-21 expression (p < 0.05) compared with cells treated with the miR-21 inhibitor alone ().
Figure 6. Downregulation of KLF5 promotes PCa carcinogenesis.
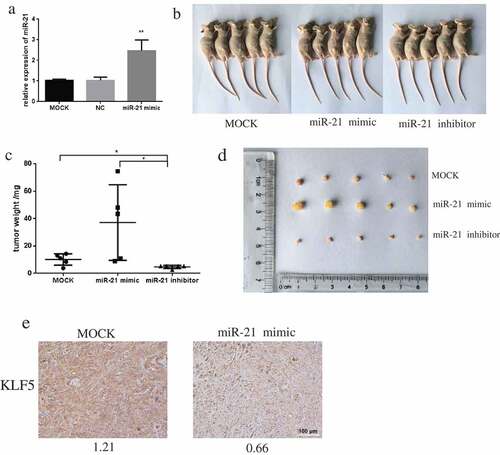
MiR-21 promotes tumor growth in xenograft mice
The mouse tumor xenograft model was successfully established by injection of mice with DU145 cells transfected with MOCK, miR-21 mimic, or miR-21 inhibitor (). Growth of miR-21–overexpressing tumors was significantly greater than that of the miR-21 inhibition or the MOCK tumors (–)). Immunohistochemistry revealed that expression of KLF5 was reduced in mouse tumors overexpressing miR-21 ().
Figure 7. miR-21 promotes PCa tumor growth in xenograft mice.
Discussion
MiR-21 has been shown to be upregulated in many types of cancer, including PCa.Citation9,Citation16,Citation17 Finding new miR-21 target genes may help us better understand the role of miR-21 in tumorigenesis and offer a potential therapeutic target for PCa. Previous work on miR-21 in PCa focused mainly on genes that have been validated in other cancers.Citation18,Citation19 Little is known about novel miR-21 target genes whose expression is significantly altered in PCa.
In this study, we demonstrated that miR-21 is upregulated in PCa and is associated with higher pathologic grade (Gleason score) and that high levels of miR-21 are associated with lower survival rate in PCa patients. The association between miR-21 and Gleason score and survival rate suggest that miR-21 might be a useful biomarker to predict the outcome of PCa. Our results are in line with previous studies whose results also suggest that miR-21 could serve as a prognostic marker for PCa.Citation20
The results of our in vitro experiments suggest that miR-21 promotes PCa cell proliferation, migration, invasion, and resistance to apoptosis. The promoting effect of miR-21 on PCa growth was supported by the in vivo experiment and was consistent with reported findings.Citation21 It reinforces the idea that miR-21 induces PCa cell growth not only in androgen-independent cells but also in androgen-dependent cells. It has been proven that miR-21 can induce cancer cell metastasis and resistance to apoptosis,Citation22 and our results support this. Increased apoptosis of cancer cells is induced by the increased pro-apoptotic proteins (Bax, Bak) and the decreased anti-apoptotic proteins (Bcl-2,Bcl-w,Bcl-XL,Bfl-1,Mcl-1), Bcl-2 and Bax are well demonstrated by many studies.Citation23–Citation25 In our study, the increased expression of Bax and the decreased expression of Bcl-2 were in line with the changes in apoptosis induced by downregulation of miR-21in the corresponding cancer cells.
A number of targets of miR-21 have been reported in PCa including PTEN.Citation26–Citation28 In this study, we identified nine genes differentially expressed in PCa tumors and normal tissue which could be potential targets of miR-21. Among these candidates, four genes (SOX5, RECK, TGFBI, and NFIB) have been reported previously as targets of miR-21.Citation29–Citation32 The relationships of the other five genes (PLEKHA1, KLF5, STAG2, KRIT1, and FRS2) with miR-21 are still unknown; only FRS2 and KLF5 have been reported to have a role in regulation of PCa biologic function.Citation33,Citation34 KLF5 is a transcription factor located in the cell nucleus that can regulate a wide range of gene expression, affecting various cellular functions in cancer.Citation35 KLF5 was reported to play contrary roles in cancer development, as not only a suppressor but also a promoter.Citation36,Citation37 Our results show that KLF5 is a target of miR-21, functioning as a tumor suppressor in PCa.
Supporting this conclusion, first, is the negative correlation between miR-21 and KLF5 expression in PCa patients. Overexpression of miR-21 downregulated KLF5 mRNA and protein levels. These findings are supported and confirmed by the results of the TargetScan database analysis and luciferase reporter assay, which show that miR-21 directly targets KLF5. Several studies have shown that KLF5 can be regulated by long non-coding RNAs, miRNAs, and drugs.Citation38–Citation40 Our results provide the first known evidence that miR-21 can directly target KLF5 in PCa. Knocking down KLF5 in miR-21–downregulated cells promoted cancer cell proliferation, migration, invasion, and resistance to apoptosis. These findings are consistent with previous studies demonstrating that deletion of KLF5 promoted cancer growth and led to molecular alterations in cells.Citation35,Citation41 Most importantly, we found that KLF5 and miR-21 form a feedback loop, in that reduced KLF5 expression can upregulate miR-21 expression. This can partly explain why androgen-independent PCa cells express high levels of miR-21.
We also found that KLF5 can regulate GSK3B, which is considered to be one of the key protein molecules involved in PCa progression. It was reported that GSK3B can reduce the transcription activity of AR.Citation42 GSK3B is activated in cancer cells that have highly metastatic properties and are resistant to chemotherapy.Citation43,Citation44 These findings indicate that GSK3B is an unfavorable factor in cancers, which is in line with our findings. In our study, GSK3B had a positive correlation with tumor grade in PCa patients. Upregulation of miR-21 promoted GSK3B expression and activated the AKT signaling pathway, inducing tumorigenesis. As previously reported, GSK3B can promote AKT phosphorylation and tumorigenesis.Citation42,Citation45 Most notably, our findings demonstrate that GSK3B plays an important role in mediating miR-21 activation of the AKT signaling pathway in PCa. GSK3B could be a new prognostic marker. However, further validation in a larger cohort of cancer patients is needed.
In summary, our study using bioinformatic approach identify new targets of miR-21 and verify it`s biological functions in PCa cell lines (androgen-dependent and androgen-independent), xenografts mice and patients. The results provide convincing evidence that miR-21 could directly target KLF5 and promote PCa tumorigenesis. In particular, the feedback loop between miR-21 and KLF5 might account for the aggressive progress of androgen-independent PCa. Our findings provide insight into the mechanism through which miR-21 and KLF5 are involved in androgen-dependent and androgen-independent PCa, suggesting that MiR-21 and KLF5 play a crucial role in PCa progression. However, there are some limitations in our present study. Firstly, apart from KLF5, other candidate target genes of miR-21 we discovered remain to be validated. Secondly, the interaction between KLF5 and AKT signaling downstream molecules remain to be investigated. Thirdly, the preliminary findings need to be demonstrated in other cell lines, animal models, and most importantly in human subjects. Future work focusing on these molecules could eventually lead to better understanding of the role of miR-21 in the tumorigenesis of PCa and potential therapeutic approach.
Conclusions
In this study, we demonstrate that miR-21 directly targets KLF5 and promotes the proliferation, migration, invasion, and resistance to apoptosis of androgen-dependent and -independent PCa cells. The underlying mechanism is mediated through upregulation of GSK3B and activation of the AKT signaling pathway. These findings offer a new, deeper understanding of the miR-21’s role and in particular, miR-21 related therapeutic targets in PCa.
Materials and methods
Cell culture and plasmid transfection
Human PCa cells, including androgen-dependent VCaP and LNCaP and androgen-independent DU145 and PC-3, were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. LNCaP and PC-3 cells were maintained in F12 medium (Gibco, Carlsbad, CA) containing 10% fetal bovine serum (FBS) (Gibco), while DU145 and VCaP cells were maintained in DMEM (Gibco) containing 10% FBS. All cell lines were cultured at 37°C in a humidified 5% CO2 atmosphere.
To establish stable transfectants, cells were treated according to our published study protocol.Citation46 The pcDNA3.1-hsa-miR-21 inhibitor sponges (miR-21 inhibitor), pGCMV-has-miR-21 (miR-21 mimic), and pGPH1-KLF5 (shKLF5) vectors, along with corresponding negative control (NC) vectors, were synthesized by GenePharma (Shanghai, China). Untransfected cells (MOCK) served as controls.
Bioinformatic analyses
The Cancer Genome Atlas (TCGA) prostate cancer datasets (PRAD) containing miRNA mature strand expression RNAseq and gene expression RNAseq were downloaded from UCSC Xena (https://xenabrowser.net). The mRNA expression level and relative clinical data (Gleason score, sample type, sample ID, survival days) were matched and analyzed in SPSS17.0 (Chicago, IL). The survival rates of different groups of PCa patients were displayed in Kaplan-Meier survival curves. MiR-21 targets were predicted by TargetScan,Citation47 PITA,Citation48 DIANA,Citation49 and MiRTargetLink.Citation50 Analyses of the protein-protein interaction network and functional enrichment of Krüppel-like factor 5 (KLF5)–related proteins were performed in STRING (https://string-db.org).Citation51 Correlations of glycogen synthase kinase 3 beta (GSK3B), tumor protein P53 (TP53), and KLF5 mRNA expression with the reviewed Gleason category in prostate adenocarcinoma were obtained from cBioPortal (http://www.cbioportal.org).Citation52
Cell proliferation assay
Cell proliferation was assessed by using the Cell Counting Kit (CCK-8; Dojindo, Kumamoto, Japan). PCa cells were seeded into 96-well microplates (LNCaP 5 × 103 cells, DU145 3 × 103 cells, PC-3 2 × 103 cells per well) and incubated for various periods of time (1–4 days). After this incubation, 10 μL of CCK-8 reagent was added to each well and the plates incubated at 37ºC for 1 h. Absorbance was measured at 450 nm by a Multimode Microplate Reader-Varioskan Flash (Thermo Scientific, Waltham, MA).
Colony formation assay
PCa cells were seeded into 6-well plates (LNCaP 3 × 103 cells, DU145 1 × 103 cells, PC-3 2 × 103 cells) and incubated for 2 weeks, during which the medium was changed every 3 days. The plates were rinsed with 1× phosphate-buffered saline solution (PBS). Cells were fixed in 4% paraformaldehyde and stained with crystal violet for 15 min. The plates were rinsed with PBS three times to remove the residual crystal violet solution, and only the clearly visible colonies (diameter >50 μm) were counted. The colonies were photographed with an Eclipse TS100 microscope (Nikon, Tokyo, Japan) at ×100 magnification.
Apoptosis assay
To quantify cell apoptosis, PCa cells (1 × 105 PCa cells per well) were seeded into 6-well plates and the cells were incubated in serum-free medium for 24 h to induce apoptosis. Cells were harvested and collected by centrifugation and stained with Annexin V-allophycocyanin and propidium iodide according to the protocol of the apoptosis detection kit (KeyGEN, Nanjing, China). Cells (2 × 104 per well) were then incubated at room temperature for 15 min in the dark. The apoptosis rates of cells were quantified by a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA).
Wound healing assay
A wound healing assay was used to measure cell migration. DU145 and PC-3 cells were plated in 12-well plates (DU145 1.5 × 105 cells, PC-3 2 × 105 cells). When cells reached 80% confluence, a wound line was scratched into the surface of each well’s contents with a 200 μL pipette tip. Cells were gently rinsed twice with PBS and incubated in a growth medium containing 2% FBS. Beginning at 72 h of incubation, the wound width was measured every 24 h at ×100 magnification under a Nikon Eclipse TS100 microscope. The wound closure rate was calculated from these measurements, as follows: percentage of wound closure = 1 -(widtht/width0) × 100%.
Cell migration and invasion assays
PCa cells were washed twice in serum-free culture medium and seeded (LNCaP 5 × 104 cells, DU145 2 × 104 cells, PC-3 2 × 104 cells) into transwell chambers with an 8-μm pore polycarbonate membrane (Corning, NY). Medium containing 20% FBS in the lower chamber served as a chemoattractant. After incubation at 37°C for 48 h, cells in the upper chamber were removed using cotton swabs. Cells that penetrated the membrane were fixed in 4% paraformaldehyde and stained with crystal violet for 15 min. The cells that migrated through the membrane were counted at a × 100 magnification under the Nikon Eclipse TS100 microscope. The invasion assay was almost identical to the migration assay except that, in the invasion assay, the transwell chamber was pre-coated with 10% Matrigel (BD Biosciences).
Luciferase reporter assay
LNCaP cells (NC, miR-21 mimic) were transfected with GP-miRGLO-KLF5-wt or GP-miRGLO-KLF5-mut vector (GenePharma). Luciferase was measured according to the instructions of the Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI). Briefly, cells were subjected to lysis in passive lysis buffer, and the LAR II and Stop&Glo Reagents were added to the lysates. The firefly and Renilla luciferase activities were measured by the Multimode Microplate Reader-Varioskan Flash. The sequences that were inserted into the vectors were as follows: 5‘- TACAAAAGTTCAACCTCTTACAATAAGCTAAACGCAATG TCATTT-3‘ (KLF5-wt) and 5‘-TACAAAAGTTCAACCTCTTACATATTCGAAAAC GCAATGTCATTT TTA AA-3‘ (KLF5-mut).
RNA extraction and qRT-PCR
Total RNA was extracted by TRIzol (Invitrogen, Carlsbad, CA) following the manufacturer’s protocols. For miRNA reverse-transcription, 500 ng of total RNA was reverse-transcribed to cDNA with miRNA-specific reverse-transcription primers (RiboBio, Guangzhou, China) by using a PrimeScript RT reagent kit (Takara, Shiga, Japan). Gene expression was quantified by a SYBR Green PCR Kit (QIAGEN, Hilden, Germany) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The relative miRNA expression levels were normalized to U6 expression. Relative fold-change of miRNA expression was calculated using the 2−ΔΔCt method. mRNA was quantified in accordance with our previously published study’s protocol.Citation46 Primers for KLF5 were 5‘-ACACCAGACCGCAGCTCCA-3‘ (forward) and 5‘-TCCATTGCTGCTGTCTGATTTGTAG-3‘ (reverse), and primers for GAPDH were 5‘-GCCAGTGGACTCCACGAC-3‘(forward) and 5‘-CAACTACATGGTTTA CATGTTC-3‘ (reverse). The experiment was repeated at least three times.
Western blot analysis
PCa cells were harvested and subjected to lysis in the presence of a protease inhibitor cocktail. The supernatants were collected after centrifugation at 14,000 rpm for 15 min at 4°C. The protein concentration of each was measured by the bicinchoninic acid protein assay kit (Beyotime, Shanghai, China). An aliquot of 20 μg of denatured protein from each sample was applied to 10% sodium dodecyl sulfate-polyacrylamide gel for electrophoresis and transferred onto nitrocellulose membranes. Membranes were blocked with 5% skim milk at ambient temperature for 1 h and incubated with the following primary antibodies: KLF5 (R&D Systems, Minneapolis, MN), GSK3B, AKT, p-AKT, Bcl2, Bax, and β-actin (all, Abcam, Cambridge, UK) at 4°C overnight. They then were incubated with horseradish peroxidase–conjugated secondary antibody (1:1000 dilution; MultiSciences, Hangzhou, China) at room temperature for 1 h. After being washed four times with TBST for 10 min each, blots were incubated with enhanced chemiluminescence solution for 3 min. The signals were detected and quantified by densitometry using Image Lab Software (Bio-Rad). β-actin was used as an endogenous control.
Tumor xenografts in mice
Male nude mice (4 weeks old) were purchased from Slack Laboratory Animal Company (Shanghai, China). To establish the tumor xenograft models, fifteen mice were divided into three groups and were injected subcutaneously into the right flank with DU145 cells (4 × 106 cells per mouse) transfected with MOCK, miR-21 inhibitor, or miR-21 mimic harvested from culture. Forty days after this injection, the mice were killed and tumors were dissected for further analysis. All animal procedures and experimental protocols were approved by the Laboratory Animal Ethics Committee of Wenzhou Medical University.
Immunohistochemical assay
Tumor tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm. To unmask the antigen, each section was immersed in sodium citrate buffer after deparaffinization and rehydration. The sections were then blocked with 5% BSA and incubated with the KLF5 primary antibody (3 μg/mL, Abcam) at 4°C overnight. The sections were stained using an immunohistochemical kit (Boster, Wuhan, China) and counterstained with hematoxylin. Sections were treated with 5% BSA alone as the negative control. The sections were photographed under a Nikon microscope. The mean optical density of sections (integrated optical density sum/area sum) was quantified by Image Pro Plus (Cybernetics, Bethesda, MD).
Statistical analysis
All statistical analyses were performed with Prism 6.0 (GraphPad Software, La Jolla, CA) and SPSS 17.0 (Chicago, IL) software. Data are expressed as the mean ± standard deviation (SD). Statistical significance was determined by analysis of variance (ANOVA) or two-tailed Student t-test. A log-rank test was used for Kaplan–Meier survival analysis. All experiments were performed at least three times, unless otherwise indicated. A p-value <0.05 was considered to indicate statistical significance.
Disclosure of interest
The authors confirm that there are no conflicts of interest.
Ethics approval
All animal procedures and experimental protocols were approved by the Laboratory Animal Ethics Committee of Wenzhou Medical University.
Supplemental Material
Download MS Power Point (226.1 KB)Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492.
- Cancer Genome Atlas Research N. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi:10.1016/j.cell.2015.10.025.
- Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–238. doi:10.1016/j.pharmthera.2013.07.003.
- Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi:10.1038/35094009.
- Culig Z, Santer FR. Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev. 2014;33:413–427. doi:10.1007/s10555-013-9474-0.
- Waltering KK, Porkka KP, Jalava SE, Urbanucci A, Kohonen PJ, Latonen LM, Kallioniemi OP, Jenster G, Visakorpi T. Androgen regulation of micro-RNAs in prostate cancer. Prostate. 2011;71:604–614. doi:10.1002/pros.21276.
- Whitington T, Gao P, Song W, Ross-Adams H, Lamb AD, Yang Y, Svezia I, Klevebring D, Mills IG, Karlsson R, et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat Genet. 2016;48:387–397. doi:10.1038/ng.3523.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297.
- Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9. doi:10.1016/j.jbior.2014.09.013.
- Cui SY, Wang R, Chen LB. MicroRNAs: key players of taxane resistance and their therapeutic potential in human cancers. J Cell Mol Med. 2013;17:1207–1217. doi:10.1111/jcmm.12131.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi:10.1038/nature03702.
- Nguyen HC, Xie W, Yang M, Hsieh CL, Drouin S, Lee GS, Kantoff PW. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate. 2013;73:346–354. doi:10.1002/pros.22572.
- Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S, Salvioni R, et al. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi:10.1186/1476-4598-9-254.
- Gao Y, Guo Y, Wang Z, Dai Z, Xu Y, Zhang W, Liu Z, Li S. Analysis of circulating miRNAs 21 and 375 as potential biomarkers for early diagnosis of prostate cancer. Neoplasma. 2016;63:623–628. doi:10.4149/neo_2016_417.
- Li T, Li RS, Li YH, Zhong S, Chen YY, Zhang CM, Hu -M-M, Shen Z-J. miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol. 2012;187:1466–1472. doi:10.1016/j.juro.2011.11.082.
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006; 103:2257–2261. doi:10.1073/pnas.0510565103
- Bonci D, Coppola V, Patrizii M, Addario A, Cannistraci A, Francescangeli F, Pecci R, Muto G, Collura D, Bedini R, et al. A microRNA code for prostate cancer metastasis. Oncogene. 2016;35:1180–1192. doi:10.1038/onc.2015.176.
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi:10.1053/j.gastro.2007.05.022.
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi:10.1038/onc.2008.72.
- Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu Y-P, Shen Y-J, Shi G-H, Ye D-W. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326–331. doi:10.1002/pros.21246.
- Guan Y, Wu Y, Liu Y, Ni J, Nong S. Association of microRNA-21 expression with clinicopathological characteristics and the risk of progression in advanced prostate cancer patients receiving androgen deprivation therapy. Prostate. 2016;76:986–993. doi:10.1002/pros.23187.
- Song B, Wang C, Liu J, Wang X, Lv L, Wei L, Xie L, Zheng Y, Song X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res. 2010;29:29. doi:10.1186/1756-9966-29-29.
- Yan X, Li W, Yang L, Dong W, Chen W, Mao Y, Xu P, Li D, Yuan H, Li Y-H. MiR-135a protects vascular endothelial cells against ventilator-induced lung injury by inhibiting PHLPP2 to activate PI3K/Akt pathway. Cell Physiol Biochem. 2018;48:1245–1258. doi:10.1159/000492010.
- Yang Z, Chen JS, Wen JK, Gao HT, Zheng B, Qu CB, Liu K-L, Zhang M-L, Gu J-F, Li J-D, et al. Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. J Exp Clin Cancer Res. 2017;36:178. doi:10.1186/s13046-017-0649-3.
- Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20:175–193. doi:10.1038/s41580-018-0089-8.
- Mishra S, Deng JJ, Gowda PS, Rao MK, Lin CL, Chen CL, Huang T, Sun L-Z. Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene. 2014;33:4097–4106. doi:10.1038/onc.2013.374.
- Mishra S, Lin CL, Huang TH, Bouamar H, Sun LZ. MicroRNA-21 inhibits p57Kip2 expression in prostate cancer. Mol Cancer. 2014;13:212. doi:10.1186/1476-4598-13-212.
- Yang Y, Guo JX, Shao ZQ. miR-21 targets and inhibits tumor suppressor gene PTEN to promote prostate cancer cell proliferation and invasion: an experimental study. Asian Pac J Trop Med. 2017;10:87–91. doi:10.1016/j.apjtm.2016.09.011.
- Zhao W, Dong Y, Wu C, Ma Y, Jin Y, Ji Y. MiR-21 overexpression improves osteoporosis by targeting RECK. Mol Cell Biochem. 2015;405:125–133. doi:10.1007/s11010-015-2404-4.
- Choi SI, Jin JY, Maeng YS, Kim TI, Kim EK. TGF-beta regulates TGFBIp expression in corneal fibroblasts via miR-21, miR-181a, and Smad signaling. Biochem Biophys Res Commun. 2016;472:150–155. doi:10.1016/j.bbrc.2016.02.086.
- Ho CY, Bar E, Giannini C, Marchionni L, Karajannis MA, Zagzag D, Gutmann DH, Eberhart CG, Rodriguez FJ. MicroRNA profiling in pediatric pilocytic astrocytoma reveals biologically relevant targets, including PBX3, NFIB, and METAP2. Neuro-oncol. 2013;15:69–82. doi:10.1093/neuonc/nos269.
- Wang P, Zhao Y, Fan R, Chen T, Dong C. MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting Sox5 in mouse skin melanocytes. Int J Mol Sci. 2016;17:959. doi:10.3390/ijms17070959.
- Liu J, You P, Chen G, Fu X, Zeng X, Wang C, Huang Y, An L, Wan X, Navone N, et al. Hyperactivated FRS2alpha-mediated signaling in prostate cancer cells promotes tumor angiogenesis and predicts poor clinical outcome of patients. Oncogene. 2016;35:1750–1759. doi:10.1038/onc.2015.239.
- Nakajima Y, Osakabe A, Waku T, Suzuki T, Akaogi K, Fujimura T, Homma Y, Inoue S, Yanagisawa J. Estrogen exhibits a biphasic effect on prostate tumor growth through the estrogen receptor beta-KLF5 pathway. Mol Cell Biol. 2016;36:144–156. doi:10.1128/MCB.00625-15.
- Gao Y, Ding Y, Chen H, Chen H, Zhou J. Targeting Kruppel-like factor 5 (KLF5) for cancer therapy. Curr Top Med Chem. 2015;15:699–713.
- Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi:10.1002/pros.10205.
- Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong XY, Bao Y, Zhou Z, Cheng X, Simons JW, et al. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346–1355. doi:10.1002/ijc.21533.
- Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W, Feng H. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37:4723–4734. doi:10.1038/s41388-018-0310-4.
- Jiang Z, Zhang Y, Cao R, Li L, Zhong K, Chen Q, Xiao J. miR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res. 2017;25:1081–1087. doi:10.3727/096504016X14831120463349.
- Shi P, Liu W, Tala WH, Li F, Zhang H, Zhang H, Wu Y, Kong Y, Zhou Z, Wang C, et al. Metformin suppresses triple-negative breast cancer stem cells by targeting KLF5 for degradation. Cell Discovery. 2017;3:17010. doi:10.1038/celldisc.2017.10.
- Xing C, Ci X, Sun X, Fu X, Zhang Z, Dong EN, Hao -Z-Z, Dong J-T. Klf5 deletion promotes Pten deletion-initiated luminal-type mouse prostate tumors through multiple oncogenic signaling pathways. Neoplasia. 2014;16:883–899. doi:10.1016/j.neo.2014.09.006.
- Darrington RS, Campa VM, Walker MM, Bengoa-Vergniory N, Gorrono-Etxebarria I, Uysal-Onganer P, Kawano Y, Waxman J, Kypta RM. Distinct expression and activity of GSK-3alpha and GSK-3beta in prostate cancer. Int J Cancer. 2012;131:E872–83. doi:10.1002/ijc.27620.
- Domoto T, Pyko IV, Furuta T, Miyashita K, Uehara M, Shimasaki T, Nakada M, Minamoto T. Glycogen synthase kinase-3beta is a pivotal mediator of cancer invasion and resistance to therapy. Cancer Sci. 2016;107:1363–1372. doi:10.1111/cas.13028.
- Chikano Y, Domoto T, Furuta T, Sabit H, Kitano-Tamura A, Pyko IV, Takino T, Sai Y, Hayashi Y, Sato H, et al. Glycogen synthase kinase 3beta sustains invasion of glioblastoma via the focal adhesion kinase, Rac1, and c-Jun N-terminal kinase-mediated pathway. Mol Cancer Ther. 2015;14:564–574. doi:10.1158/1535-7163.MCT-14-0479.
- Zhou A, Lin K, Zhang S, Chen Y, Zhang N, Xue J, Wang Z, Aldape KD, Xie K, Woodgett JR, et al. Nuclear GSK3beta promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat Cell Biol. 2016;18:954–966. doi:10.1038/ncb3396.
- Li C, Zhao L, Chen Y, He T, Chen X, Mao J, Li C, Lyu J, Meng QH. MicroRNA-21 promotes proliferation, migration, and invasion of colorectal cancer, and tumor growth associated with down-regulation of sec23a expression. BMC Cancer. 2016;16:605. doi:10.1186/s12885-016-2628-z.
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 4. doi:10.7554/eLife.06416.
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi:10.1038/ng2135.
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–73. doi:10.1093/nar/gkt393.
- Hamberg M, Backes C, Fehlmann T, Hart M, Meder B, Meese E, Keller A. MiRTargetLink–miRNAs, Genes and Interaction Networks. Int J Mol Sci. 2016;17:564. doi:10.3390/ijms17040564.
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D8. doi:10.1093/nar/gkw937.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi:10.1158/2159-8290.CD-12-0095.
