ABSTRACT
Cervical cancer, as the deadliest gynecological tumor with high risk of incidence, manifests aberrantly expressed lncRNAs in the malignant cellular processes. Long intergenic non-protein coding RNA 1133 (LINC01133) has been acknowledged to actively participate in aggressive tumor phenotypes. Our study focused on the identification of the function and corresponding mechanism of the novel molecule, LINC01133 in cervical cancer. LINC01133 expression profile was validated by digging The Cancer Genome Atlas (TCGA) database and qRT-PCR analysis. A considerably up-regulated expression of LINC01133 was unveiled. The results of CCK-8, trypan blue exclusion, EdU and transwell migration assays manifested the facilitating property of LINC01133 in cervical cancer. The epithelial–mesenchymal transition (EMT) was also exacerbated by LINCO1133. Apoptotic rate of cervical cancer cells was promoted after silencing LINCO1133. Mechanically, LINC01133 functioning as a ceRNA targeted miR-4784 to augment AHDC1 expression. Finally, LINCO1133/miR-4784 aggravated the malignant growth and aggressiveness and EMT of cervical cancer in an AHDC1-dependant way.
KEYWORDS:
Introduction
The prevailing gynecological malignancy among females was extensively acknowledged to be cervical cancer (CC), which brings about a large proportion of mortalities in developing countries and the global incidence is still climbing.Citation1,Citation2 Worse still, the persistent exposure to diverse oncogenic substances, such as herpes simplex virus type 2 (HSV-2), cervical thymus (CT), most importantly the human papillomavirus (HPV), complicates the pathogenesis of CC.Citation3–Citation5 CC turns into a rising concern worldwide with a depressing 5-year survival rate in spite of the current available therapeutic methods such as chemical treatment, radiotherapy and surgery operation.Citation6,Citation7 Therefore, effective identification of genetic diagnostics or prognosis prediction strategies by exploring potential mechanism may offer new opportunities to alleviate the physical pain or mental stress of CC patients.
As introduced, approximately 98% of genome coding no protein is composed by long noncoding RNAs (lncRNA), which are defined to be at least 200 bases long and transcribed by RNA polymerase II (RNA pol II) and has been neglected as the ‘dark matter’ and ‘noises’ of the transcriptome previously.Citation8 However, multiple biological and pathological processes such as migration, proliferation, invasion, apoptosis and epithelial–mesenchymal transition (EMT) were unveiled to critically associate with lncRNAs in tumor progression.Citation9–Citation12 For example, lncRNA OTUD6B-AS1 inhibits clear cell renal cell carcinoma proliferation, indicating poor prognosis.Citation13 LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway.Citation14 EMT was increasingly demonstrated to act as a crucial contributor to intensify the cell metastatic dissemination activities and malignant aggressiveness through empowering cancer cells migratory and invasive properties and loss of cell-cell junctions.Citation15,Citation16 For instance, lncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer.Citation17 Mechanistically, a well-established mechanism of lncRNAs-modulated oncogenesis is that lncRNAs, as a competing endogenous RNA (ceRNA) for microRNAs, sponge miRNAs to post-transcriptionally regulate downstream gene expression by binding the response element (MRE) of miRNAs.Citation18,Citation19 Additionally, this mechanism was also exemplified in CC development. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143–5p.Citation20 LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p.Citation21
Up to now, long intergenic non-protein coding RNA 1133 (LINC01133) was certificated to be valuable in colorectal cancer metastasis,Citation22 oral squamous cell carcinoma metastasis,Citation23 osteosarcoma tumorigenesis,Citation24 gastric cancer progression,Citation25 non-small cell lung cancer oncogenesisCitation26 and hepatocellular carcinoma aggravation.Citation27 Available documentation of LINC01133 about CC suggests its prognostic value in cervical squamous cell carcinoma.Citation28 Furthermore, the remarkable up-regulation of LINC01133 in CC cells was unmasked relative to the matched normal cells. However, the precise function and mechanism of LINC01133 remain to be certified in CC.
The priority of our work is to annotate the potential role of LINC01133 in CC and may provide measurable clinical utility for the CC treatment.
Materials and methods
Cell lines and cell culture
The following immortalized cervical epithelium NC104 and four cervical carcinoma cell lines Hela, ME-180, C33A and MS751 (Chinese Academy of Sciences, Shanghai, China) were needed for the study and the cultivation of them was conducted in the Roswell Park Memorial Institute-1640 (RPMI-1640) medium incorporating 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA). One percent penicillin-streptomycin (Invitrogen, CA, USA) was also added to the culture medium. Specific condition was required in an incubator (37℃,5% CO2) until the growth of cells reached to the logarithmic phase.
Cell transfection
To downregulate LINC01133, shRNA against LINC01133 (sh-LINC01133) and its negative control shRNA were stably transfected into ME-180 and MS751 cervical cancer cells. The insertion of the full-length sequence of NAV1 into pcDNA 3.1 vector from GenePharm (Shanghai, China) as the carrier was executed at room temperature lasting 2 h for upregulating NAV1. Lipofectamine2000 (Thermo Fisher Scientific, Inc.) was applied for the transfection of constructed plasmid (pcDNA3.1/NAV1) and the negative control (pcDNA3.1) into ME-180 and MS751 cells. To augment miR-4784, miR-4784 mimics and corresponding negative control (NC-mimics) were acquired from GenePharma Co. Ltd (Shanghai, China).
Real-time quantitative PCR (qRT-PCR) assay
Different lncRNAs and miRNAs expression status were interrogated by qRT-PCR assay with GAPDH or U6 as reference gene using the 2−ΔΔCt method. A Trizol reagent (Thermo Fisher Scientific) was used for collecting total RNA from Hela, ME-180, C33A and MS751 cells. The first-strand cDNA synthesis, in line with the supplier’s protocols, was accomplished via PrimeScript Reverse Transcriptase kit (Takara, Dalian, China) and AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) was required for real-time PCR implementation. The forward and reverse primers of involved genes were as follows: LINC01133-forward, TGGTGGAGAGAATGGAGG and LINC01133-reverse, AACCCAGTTCCTTAGAATCTTC. MiR-4784-forward, TGAGGAGATGCTGGGACT and miR-4784-reverse, GAACATGTCTGCGTATCTC. AHDC1-forward, GACCTTCTCTGAGTCATCCTCC and AHDC1-reverse, GCAGATGATGCCTCGTTCCAGT. GAPDH-forward, CTCCTCCTGTTCGACAGTCAGC and GAPDH-reverse, CCCAATACGACCAAATCCGTT.
Western blot analysis
Western blot analysis of the protein content of AHDC1, EMT-related proteins (E-cadherin, Vimentin, N-cadherin, Rac1, Twist) and the reference gene β-actin were executed with the aid of RIPA lysis buffer incorporating protease inhibitor for the total cellular protein extraction from CC cells and normal cervical cells. Proteins isolated from cervical cells were subsequently electrophoresed and separated on 12.5% sodium dodecyl sulfate-polyacrylamide gel, followed by transfer onto a polyvinylidene fluoride (PVDF) membrane. Membranes were blocked by 5% defatted milk lasting more than 1 h and later blotted by specific primary anti-body overnight, which finally probed by secondary antibody. Primary antibodies were referred to in the following: anti-AHDC1 (Novus Biologicals), anti-E-cadherin (Abcam), anti-Vimentin (Abcam), anti-N-cadherin (Abcam), anti-Rac1(Abcam), anti-Twist(Abcam), anti-β-actin (Abcam). A chemiluminescent detection system (Bio-Rad) was applied for detecting the antigen-antibody complex.
Trypan blue exclusion assay
Trypan blue exclusion method on the basis of the changes in cell membrane integrity was applied for cell viability detection following transfection. In brief, treated or untreated cells (5 × 104 cells) were seeded into each well of 24-well plates. Following incubation for 48 h at 37°C, cells were rinsed, trypsinized and dyed with trypan blue dye (Beyotime Biotechnology, Shanghai, China). Viable cells were counted by a cell counting chamber.
Cell counting kit-8 assay
The cell proliferation reagent CCK-8 (Roche, Basel, Switzerland) was needed for the implementation of CCK-8 assay to evaluate transfected cell proliferation. The 96-well microtiter plates (Corning, NY, USA) were seeded with treated CC cells and maintained at 37°C with 1 × 103 distributed in each well, which were added with 10 μL of CCK8 solution (Dojindo, Japan) at indicated time point. After 1.5 h of incubation, a microplate reader (BioTek Instruments, USA) probed the absorbance at 450 nm.
Ethynyl-2-deoxyuridine (EdU) assay
5-Ethynyl-2′-deoxyuridine (EdU) assays together with CCK-8 assay were designed to assess the proliferation of CC cells after transfection with the application of the EdU DNA Proliferation in vitro Detection kit (RiboBio, China) in line with the recommendations of the manufacturer. In brief, first of all, CC cells were planted in the 96-well plates. CC cells, post-transfection of 48 h, were stained by EdU and DAPI. The ratio of the EdU positive cells reflects the proliferating rate of CC cells.
Transwell migration and migration assay
Twenty-four-hour post-transfection, cells were harvested and the preparation of single-cell suspensions with a density of 5 × 104 cells/ml was also completed. The migratory and invasive potential of CC cells was tested by the employment of transwell assay in 24-well plates which possessed poly-carbonate transwell filters (Corning, USA). For migration assay, serum-free RPMI-1640 medium (100 μl) together with CC cells was planted into the upper chamber, in contrast, 500 μl of 20% FBS-containing RPMI-1640 medium was filled into the lower chamber to induce cell migration and invasion. After culturing for 24 h, collected membranes were stained by the 0.5% crystal violet (Sigma-Aldrich; Merck KGaA) for about 20 min at room temperature. For invasion assay, it followed the same steps, except the upper chamber pre-coated with Matrigel. The number of migrated or invaded cells from five random views was measured and counted by using an inverted microscope.
Caspase-3 activity assay
The effect of LINC01133, miR-4784 and AHDC1 was probed through caspase-3 activity assay with the help of a Caspase-3 Activity Assay Kit (Beyotime Institute of Biotechnology) in line with the guide of the manufacturer’s protocols. In short, cells with the density of 1 × 105 cells/well were seeded onto 48-well plates and treated cells were harvested and lysed in the cell lysis buffer followed by the determination of protein concentration using a pierce BCA protein assay kit (Thermo Scientific). Subsequently, each protein sample was supplemented with 50 μl of 2 × Reaction buffer which contained 10mM DTT, followed by 2 h of incubation with 10 μL of 4 mM DEVD-p-NA at 37°C. After incubation, a microplate reader assessed the optical density value at 405 nm.
Luciferase reporter assay
Luciferase reporter assay was employed to interrogate a regulatory relationship between miR-4784 and LINC01133 or AHDC1. LINC01133 fragment harboring the potential miR-4784 binding site and 3ʹ-UTR of AHDC1 (Biosune, Shanghai, China) were inserted into pmirGLO vector (Promega, USA) to yield PmirGLO-LINC01133-WT and PmirGLO-AHDC1-WT plasmids. Besides, site-directed mutagenesis was employed to generate mutant type of LINC01133 and AHDC1 3ʹ-UTR and PmirGLO-LINC01133-Mut and PmirGLO-AHDC1-Mut were constructed.
The wild-type or mutant-type of LINC01133 and AHDC1 reporter plasmids and controls (empty vector) were co-transfected with miR-4784 mimics or NC mimics into CC cells, respectively. A dual-luciferase reporter assay system (Promega, USA, E1910) was applied for detecting the relative luciferase activities after normalization to Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
Ago2-RIP assay was conducted in strict accordance with the recommendations of Magna RIPTM RNA kit (Millipore, Bedford, MA, USA) to confirm the co-existence of LINC01133/miR-4784/AHDC1 in the RISC complex. Complete RNA immunoprecipitation (RIP) lysis buffer lysed ME-180 and MS751 cells and subsequently, the whole cell extracts were incubated with anti-Ago2 or anti-IgG antibody conjunct with magnetic beads. After maintaining at 4°C for 6 h, magnetic beads were removed and purified RNA was quantified by qRT-PCR analysis.
Statistical analysis
SPSS Statistics 21 was employed for the analysis of data expressed as the mean ± SD. Student’s t-test was required for comparing two treatment groups with one-way analysis of variance (ANOVA) used for comparing the differences between three or more groups. The experiments in this study were ensured to be conducted at least three times. A p-value < 0.05 was stated to have statistical significance. *denotes p < .05, **denotes p < .01.
Results
High expression of LINC01133 accelerated the malignant phenotypes of CC cells
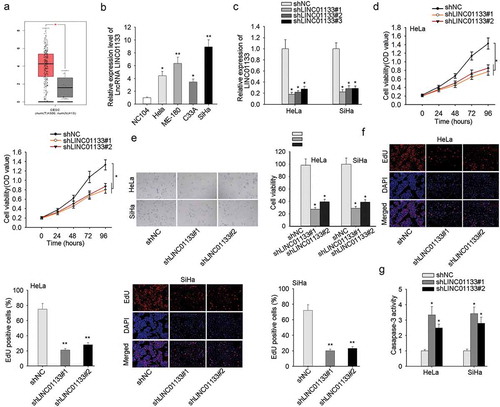
Highly expressed LINC01133 in CC tissues was detected relative to non-tumor tissues from TCGA database (). Consistently, in CC cell lines, the expression level of LINC01133 was conspicuously elevated compared with non-cancerous cell line NC104 (). To determine the function of LINC01133 CC cells, we selected ME-180 and MS751 cells for the loss-of-function assays and its knockdown efficacy was measured in . Assessment of the cellular function of down-regulating LINC01133 was conducted in ME-180 and MS751 cells and it demonstrated the repressed proliferation rate of in CCK-8 assay and EdU assay following LINC01133 depletion (). Meanwhile, LINC01133 silencing led to increased apoptosis rate of CC cells (). It collectively uncovered that LINC01133 exacerbated the malignant proliferation of CC cells.
Figure 1. High expression of LINC01133 accelerated the malignant phenotypes of CC cells. (a) Differential expression of LINC01133 in CC was obtained from The Cancer Genome Atlas (TCGA). (b) Relative to normal cervical cells, LINC01133 was markedly high-expressed in CC cells by qRT-PCR. (c) The expression of LINC01133 in cells transfected with shLINC01133#1, shLINC01133#2 and shLINC01133#3 were examined. (d-e) LINC01133 inhibition suppressed the growth of ME-180 and MS751 by CCK-8 and EdU assay. (f) LINC01133 silencing posed pro-apoptosis effect detected by caspase-3 activity assay. *P < .05, **P < .01.
LINC01133 elevated the invasive and migratory capacity of CC cells and accelerated EMT
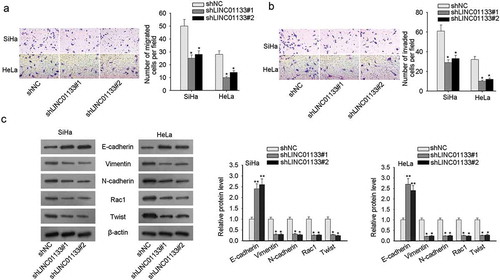
Transwell migration and invasion assays presented that LINC01133 blockade resulted in impeded cell motility and aggressiveness (-). In order to probe the impact of LINC01133 on the EMT process in CC cells, the following relevant EMT proteins such as E-cadherin (epithelial marker), Vimentin, N-cadherin (mesenchymal markers), and downstream proteins (Rac1, Twist) were evaluated by western blot. It pointed out the remarkable increase in E-cadherin and the noticeable decline in Vimentin, N-cadherin, Rac1 and Twist with LINC01133 depletion, which collectively illustrated that LINC01133 boosted EMT process in CC cells (). In summary, the facilitating effect of LINC01133 on migration, invasion, and EMT was unveiled in CC.
Figure 2. LINC01133 elevated the invasive and migratory capacity of CC cells and accelerated EMT. (a) LINC01133 down-regulation impeded migratory potential of ME-180 and MS751 by Transwell migration assay. (b) Transwell invasion assay indicated the invasion-inhibiting effect of LINC01133 suppression. (c) LINC01133 interference hindered EMT process in CC. *P < .05.
LINC01133 directly targeted miR-4784
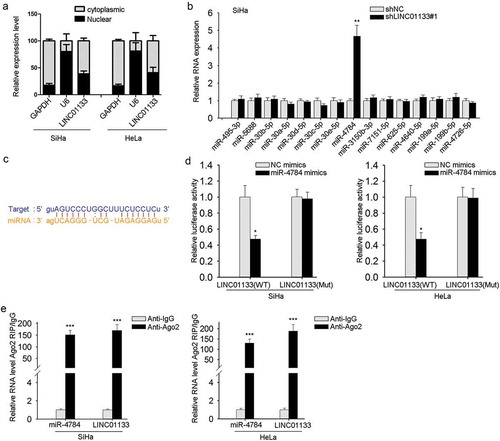
As illuminated in preliminary works, one of the well-constructed regulatory mechanisms of lncRNAs is miRNA sponging.Citation18,Citation19 First, the presence of LINC01133 was found mainly located in GC cell cytoplasm (), which is indispensable for LINC01133-regulated post-transcriptional mechanism. Next, we applied bioinformatic prediction to identify the downstream targeting miRNAs (http://starbase.sysu.edu.cn/starbase2/index.php) and 15 miRNAs were observed, among which miR-4784 was markedly up-regulated after LINC01133 inhibition as listed in and FigureS1 A. The binding sites of LINC01133 and miR-4784 predicted by starbase were also provided (). For validation of this predication, dual-luciferase reporter assay was conducted and indicated that mutations in the 3′-UTR of LINC01133 abrogated the binding of between LINC01133 and miR-4784 (). Additional Ago2-RIP assay further confirmed the direct binding of LINC01133 and miR-4784 and suggested the enrichment of miR-4784 and LINC01133 in Ago2-immunoprecipitated complex (). Altogether, the results so far indicated the interaction between LINC01133 and miR-4784.
Figure 3. LINC01133 directly targeted miR-4784. (a) The location of LINC01133 was discovered in ME-180 and MS751 cells. (b) The expression of 15 potential miRNAs predicted by bioinformatics tools was examined in ME-180 cells by qRT-PCR after LINC01133 blockade. (c) Potential binding site of LINC01133 for miR-4784 was predicted. (d) Luciferase reporter assay indicated a suppressed activity of wild type of LINC01133 3′-UTR reporter in ME-180 and MS751 cells in the presence of miR-4784 mimics. (e) Cell lysate of ME-180 and MS751 cell lysates underwent incubation with anti-IgG or anti-Ago2 antibody and the enriched LINC01133 and miR-4784 were analyzed by qRT-PCR. *P < .05, **P < .01, *** P < .001.
LINC01133 up-regulated the target gene of miR-4784, AHDC1
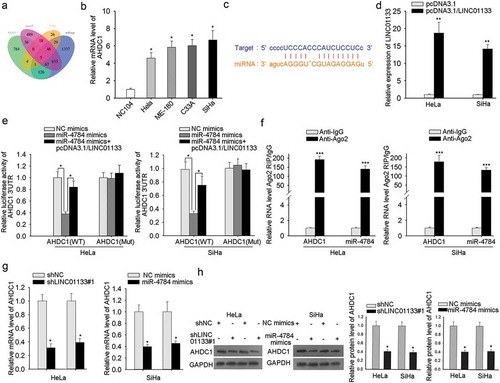
Typically, the next issue is to predict potential target mRNAs of miR-4784. As manifested in , five overlapped miRNAs were predicated by RNA22, microT, PicTar, and miRmap (). To recognize the potent candidate, the expression profiles of five miRNAs were monitored by qRT-PCR analysis. AHDC1 were observed to be highly expressed in CC cells (), while the other four exhibited no significant expression (data shown in B-D). Putative binding sites between AHDC1 and miR-4784 were provided by starbase (). To validate that potential effect that LINC01133 may pose on the binding of AHDC1 and miR-4784, overexpression efficiency of LINC01133 was examined in ME-180 and MS751 cells (). Dual-luciferase reporter assay revealed that introduction of miR-4784 mimics notably repressed the luciferase activity of wild type of AHDC1, while brought no impact on its mutant type. Furthermore, the suppressed luciferase activity of wild type of AHDC1 by miR-4784 mimics was restored after LINC01133 augmentation (). To further interrogate the interplay of AHDC1 and miR-4784, Ago2-RIP assay demonstrated the copious enrichment of AHDC1 mRNA and miR-4784 in the immunoprecipitated-Ago2 complexes, implying the co-expression of AHDC1 mRNA and miR-4784 in the RISC complex (). Finally, the impact of LINC01133 alone or the interference of miR-4784 alone on AHDC1 mRNA and protein was evaluated through qRT-PCR and western blot assays. Either LINC01133 repression or miR-4784 up-regulation strikingly reduced AHDC1 mRNA and protein level in ME-180 and MS751 cells (-). It concluded that LINC01133 competing for miR-4784 advanced the expression of AHDC1.
Figure 4. LINC01133 up-regulated the target gene of miR-4784, AHDC1. (a) Bioinformatic analysis of five potential downstream genes of miR-4784. (b) The expression of AHDC1 was detected in normal cervical cells and CC cells. (c) The predicated binding sites between AHDC1 and miR-4784. (d) The expression level of LINC01133 post-transfection of pcDNA3.1/LINC01133 into ME-180 and MS751 cells was tested. (e) The luciferase activity of AHDC1-WT reporter was monitored after transfection with specific plasmids. (f) RIP assay consolidated the binding between AHDC1 and miR-4784. (g-h) qRT-PCR and western blot disclosed the effect of LINC01133 or miR-4784 on AHDC1 mRNA and protein levels. *P < .05, **P < .01, *** P < .001.
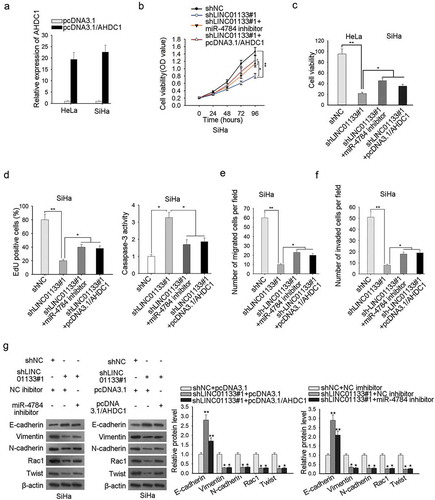
miR-4784/ahdc1 reversed the modulatory impact of LINC01133 on invasive and migratory potentials and EMT in CC
To clarify the LINC01133/miR-4784/AHDC1 on cell proliferation, migration, apoptosis, and EMT in CC, rescue experiments were designed and the AHDC1 augmentation efficiency was ensured beforehand (). CCK-8 and EdU analyses presented an inhibited proliferation capacity of ME-180 cells by LINC01133 depletion, and miR-4784 silencing or AHDC1 overexpression abolished the previous trend (-). The pro-apoptosis effect of LINC01133 depletion was countervailed by miR-4784 inhibition or AHDC1 overexpression (). The pro-migration or pro-invasion impact of LINC01133 was also counteracted by miR-4784 or AHDC1 (-). The results collectively showed that LINC01133 depletion decreased CC cell proliferation and migration and impaired EMT process, whereas, miR-4784 or AHDC1 mitigated the suppressive impact.
Figure 5. MiR-4784/AHDC1 reversed the modulatory impact of LINC01133 on invasive and migratory potentials and EMT in CC. (a) Confirmation of the overexpression efficiency of AHDC1 in ME-180 and MS751 cells was accomplished by qRT-PCR. (b-c) Post-transfection of indicated plasmids, the growth of ME-180 cells was assessed by CCK-8 and EdU assays. (d) ME-180 cell apoptosis after transfection with certain plasmids was evaluated by caspase-3 activity assay. (e-f) Transwell assay was executed to probe the migration and invasion of ME-180 cells transfected with specific plasmids. (g) Western blot was implemented to examine the effects of LINC01133/miR-4784/AHDC1 pathway on the EMT-associated protein contents. *P < .05, **P < .01.
Discussion
Most females suffered from or at a threatening risk of being attacked by cervical cancer (CC), and a mass of female mortalities were attributed to this gynecological malignancy.Citation1,Citation2 The situation becomes more intricate with diverse oncogenic infections and the 5-year survival rate remains frustrating despite the achievements we made.Citation3–Citation5 Unraveling more molecular mechanism concerning the initiation and development of cervical cancer is advantageous to the emergence of innovative and effective therapeutic targets of CC.
It has been substantially illustrated that lncRNA served as core modulators on gene expression during multiple biological processes from several facets such as transcriptional, post-transcriptional and epigenetic level.Citation29,Citation30 Recently, lncRNAs participating in cancer progression was increasingly discussed and enormously validated.Citation9–Citation11 Besides, cervical cancer-associated lncRNAs have also drawn a great deal of attention in recent years.Citation31,Citation32 For instance, lncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis.Citation33 CERNA2 serves as a potent predictor for aggravation and poor prognosis in cervical carcinoma.Citation34 Higher lncRNA CASC15 expression predicts poor prognosis and associates with tumor growth in cervical cancer.Citation35 Herein, LINC01133 was identified to be notably overexpressed in cervical carcinoma compared with non-carcinoma tissues from The Cancer Genome Atlas (TCGA), and LINC01133upregulation was elaborated to be strongly related to prognosis predication in cervical squamous cell carcinoma.Citation28 Never has the function and mechanism been revealed in CC, which certainly deserves more investigation. Consistent with previous work, the heightened level of LINC01133 in cervical cancer was validated in CC cells, while the facilitating effects of LINC01133 on CC cell proliferation, migration and EMT were first uncovered in the present study.
MicroRNAs are typically sponged and inhibited by lncRNAs acting as ceRNA and interrogation about this mechanism was also extensively carried out.Citation18,Citation19 MiR-143-5p was a direct target of lncRNA-TCONS_00026907.Citation20 LncRNA XLOC_006390 targeted miR-331-3p and miR-338-3p.Citation21 In the current study, this mechanism was also validated. Moreover, our study first unmasked that miR-4784, a microRNA associated with chondrocyte hyperplasiaCitation36 was targeted by LINC01133. Besides, the interaction between miR-4784 and AHDC1 was also revealed for the first time. Rescue experiments also first proved the modulatory network composed by LINC01133, miR-4784 and AHDC1 in CC.
In conclusion, we unveiled that LINC01133 may function as a ceRNA for miR-4784 to advance AHDC1 expression, intensifying CC cell malignant phenotypes and EMT process, which may demonstrate the implied value of LINC01133 as a therapeutic target for CC patients.
Supplemental Material
Download Zip (155.2 KB)Acknowledgments
We express our gratitude for all involved in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary materials
Supplemental data for this article can be accessed on the publisher’s website.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi:10.3322/caac.21387.
- Obel J, Souares Y, Hoy D, Baravilala W, Garland SM, Kjaer SK, Roth A. A systematic review of cervical cancer incidence and mortality in the Pacific Region. Asian Pac J Cancer Prev. 2014;15:9433–9437. doi:10.7314/apjcp.2014.15.21.9433.
- Shrestha AD, Neupane D, Vedsted P, Kallestrup P. cervical cancer prevalence, incidence and mortality in low and middle income countries: a systematic review. Asian Pac J Cancer Prev. 2018;19:319–324. doi:10.22034/APJCP.2018.19.2.319.
- Ellenson LH, Wu TC. Focus on endometrial and cervical cancer. Cancer Cell. 2004;5:533–538. doi:10.1016/j.ccr.2004.05.029.
- Schmitt M, Dalstein V, Waterboer T, Clavel C, Gissmann L, Pawlita M. Diagnosing cervical cancer and high-grade precursors by HPV16 transcription patterns. Cancer Res. 2010;70:249–256. doi:10.1158/0008-5472.CAN-09-2514.
- Derks M, van der Velden J, de Kroon CD, Nijman HW, van Lonkhuijzen L, van der Zee AGJ, Zwinderman AH, Kenter GG. Surgical treatment of early-stage cervical cancer: a multi-institution experience in 2124 cases in the Netherlands over a 30-year period. Int J Gynecol Cancer. 2018;28:757–763. doi:10.1097/IGC.0000000000001228.
- Meijer CJ, Snijders PJ. Cervical cancer in 2013: screening comes of age and treatment progress continues. Nat Rev Clin Oncol. 2014;11:77–78. doi:10.1038/nrclinonc.2013.252.
- ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi:10.1038/nature05874.
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi:10.1038/nature12451.
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi:10.1016/j.cell.2010.06.040.
- Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17:61. doi:10.1186/s12943-018-0812-2.
- Li J, Hao Y, Mao W, Xue X, Xu P, Liu L, Yuan J, Zhang D, Li N, Chen H, et al. LincK contributes to breast tumorigenesis by promoting proliferation and epithelial-to-mesenchymal transition. J Hematol Oncol. 2019;12:19. doi:10.1186/s13045-019-0707-8.
- Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W, Wang TD, Wu C, Zhou Q, Hu W, Wu C, et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/beta-catenin signaling pathway. Mol Cancer. 2019;18:15. doi:10.1186/s12943-019-1010-6.
- Yu J, Han Z, Sun Z, Wang Y, Zheng M, Song C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through beta-catenin-dependent Wnt pathway. J Exp Clin Cancer Res. 2018;37:222. doi:10.1186/s13046-018-0896-y.
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi:10.1016/j.cell.2009.11.007.
- Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–654. doi:10.1007/s10555-016-9648-7.
- Liang H, Yu T, Han Y, Jiang H, Wang C, You T, Zhao X, Shan H, Yang R, Yang L, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. 2018;17:119. doi:10.1186/s12943-018-0870-5.
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi:10.1016/j.cell.2011.09.028.
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi:10.1016/j.cell.2011.07.014.
- Jin X, Chen X, Hu Y, Ying F, Zou R, Lin F, Shi Z, Zhu X, Yan X, Li S, et al. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143-5p. Cancer Med. 2017;6:1409–1423. doi:10.1002/cam4.1084.
- Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29:e95. doi:10.3802/jgo.2018.29.e95.
- Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, Xu E, Zhang H, Lai M. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380:476–484. doi:10.1016/j.canlet.2016.07.015.
- Kong J, Sun W, Zhu W, Liu C, Zhang H, Wang H. Long noncoding RNA LINC01133 inhibits oral squamous cell carcinoma metastasis through a feedback regulation loop with GDF15. J Surg Oncol. 2018;118:1326–1334. doi:10.1002/jso.25278.
- Zeng HF, Qiu HY, Feng FB. Long Noncoding RNA LINC01133 Functions as an miR-422a Sponge to Aggravate the Tumorigenesis of Human Osteosarcoma. Oncol Res. 2018;26:335–343. doi:10.3727/096504017X14907375885605.
- Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao Q-X, Zhang H, Zeng L-S, Cui S-Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126. doi:10.1186/s12943-018-0874-1.
- Zang C, Nie FQ, Wang Q, Sun M, Li W, He J, Zhang M, Lu K-H. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget. 2016;7:11696–11707. doi:10.18632/oncotarget.7077.
- Zheng YF, Zhang XY, Bu YZ. LINC01133 aggravates the progression of hepatocellular carcinoma by activating the PI3K/AKT pathway. J Cell Biochem. 2019;120:4172–4179. doi:10.1002/jcb.27704.
- Mao X, Qin X, Li L, Zhou J, Zhou M, Li X, Xu Y, Yuan L, Liu Q-N, Xing H. A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol Oncol. 2018;149:181–187. doi:10.1016/j.ygyno.2017.12.011.
- Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinf. 2017;15:177–186. doi:10.1016/j.gpb.2016.12.005.
- Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216–217:105–110. doi:10.1016/j.cancergen.2017.06.003.
- Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 2016;37:2779–2788. doi:10.1007/s13277-015-4663-9.
- Taheri M, Ghafouri-Fard S. Long non-cod-ing RNA signature in cervical cancer. Klin Onkol. 2018;31:403–408. doi:10.14735/amko2018403.
- Gao F, Feng J, Yao H, Li Y, Xi J, Yang J. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artificial Cells Nanomedicine Biotechnol. 2019;47:776–782. doi:10.1080/21691401.2019.1577883.
- Wang M, Ouyang J, Li H. CERNA2: a predictor for clinical progression and poor prognosis in cervical carcinoma. J Cell Biochem. 2019.
- Shan S, Li HF, Yang XY, Guo S, Guo Y, Chu L, Xu M-J, Xin D-M. Higher lncRNA CASC15 expression predicts poor prognosis and associates with tumor growth in cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23:507–512. doi:10.26355/eurrev_201901_16862.
- Liu J, Yu Q, Ye Y, Yan Y, Chen X. Abnormal expression of miR-4784 in chondrocytes of osteoarthritis and associations with chondrocyte hyperplasia. Exp Ther Med. 2018;16:4690–4694. doi:10.3892/etm.2018.6739.
