ABSTRACT
CD16a (FcγRIIIa) mediates the antibody dependent cellular cytotoxicity (ADCC) and is important for anti-tumor activities of many therapeutic antibodies. Bispecific antibody targeting natural killer (NK) cells has been studied for cancer therapy. In this work, anti-CD16a single-domain antibodies were identified from hCD16a immunized camel. Bispecific antibodies are then constructed by fusing these single domain antibodies with an anti-CEA single domain antibody. These bispecific antibodies can recruite NK cells to kill CEA-positive tumor cells, and inhibit tumor growth in vivo, suggesting that these anti-CD16a single domain antibodies are powerful tools to engaging NK cells for cancer therapy.
Background
CD16a (FcγRIIIa), which is a low-affinity receptor for the IgG Fc domain, is involved in the antibody-dependent cellular cytotoxicity (ADCC) and responsible for triggering of cytolysis of target cells by natural killer (NK) cells.Citation1,Citation2 ADCC is one of the dominant cytotoxic mechanisms employed by FcγR-expressing effector cells to eliminate tumor cells.Citation1 Several tumor antigen-specific antibodies, such as Herceptin targeting Her2,Citation3 Rituximab targeting CD20,Citation4 have been shown dependent on ADCC to kill tumor cells.Citation3–Citation5 However, as the binding affinity of IgG1 Fc to CD16a on NK cells is low, methods to improve the binding of tumor antigen-specific antibodies to NK cells have been studied,Citation6,Citation7 including IgG1 Fc mutations to enhance IgG1 Fc affinity to CD16a (FcγRIIIA).Citation8,Citation9 Another approach is to use bispecific antibody targeting both tumor cells and immune cells to improve effector cell recruitment.Citation10,Citation11 Using different targeting molecule, immune effector cells, e.g. T cells, NK cells, macrophages, or monocytes, can be recruited to kill tumor cells in a non-MHC-restricted manner by redirecting effector cells to the tumor cells.Citation10 Bispecific antibodies to engage NK cells are attractive candidates for cancer immunotherapy as NK cells are potent in killing tumor cells.Citation12,Citation13 To actively engage NK cells, anti-CD16a antibodies have been studied and employed to build bispecific antibodies.Citation1,Citation14,Citation15
To recognize cancer cells in the bispecific format, many different tumor antigens have been studied, including CD19,Citation16 EpcamCitation17 and many others.Citation18 One of the well-studied tumor antigen is carcinoembryonic antigen (CEA; CEACAM5; CD66e), which is a glycosylated human oncofetal antigen that belongs to the CEA-related cell adhesion (CEACAM) superfamily.Citation19,Citation20 In normal tissues, CEA is expressed at low levels on epithelial cells in a polarized manner, while overexpressed and not polarized in many malignant cancers, including colorectal, gastric, breast and other cancers.Citation20,Citation21 Thus, CEA has been frequently targeted for cancer therapy. Clinical studies have demonstrated that radiolabeled anti-CEA antibodies or antibody fragments can be used as imaging reagents to localize CEA-expressing solid cancers, such as anti-CEA arcitumomab (CEA-Scan).Citation22–Citation24 MEDI-565, a BiTE antibody mediating T cell-directed cytotoxicity toward CEA positive tumor cells, is currently in clinical development.Citation20 Bispecific antibodies using anti-CEA single domain antibody have also shown potent anti-tumor activities in pre-clinical studies.Citation15,Citation25–Citation27
Single domain antibodies, also referred to as nanobodies or VHHs, were derived from the variable domain of the heavy-chain-only antibodies found in camelids.Citation28,Citation29 Single domain antibodies recognize the antigens with high specificity and affinities similar to IgG antibodies, but can penetrate into tumor tissues better due to smaller size (~15 kD).Citation30 In addition, single domain antibodies are resistant to extreme pH, heat denaturation, proteolysis, solvents and detergents.Citation31 They can be expressed and produced in high yields with high solubility.Citation32
Previous reports have shown that anti-CD16a VHHs can be used in the bispecific format to engage NK cells and kill tumor cells.Citation14,Citation15,Citation25,Citation33,Citation34 To generate more diverse anti-CD16a VHH for bispecific antibody studies, we performed phage display screening after immunizing a camel with human CD16a protein. We then constructed different bispecific antibodies by linking an anti-CEA single domain antibody with the selected anti-CD16a VHHs. The bispecific antibodies were expressed and produced in E. coli in high yield, and showed high affinity to CEA and CD16a. The bispecific antibodies can recruit NK cells to kill CEA-positive tumor cells in vitro with high potency. In vivo studies also demonstrated potent anti-tumor activities. Those studies demonstrated that these anti-CD16a VHHs are powerful tools to engaging NK cells for cancer therapy.
Methods and materials
Immunized VHH phage display library construction and screen
To generate anti-CD16a single domain antibody, the CD16a-His (Acrobiosystems, Cat# CDA-H5220) protein was used to immunize camel as described previously.Citation35 Briefly, after 4 rounds immunizations of one camel, which achieved high titer by Elisa, peripheral blood cells were extracted and isolated with gradient centrifugation. The RNA was isolated from lymphocytes entirely by Trizol Reagent (Invitrgoen). After reverse-transcript into the first strand of cDNA, the VHH fragments were amplified and ligated to pMECS phagemid vector. VHH phage library was created by transforming ligation products into XL1-Blue E.coli cells.Citation36,Citation37
To amplify the phage library, 200 µL of CD16a-VHH phage library was cultured in 40 mL of super broth medium (10 g MOPS Sigma, 30 g tryptone BD-Bioscience, 20 g yeast extract BD-Bioscience, 1 liter total volume with ddH2O) containing 100 µg/mL ampicillin and 10 µg/mL tetracycline at 37°C and 220 rpm/min until the OD600 to 0.6 − 0.8. ~1.4 × 1012 cfu (colony-forming unit) helper phage VCSM13 were then added and incubated at 37°C without agitation for 15 minutes, and then cultured at 220 rpm/min about 1.5–2 hours. Bacteriophages were collected by centrifuging at 4000 rpm/min for 10 minutes after the incubation, then re-suspended in 40 mL of the fresh super broth medium with 100 µg/mL ampicillin, 10 µg/mL tetracycline and 50 µg/mL kanamycin, and cultured at 30°C overnight. Bacterial cells were then discarded by centrifugation at 4000 rpm/min for 10 minutes at 4°C. Phages were precipitated from the supernatant with 5× PEG/NaCl (20% PEG/2.5 M NaCl) and re-suspended in 1 mL PBS and then precipitated again to remove bacterial cells completely. The phage was then re-suspended in 100–200 µL PBS+1%BSA.Citation37
Plate panning was used to select CD16a-specific VHH binder as described.Citation34 Briefly, the human CD16a-His antigen was coated on 96 well microplate. Then the phage library containing approximately 1012 cfu phages, which was named Input, was incubated with the coated plates for 15 minutes at 37°C. The weakly bound phages or excess of non-binding phages were washed by 0.1%PBST for 5 times. Specific binder phages were eluted with Glycine-BSA buffer (pH 2.2) and neutralized with 2 M Tris buffer (pH 9.0) immediately. The resulting phage collection was named Output. The eluted phage binders were used to infect competent E. coli XL1-blue (OD600 = 0.6) and amplified for next panning rounds. To enrich the positive binders, the panning was processed for 4 cycles by coating lower CD16a concentration from 1 μg to 100 ng in each cycle, which increased stringency. Through 4 round phage panning, positive clones were randomly picked and expanded in a 96-well deep block and rescued by the addition of VCSM13 helper phage. Phage ELISA was proceeded with phage-containing medium supernatant to further confirm the positive clones. Then the positive phage clones were precipitated by PEG/NaCl solution and re-suspended in PBS. Phage concentration was determined by measuring the OD at A280.Citation38
Production of anti CEA-CD16a VHH protein
Selected phage clones were sequenced and cloned, then fused with anti CEA-VHH (GenBank: ABS29544.1), and sub-cloned into pET26b vector (Novagen) as described previously.Citation15 The periplasmic protein purification method was performed as described previously.Citation15 In brief, the anti CEA-CD16a VHH plasmids were transformed into E. coli strain BL21 (DE3) competent cells for induced expression with 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG). The CEA-CD16a VHH antibodies were purified with Ni-NTA affinity chromatograph and analyzed by SDS-PAGE.
Gel filtration was performed by Superdex 75 10/300 GL(GE health, Cat#17-5174-01) with a flow rate of 0.5 ml/min. Protein markers(Sigma Aldrich, Cat#MWGF200) were loaded as standard controls for gel filtration analysis.
Dynamic light scattering (DLS)
DLS measurements were performed using an DynaPro Plate Reader (Wyatt, USA) operating at a light source wavelength of 830 nm and a fixed scattering angle of 150°. Approximately 50 µL of samples were measured as a function of temperature from 25°C to 75°C.
Cell culture and animals
The CEA-positive cancer cell lines LS174T and HT29 (human colon cancer cell lines), and the CEA-negative cancer cell lines SKOV3 (human ovarian cancer cell line) were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. HT29 and SKOV3 were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Life Technologies, China) with 10% HI fetal bovine serum (Gibco, Life Technologies, USA) and 1% Penicillin/Streptomycin (HyClone); LS174T was cultured in RPMI-1640 medium (Gibco, Life Technologies, China) also with 10% HI fetal bovine serum and 1% Penicillin/Streptomycin.
Fresh human peripheral blood mononuclear cells (PBMCs) were prepared from healthy donors by gradient centrifugation method with Ficoll-Plaque Plus (GE health) as described previously.Citation15,Citation27 Fresh NK cells were isolated from the PBMCs using an EasySep Human NK cell enrichment Kit (Stem cell Co.Ltd, Vancouver, Canada) according to the manufacturer’s instructions.
Non-obese diabetic-severe combined immunodeficiency disease (NOD/SCID), female, 4–5 weeks old, 18–22g mice were purchased from the Vital River Laboratory Animal Technology Co., Ltd. (Beijing) and housed in the animal experiment center of Sun Yat-sen University (20–26°C room temperature, 40%-70% relative humidity, and 12 hours light-dark rhythm).
Flow cytometry analysis
For flow cytometry analysis, LS174T and SKOV3 cells were digested with 0.25% trypsin and collected. 5 × 105 cells per sample were collected and then washed with 1 mL of ice cold PBS+0.2% BSA twice. The pellet was re-suspended in 200 µL of ice-cold PBS+0.2% BSA. In each tube, anti CEA-CD16a VHHs as primary antibody was added. Anti-His PE (BioLegend, Cat#652504) was used as secondary antibody. Flow cytometry analysis was then performed on FC500 (Beckman Colter).
ELISA assay
ELISA method was performed to determine the interaction between bispecific antibodies and hCD16a. Briefly bispecific antibodies SBC75-79 and BiSS were coated 5µg per well on 96-well plates in 100 µL PBS (pH 7.4) buffer overnight, then incubated with blocking buffer (PBS+0.2% BSA) at 37°C for 2 hours. After washing three times with PBS containing 0.05% Tween-20 (pH 7.2), CD16a-Avi (Sino Biological, 10389-H27H1-B) was added and incubated. After washing three times, anti-Streptavidin-HRP (R&D, 1:40 v/v) was used for detection. The absorbance was measured on 450nm. Data was analyzed with Graphpad Prism 5.
In vitro cytotoxic assays
In vitro cytotoxicity assays were performed as described previously.Citation27 Briefly, tumor cells were plated into 96-well plates at approximately 2500 cells per well and incubated 6 hours at 37°C, 5% CO2. 25000 NK cells and different concentrations of antibodies were then mixed with growth medium and added to each well. After 72 hrs, the Cell Counting Kit-8 reagent (Dojindo, CK04) was applied. After 1–4 hours incubation, OD450nm was measured by a TECAN microplate reader. The survival rate (%) of target cells was calculated using the following formula: [(live target cells (sample)- medium)/(live target cells (control) -medium)].
In vivo tumor growth inhibition assay
In vivo tumor growth studies were performed as described previously.Citation15 In the co-transplantation model, LS174T human colon carcinoma cells were harvested, washed twice and re-suspended in PBS. The cells were then mixed with freshly isolated human peripheral blood monocytes (PBMCs) and injected subcutaneously into the right flank of NOD/SCID mice containing 1 × 106 LS174T cells and 5 × 106 PBMC cells in a total volume of 200 µL per mouse. After transplantation, the antibody (20 µg/mice or 5 µg/mice) or vehicle control (PBS) were administered intraperitoneally (i.p.) (n = 5 each group). The animals were then treated every two days over the following 10 days.
In another study, freshly prepared PBMCs were depleted with NK cells using an EasySep Human NK cell enrichment Kit (Stem cell Co. Ltd, Vancouver, Canada). The cells (NK negative cells) were then mixed with LS174T cells (5 × 106 NK negative PBMC cells and 1 × 106 LS174T cells in a total volume of 200 µL per mouse) and injected subcutaneously into the right flank of NOD/SCID. When the tumor volumes reach to 50–100 mm,Citation3 SBC77 (20 µg/mice) or PBS was administered intraperitoneally (i.p.) every 2 days (n = 5 each group). Animal weight and tumor volume were measured every two days. The tumor volume was calculated using the formula (width2× length)/2.
Results
Phage display library construction and anti-hCD16a antibody screening
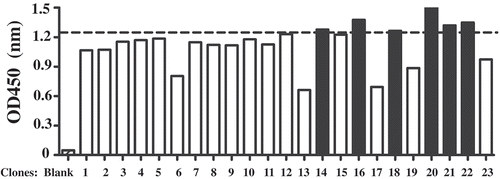
To obtain single domain anti-hCD16a antibodies, a camel was immunized with 300 µg hCD16a-His protein for 4 times and the phage library were constructed from lymphocytes isolated from the immunized camel. After 4 round of panning with increased stringency by decreasing the coating antibody from 1 µg to 100 ng (), 54 positive clones were obtained. Positive phage clones were then precipitated by PEG/NaCl solution and re-suspended in PBS. Phage concentration was determined by measuring the OD at A280. These purified and quantified 54 clones were then used to do another quantitative ELISA. ELISA experiments demonstrated that 34 of these phages can specifically recognize CD16a-His protein. After analyzing the sequences, 23 different sequences have been obtained. Based on the diversity and Elisa results, 6 of 23 clones were further analyzed ().
Table 1. Enrichment data for the hCD16a-VHH library.
Expression and purification of anti CEA-CD16a bispecific antibodies
We have previously constructed a bispecific antibody BiSS by linking an anti-CD16a VHH with anti-CEA VHH.Citation15 The BiSS bispecific antibody exhibited potent anti-tumor activities in vitro and in vivo,Citation15 suggesting that this bispecific antibody format can be used to evaluate anti-CD16a VHH activity in bispecific format. Thus, the six different anti-CD16a VHHs () were used to construct bispecific antibodies (SBC74-SBC79) by replacing the anti-CD16a VHH in BiSS with the selected anti-CD16a VHH respectively with a flexible linker, (GGGGS)3, between anti-CD16a VHH and anti-CEA VHH ()).
Figure 2. Biochemical characterization of anti CEA-CD16a VHHs. (a) The anti CEA-CD16a VHH SBC74-79 were constructed by fusing an anti-CEA and anti-CD16a single domain antibodies with a (GGGGS)3 linker. A His-tag was added to the C-terminal end to protein detection and purification. (b) Coomassie blue staining of purified proteins (SBC74, SBC75, SBC76, SBC77, SBC78, SBC79) after Ni-NTA affinity chromatography. (c) Gel filtration of protein marker (top panel) and SBC77 (bottom panel). (d) DLS experiment was performed as described in the Materials and Methods using 0.5 mg/ml of purified protein from 25°C–75°C.
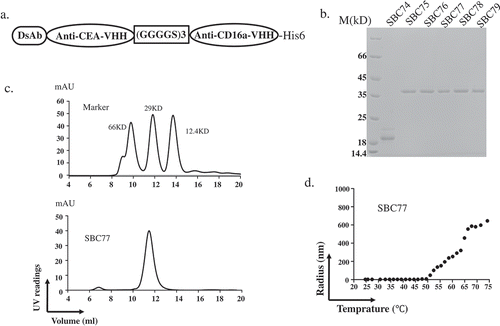
The antibodies were purified by Ni-NTA affinity chromatography and exchanged in phosphate-buffered saline (PBS, pH 7.4) at 4°C for at least 12h. SBC75-79 ran as a single band at approximately ~36KD on SDS-PAGE, which was the expected molecular weight of the monomer. The size of SBC74 was the almost half of expected size, suggesting degradations ()). Thus, SBC74 was not characterized further.
To further characterize the purified SBC75-79, gel filtration was performed. The bispecific antibody SBC77 ran a single peak with a molecular size of approximately ~36KD, suggesting the majority of SBC77 is in the form of monomer ()). Similar results were observed for bispecfic antibodies SBC75, SBC76, SBC78, SBC79 and BiSS (data not shown).
To further characterize SBC75-79, the melting temperature of bispecific antibodies was measured by DLS. The radius of SBC77 showed no variation under 50°C, while increased significantly above 50°C ()). Similar results were observed for other bispecfic antibodies SBC75, SBC76, SBC78, SBC79 and BiSS (data not shown).
Anti CEA-CD16a bispecific antibodies could recognize CEA and CD16 antigen
To determine whether the anti CEA-CD16a (SBC75-SBC79) bispecific antibodies can bind to the tumor antigen CEA, flow cytometry analysis was performed using the CEA-positive cell lines LS174T and CEA-negative cell line SKOV3. The positive control BiSS and the anti CEA-CD16a (SBC75-SBC79) demonstrated positive staining on CEA positive cell line LS174T and no staining on CEA-negative cell line SKOV3 ()).
Figure 3. Anti CEA-CD16a bispecific antibodies can bind CEA and CD16a antigen. a) Flow cytometry analysis of anti CEA-CD16a bispecific antibodies were performed using LS174T and SKOV3 cells. Light gray area (blank), indicating LS174T cells with no staining; Dark gray area (blank), indicating SKOV3 cells with no staining; dotted line, cells with anti-His-PE staining; Solid line, anti CEA-CD16a VHHs and then anti-His-PE staining. b) Elisa analysis of different bispecific antibodies binding to CD16a antigen, BiSS (gray, dashed line); Anti CEA-CD16a bispecfic antibodies SBC75-79 (black, solid line); No CD16a(gray, solid line). The data are the mean of triplicates with error bars representing the standard deviation.
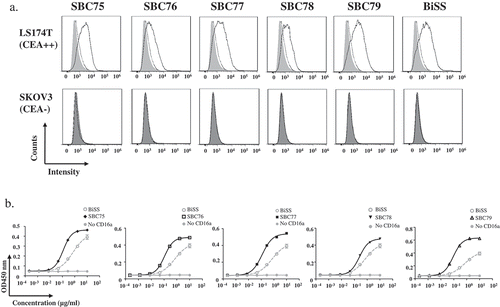
Elisa assay was then performed to check whether the bispecific antibodies can bind the CD16a protein ()). All bispecific antibodies, SBC75, SBC76, SBC77, SBC78, and SBC79 can bind CD16a with similar strength ()). As a negative control, no CD16a coating showed no binding for each of the bispecific antibodies, suggesting specific binding of the bispecific antibodies. As a positive control, BiSS, whose anti-CD16a VHH part has Kd of 10 × 10−9 mol/L to hCD16a,Citation15,Citation34 has lower binding strength than SBC75-79, suggesting SBC75-79 have stronger binding than the anti-CD16 VHH in BiSS.
Anti CEA-CD16a bispecific antibodies mediated potent cytotoxic activities in CEA-expressing cells
To evaluate whether the anti CEA-CD16a bispecific antibodies can mediate tumor cell killing, cytotoxic assays were performed. No cytotoxicity was observed in the CEA-negative cell SKOV3 with NK cells (). No cytotoxicity was observed in the CEA-positive cells LS174T when respective anti-CD16a VHHs were used with NK cells (). For the CEA-positive cells LS174T, all bispecific antibodies exhibit potent tumor cell killing in the presence of NK cells, while no cell killing was observed in the absence of NK cells ().
Figure 4. Anti CEA-CD16a bispecific antibodies induce NK cell-mediated cytotoxicity. a) Different cell lines were treated with anti CEA-CD16a bispecific antibodies or anti-CD16a VHHs with fresh isolated NK cells. The effector (NK cells) (25000 cells/well) and target cells LS174T and SKOV3 (2500 cells/well) at ratio of 10:1. All data are the mean of triplicates with error bars representing the standard deviation. (***P < .001, t test, compared with Anti-CD16a VHHs).
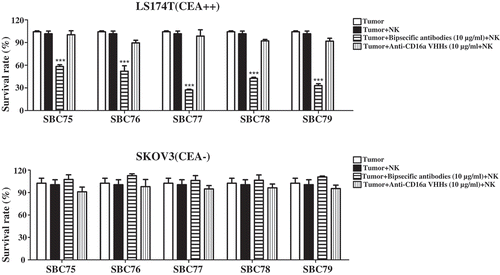
Figure 5. Anti CEA-CD16a bispecific antibodies mediates NK cell-dependent tumor cell killing. (a) Different cell lines were treated with anti CEA-CD16a bispecific antibodies with or without fresh isolated NK cells. The effector (NK cells) (25000 cells/well) and target cells LS174T, HT29 and SKOV3 (2500 cells/well) at ratio of 10:1. All data are the mean of triplicates with error bars representing the standard deviation. (***P < .001, t test, compared with Tumor+NK). (b) SBC77 induce NK cell-mediated cytotoxicity in a dose dependent manner. The concentration of BiSS, SBC77 and Anti-CD16a VHH (D5) are from 0.001 ng/mL to 10 µg/mL. All data are the mean of triplicates with error bars representing the standard deviation.
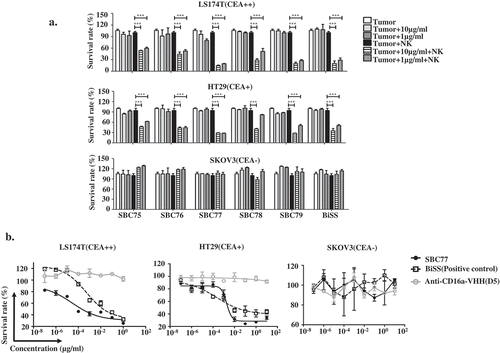
To further evaluated the cytotoxicity of anti CEA-CD16a bispecific antibodies, the dose dependent cell killing were studied (). No cytotoxicity was observed in the CEA-negative cell SKOV3 regardless of the concentrations of bispecfic antibodies SBC75-SBC79 (,)). For the CEA-positive cells LS174T and HT29, all the bispecific antibodies exhibit potent tumor cell killing in the presence of NK cells ()). Among the 5 different bispecific antibodies, SBC77 showed the strongest killing capacity. Thus, it was further evaluated for the dose dependent cell killing ()). Comparing with the positive control BiSS antibody, SBC77 showed higher cytotoxicity against both high CEA expression LS174T cells and lower CEA expression HT29 cells ()). No cell killing was observed in the LS17T and HT29 cells for anti-CD16a VHHs with NK cells ()).
Anti CEA-CD16a bispecific antibody SBC77 inhibits tumor growth in vivo
To further investigate whether the anti CEA-CD16a bispecific antibodies can inhibit tumor cell growth in vivo, SBC77 was used to evaluate anti-tumor activity in Nod/Scid mice. After Nod/Scid mice were transplanted with LS174T cells and freshly isolated human PBMCs. Rapid tumor growth was observed. SBC77 exhibited minimal tumor growth inhibition at lower dosage (5 μg per animal) but potent tumor growth inhibition at high dosage (20 μg per animal) ()). No significant weight loss and toxicity were observed in these mice treated with either low or high dosage of SBC77. These data suggest that SBC77 can inhibit tumor growth potently in the xenograft mouse model.
Figure 6. SBC77 inhibits tumor growth in vivo. (a) NOD/SCID mice (n = 5/group) were engrafted subcutaneously with LS174T cells (1 × 106 per animal) with freshly isolated human PBMCs (5 × 106 per animal). The mice then treated intraperitoneally with vehicle PBS (gray line), SBC77(20 µg/mice) (triangle, solid line), or SBC77 (5 µg/mice)(triangle, dashed line) every two days. The tumor volume was then measured. The data represent the average tumor volume of 5 mice. Error bars represent the standard deviation (***P < .001, t test, vehicle vs SBC77(20 µg, 5 µg)). (b) NOD/SCID mice (n = 5/group) were engrafted subcutaneously with LS174T cells (1 × 106 per animal) with freshly isolated human NK negative PBMC cells (5 × 106 per animal). The mice were then treated with SBC77 (20 µg/mice) or PBS as described in the Materials and Methods.
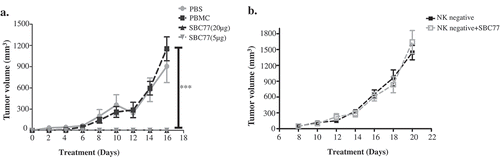
To further analyze whether NK cells are needed in the SBC77 in vivo efficacy. NK cells were depleted from the fresh isolated human PBMCs. LS174T cells and NK negative PBMC cells were then grafted onto NOD/SCID mice. The mice were treated with SBC77 or vehicle PBS every two days. No significant tumor growth inhibition was observed with SBC77 treatment ()). These data confirmed that NK cells are needed to confer tunor inhibition for SBC77 in vivo.
Discussion
Redirecting the cytotoxic potential of leukocytes to eliminate tumor cells has been a major promise for the development of bispecific antibodies for cancer immunotherapy.Citation25 CD16a is an attractive candidate to recruit NK cells, as ADCC mediated by CD16a is one of the major modes of action of antitumor antibodies.Citation34 Anti-CD16a bispecific antibodies have the potential to avoid several important issues faced by Fc containing antibodies, such as variations in glycosylation from cell line to cell line,Citation34 polymorphism at position 158 of CD16aCitation9, and low affinity of Fc to CD16a.
In the present work, we have isolated anti-CD16a single domain antibodies from a phage library of VHH fragments derived from an immunized camel. Six candidates with diverse CDR sequences have been selected to construct bispecific antibodies. Consistent with the previous studies, the selected anti-CD16a VHH linked the anti-CEA VHH (SBC75-79) were highly soluble and stable with high yields from E.coli. SBC75-SBC79 also displayed high specificity to CEA and CD16a, potent and specific NK-mediated lysis of CEA-expressing tumor cells at low concentrations. These results suggest that a high affinity for CD16a translates into improved cytotoxic activity of effector cells as previous studies reported.Citation25 In mouse model, significant inhibition of tumor growth was also observed.
To further explore the application of these antibodies in patient, improved the half-life in vivo of those bispecific antibodies is likely needed as single domain antibody has short half-life in vivo due to the small size (~15 kD).Citation31 Conjugation with PEG, fusion with Fc, or serum albumin, or other approaches that can extend serum half-life of the single domain antibodies can be explored in future studies. Other factors, such as the linker between VHHs either in the format of single domain fusion, or Fc format, may also influence the recruitment or formation of immune cell-tumor cell complex and thus lead to different potency of cell killing. The flexibility of anti-CD16a VHH will accelerate these studies considering its size and easy to reformat.
In summary, this study demonstrated that the anti-CD16a VHHs can be produced with high stability and high solubility. These antibodies can be used to recruit NK cells and mediate tumor cell lysis. These antibodies will provide valuable tools for cancer immunotherapy.
Conflict of interest
The authors declare no competing financial interest.
References
- Reusch U, Burkhardt C, Fucek I, Le Gall F, Le Gall M, Hoffmann K, Knackmuss SH, Kiprijanov S, Little M, Zhukovsky EA. 2014. A novel tetravalent bispecific tandab (cd30/cd16a) efficiently recruits nk cells for the lysis of cd30+ tumor cells. MAbs. 6(3):728–739. doi:10.4161/mabs.28591.
- Mandelboim OMP, Davis DM, Jo CH, Boyson JE, Strominger JL. Human cd16 as a lysis receptor mediating direct natural killer cell cytotoxicity. Proc Natl Acad Sci USA. 1999;96:5640–5644. doi:10.1073/pnas.96.10.5640.
- Arnould L, Gelly M, Penault-Llorca F, Benoit L, Bonnetain F, Migeon C, Cabaret V, Fermeaux V, Bertheau P, Garnier J, et al.. 2006. Trastuzumab-based treatment of her2-positive breast cancer: an antibody-dependent cellular cytotoxicity mechanism?. Br J Cancer. 94(2):259–267. doi:10.1038/sj.bjc.6602930
- Ejaz AA, Asmar A, Alsabbagh MM, Ahsan N. 2012. Rituximab in immunologic glomerular diseases. MAbs. 4(2):198–207. doi:10.4161/mabs.4.2.19286.
- Alderson KL, Sondel PM. 2011. Clinical cancer therapy by nk cells via antibody-dependent cell-mediated cytotoxicity. J Biomed Biotechnol. 2011:379123. doi:10.1155/2011/379123.
- Bowles JA, Wang SY, Link BK, Allan B, Beuerlein G, Campbell MA, Marquis D, Ondek B, Wooldridge JE, Smith BJ, et al.. 2006. Anti-cd20 monoclonal antibody with enhanced affinity for cd16 activates nk cells at lower concentrations and more effectively than rituximab. Blood. 108(8):2648–2654. doi:10.1182/blood-2006-04-020057
- Oppenheim DE, Spreafico R, Etuk A, Malone D, Amofah E, Pena-Murillo C, Murray T, McLaughlin L, Choi BS, Allan S, et al.. 2014. Glyco-engineered anti-egfr mab elicits adcc by nk cells from colorectal cancer patients irrespective of chemotherapy. Br J Cancer. 110(5):1221–1227. doi:10.1038/bjc.2014.35
- Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. 2002. Therapeutic activity of humanized anti-cd20 monoclonal antibody and polymorphism in igg fc receptor fcgammariiia gene. Blood. 99(3):754–758. doi:10.1182/blood.V99.3.754.
- Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M. 1997. Fc gammariiia-158v/f polymorphism influences the binding of igg by natural killer cell fc gammariiia, independently of the fc gammariiia-48l/r/h phenotype. Blood. 90(3):1109–1114. doi:10.1016/S0165-2478(97)85823-3.
- Fan G, Wang Z, Hao M, Li J. 2015. Bispecific antibodies and their applications. J Hematol Oncol. 8:130. doi:10.1186/s13045-015-0227-0.
- van Spriel AB, van Ojik HH, van De Winkel JG. 2000. Immunotherapeutic perspective for bispecific antibodies. Immunol Today. 21(8):391–397. doi:10.1016/S0167-5699(00)01659-5.
- Arndt MA, Krauss J, Kipriyanov SM, Pfreundschuh M, Little M. A bispecific diabody that mediates natural killer cell cytotoxicity against xenotransplantated human hodgkin’s tumors. Blood. 1999;94(8):2562–2568.
- McCall AM, Shahied L, Amoroso AR, Horak EM, Simmons HH, Nielson U, Adams GP, Schier R, Marks JD, Weiner LM. 2001. Increasing the affinity for tumor antigen enhances bispecific antibody cytotoxicity. J Immunol. 166(10):6112–6117. doi:10.4049/jimmunol.166.10.6112.
- Li Y, Zhou C, Li J, Liu J, Lin L, Li L, Cao D, Li Q, Wang Z. 2018. Single domain based bispecific antibody, muc1-bi-1, and its humanized form, muc1-bi-2, induce potent cancer cell killing in muc1 positive tumor cells. PLoS One. 13(1):e0191024. doi:10.1371/journal.pone.0191024.
- Dong B, Zhou C, He P, Li J, Chen S, Miao J, Li Q, Wang Z. 2016. A novel bispecific antibody, biss, with potent anti-cancer activities. Cancer Biol Ther. 17(4):364–370. doi:10.1080/15384047.2016.1139266.
- Oak E, Bartlett NL. 2015. Blinatumomab for the treatment of b-cell lymphoma. Expert Opin Investig Drugs. 24(5):715–724. doi:10.1517/13543784.2015.1021415.
- Amann M, Friedrich M, Lutterbuese P, Vieser E, Lorenczewski G, Petersen L, Brischwein K, Kufer P, Kischel R, Baeuerle PA, et al.. 2009. Therapeutic window of an epcam/cd3-specific bite antibody in mice is determined by a subpopulation of epcam-expressing lymphocytes that is absent in humans. Cancer Immunol Immunother. 58(1):95–109. doi:10.1007/s00262-008-0529-y
- Clynes RA, Desjarlais JR. 2018. Redirected t cell cytotoxicity in cancer therapy. Annu Rev Med. doi:10.1146/annurev-med-062617-035821
- Oikawa S, Nakazato H, Kosaki G. 1987. Primary structure of human carcinoembryonic antigen (cea) deduced from cdna sequence. Biochem Biophys Res Commun. 142(2):511–518. doi:10.1016/0006-291X(87)90304-4.
- Peng L, Oberst MD, Huang J, Brohawn P, Morehouse C, Lekstrom K, Baeuerle PA, Wu H, Yao Y, Coats SR, et al.. 2012. The cea/cd3-bispecific antibody medi-565 (mt111) binds a nonlinear epitope in the full-length but not a short splice variant of cea. PLoS One. 7(5):e36412. doi:10.1371/journal.pone.0036412
- Hammarstrom S. 1999. The carcinoembryonic antigen (cea) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 9(2):67–81. doi:10.1006/scbi.1998.0119.
- Hong H, Sun J, Cai W. 2008. Radionuclide-based cancer imaging targeting the carcinoembryonic antigen. Biomark Insights. 3:435–451. doi:10.4137/BMI.S1124.
- Goldenberg DM, Wlodkowski TJ, Sharkey RM, Silberstein EB, Serafini AN, Garty II, Van Heertum RL, Higginbotham-Ford EA, Kotler JA, Balasubramanian N, et al.. 1993. Colorectal cancer imaging with iodine-123-labeled cea monoclonal antibody fragments. J Nucl Med. 34(1):61–70. doi:10.1097/00004728-199301000-00010
- Moffat FL Jr., Pinsky CM, Hammershaimb L, Petrelli NJ, Patt YZ, Whaley FS, Goldenberg DM. 1996. Clinical utility of external immunoscintigraphy with the immu-4 technetium-99m fab’ antibody fragment in patients undergoing surgery for carcinoma of the colon and rectum: results of a pivotal, phase iii trial. The immunomedics study group. J Clin Oncol. 14(8):2295–2305. doi:10.1200/JCO.1996.14.8.2295.
- Rozan C, Cornillon A, Petiard C, Chartier M, Behar G, Boix C, Kerfelec B, Robert B, Pelegrin A, Chames P, et al.. 2013. Single-domain antibody-based and linker-free bispecific antibodies targeting fcgammariii induce potent antitumor activity without recruiting regulatory t cells. Mol Cancer Ther. 12(8):1481–1491. doi:10.1158/1535-7163.MCT-12-1012
- Kuroki M, Hachimine K, Huang J, Shibaguchi H, Kinugasa T, Maekawa S, Kuroki M. 2005. Re-targeting of cytotoxic t lymphocytes and/or natural killer cells to cea-expressing tumor cells with anti-cea antibody activity. Anticancer Res. 25(6A):3725–3732. doi:10.1063/1.1146580.
- Li L, He P, Zhou C, Jing L, Dong B, Chen S, Zhang N, Liu Y, Miao J, Wang Z, et al.. 2015. A novel bispecific antibody, s-fab, induces potent cancer cell killing. J Immunother. 38(9):350–356. doi:10.1097/CJI.0000000000000099
- Rossotti MA, Gonzalez-Techera A, Guarnaschelli J, Yim L, Camacho X, Fernandez M, Cabral P, Leizagoyen C, Chabalgoity JA, Gonzalez-Sapienza G. 2015. Increasing the potency of neutralizing single-domain antibodies by functionalization with a cd11b/cd18 binding domain. MAbs. 7(5):820–828. doi:10.1080/19420862.2015.1068491.
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. 1993. Naturally occurring antibodies devoid of light chains. Nature. 363(6428):446–448. doi:10.1038/363446a0.
- Nanobodies: MS. 2013. Natural single-domain antibodies. Annu Rev Biochem. 82:775–797. doi:10.1146/annurev-biochem-063011-092449.
- Arezumand R, Alibakhshi A, Ranjbari J, Ramazani A, Muyldermans S. 2017. Nanobodies as novel agents for targeting angiogenesis in solid cancers. Front Immunol. 8:1746. doi:10.3389/fimmu.2017.01746.
- Muyldermans S, Baral TN, Retamozzo VC, De Baetselier P, De Genst E, Kinne J, Leonhardt H, Magez S, Nguyen VK, Revets H, et al.. 2009. Camelid immunoglobulins and nanobody technology. Vet Immunol Immunopathol. 128(1–3):178–183. doi:10.1016/j.vetimm.2008.10.299
- Li J, Zhou C, Dong B, Zhong H, Chen S, Li Q, Wang Z. 2016. Single domain antibody-based bispecific antibody induces potent specific anti-tumor activity. Cancer Biol Ther. 17(12):1231–1239. doi:10.1080/15384047.2016.1235659.
- Behar G, Siberil S, Groulet A, Chames P, Pugniere M, Boix C, Sautes-Fridman C, Teillaud JL, Baty D. 2008. Isolation and characterization of anti-fcgammariii (cd16) llama single-domain antibodies that activate natural killer cells. Protein Eng Des Sel. 21(1):1–10. doi:10.1093/protein/gzm064.
- Stewart CS, MacKenzie CR, Hall JC. 2007. Isolation, characterization and pentamerization of alpha-cobrotoxin specific single-domain antibodies from a naive phage display library: preliminary findings for antivenom development. Toxicon. 49(5):699–709. doi:10.1016/j.toxicon.2006.11.023.
- Shahsavarian MA, Le Minoux D, Matti KM, Kaveri S, Lacroix-Desmazes S, Boquet D, Friboulet A, Avalle B, Padiolleau-Lefevre S. 2014. Exploitation of rolling circle amplification for the construction of large phage-display antibody libraries. J Immunol Methods. 407:26–34. doi:10.1016/j.jim.2014.03.015.
- Vincke C, Gutierrez C, Wernery U, Devoogdt N, Hassanzadeh-Ghassabeh G, Muyldermans S. 2012. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol Biol. 907:145–176. doi:10.1007/978-1-61779-974-7_8.
- Sharifzadeh Z, Rahbarizadeh F, Shokrgozar MA, Ahmadvand D, Mahboudi F, Rahimi Jamnani F, Aghaee Bakhtiari SH. 2013. Development of oligoclonal nanobodies for targeting the tumor-associated glycoprotein 72 antigen. Mol Biotechnol. 54(2):590–601. doi:10.1007/s12033-012-9601-0.