ABSTRACT
Acute myeloid leukemia (AML) is a prevalent class of blood disease with a high occurrence rate and relapse rate. The role of dysregulated microRNAs (miRNAs) in AML is emerging. MiR-4260 was identified to be a carcinogenic miRNA in colorectal cancer, but never has it been reported in AML. We aimed to study the function and mechanism of miR-4260 in AML. The miR-4260 level was higher in AML cell lines than the normal cell lines. Inhibition of miR-4260 hindered proliferation and increased apoptosis of AML cells. Mechanistically, long intergenic non-protein coding RNA 1128 (LINC01128) competed with nuclear receptor subfamily 3 group C member 2 (NR3C2) for miR-4260 so as to upregulate NR3C2. We identified the reduced levels of LINC01128 and NR3C2 in AML and it was suggested through rescue assays that LINC01128 repressed AML progression through regulating miR-4260/NR3C2 axis. In conclusion, we firstly uncovered that LINC01128 resisted acute myeloid leukemia through regulating miR-4260/NR3C2, providing novel clues for the treatment improvement of AML.
Introduction
Acute myeloid leukemia (AML) is the commonest blood disease with high morbidity and relapse rate in adults.Citation1 AML is featured with reduced proliferation of the functional blood cells as well as surging growth and accumulation of the immature leukemic blasts.Citation2 The pathogenesis of AML could result from genetic or epigenetic alterations.Citation3,Citation4 However, the detailed mechanism underlying AML remains incompletely understood.
MicroRNAs (miRNAs), a set of small, conserved endogenous RNAs, are emerging as pivotal modulators of human cancers and diseases.Citation5,Citation6 Numerous works have delineated the essential functions of miRNAs in AML. For instance, miR-4319 exerted anti-tumor functions in AMLCitation7; miR-34a promoted apoptosis and hindered autophagy in AML through targeting HMGB1.Citation8 miR-4260 has been reported as an oncogenic miRNA in colorectal cancer (CRC), and its overexpression promoted cell proliferation and migration, reduced apoptosis in CRC.Citation9 However, the regulation of miR-4260 on AML has not been explored.
Also, recent years have witnessed the implication of long non-coding RNAs (lncRNAs) in the development of the hematopoietic system.Citation10 As a class of RNA transcripts without coding proteins,Citation11,Citation12 lncRNAs are reported to regulate the growth and death of AML cells by mounting studies. For example, long intergenic non-protein coding RNA 319 (LINC00319) was induced by MYC and promoted AML cell growth through stabilizing SIRT6.Citation13 LncRNA H19 overexpression facilitated the leukemogenesis and indicated poor prognosis in AML.Citation14 LncRNA CCAT1 performed as a competitive endogenous RNA (ceRNA) to regulate the growth and differentiation of AML cells.Citation15 Long intergenic non-protein coding RNA 1128 (LINC01128) is newly discovered by our study to potentially interact with microRNA-4260 (miR-4260) through miRDB, and it has never been explored in AML before.
Nuclear receptor subfamily 3 group C member 2 (NR3C2) is a mineralocorticoid receptor gene whose translating protein is mineralocorticoid receptor (MR), which can regulate the electrolyte balance.Citation16 To date, NR3C2 has been demonstrated to suppress the progression of cancers by inhibiting proliferation, migration, invasion, and epithelial to mesenchymal transition (EMT).Citation17,Citation18 Importantly, NR3C2, together with some other nuclear receptors (NRs), is reported to be downregulated in AML samples according to a pan-cancer analysis.Citation19 However, the function of NR3C2 in AML still needs to be further investigated.
Materials and methods
Cell culture
Human marrow stromal cells (HS-5), human renal epithelial cells (293T) and human AML cells (U937, THP-1, KG-1a, HL60) were all purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HS-5 and 293T cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA). AML cells were grown in Roswell Park Memorial Institute 1640 medium (RPMI 1640; Thermo Fisher Scientific). All culture media contained 1% penicillin/streptomycin (Thermo Fisher Scientific) and 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) and maintained at 37°C in 5% CO2.
Cell transfection
HL60 or KG-1a cells were plated into 6-well plates the moment the confluent of cells was up to 50–80%. Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) was applied for transfection following its instructions. The miR-4260 mimic and miR-4260 inhibitor, together with NC mimic and NC inhibitor were designed by Genepharma (Shanghai, China) and transfected into HL60 or KG-1a cells, respectively. The pcDNA3.1 vector inserted with LINC01128 or NR3C2 and the empty vector were purchased from Genechem (Shanghai, China) and separately transfected into HL60 or KG-1a cells. At 48 h post-transfection, cells were reaped.
Quantitative real-time PCR
Extraction of total RNA was accomplished via Trizol (Invitrogen) in this study in line with the manufacturer’s guide. Concentration was measured by an ultraviolet spectrophotometer (PerkinElmer, Shanghai, China). 5 μg RNA was reverse transcribed via PrimeScript™ RT reagent Kit (Takara, Tokyo, Japan) as per the supplier’s instructions. Subsequently, SYBR_Green PCR Master Mix™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was used for conducting RT-qPCR. Relative gene expression levels were subjected to 2−ΔΔCT analysis. GAPDH served as an internal reference for LINC01128 and NR3C2, and U6 for miR-4260.
Cell counting Kit-8 assay
Cell proliferation was detected via Cell Counting Kit-8 assay. Transfected HL60 or KG-1a cells were first added into 96-well plates at a concentration of 2000 cells/well. CCK8 reagents (10 μL; Beyotime, Shanghai, China) were added to each well in indicated time and cells were cultivated for another 4 h. The absorbance at the wavelength of 450 nm was eventually examined by a microplate reader (Molecular Devices, Sunnyvale, USA).
Colony formation assay
HL60 or KG-1a cells after transfection were put into 6-well plates. Fourteen days later, colonies were formed and fixed in 10% formaldehyde (JM, Aliso Viejo, CA) for 30 min and stained with 0.5% crystal violet (Sigma-Aldrich, St. Louis, USA) for 5 min. Data analysis was carried out with Image-Pro Plus 6.0 (Silver Springs, MD, USA).
Trypan blue exclusion assay
According to the changes in cell membrane integrity, trypan blue exclusion assay was applied to examine cell proliferation. To begin with, 1 × 103 cells were cultured in 96-well plates for 48 h at 37°C. Then, cells were rinsed, trypsinized, and dyed with trypan blue dye (Beyotime Biotechnology, Shanghai, China). A cell counting chamber was employed to count viable cells.
Caspase-3 activity
Activity of caspase-3 was carried out by using a caspase-3 activity kit (Beyotime). The SpectraMax M2 microplate reader (Molecular Devices, San Jose, CA, USA) was used for monitoring caspase-3 activity at 405 nm.
TUNEL assay
TUNEL assay was conducted for assessing the apoptotic cells by applying an In Situ Cell Death Detection kit (Roche, Mannheim, Germany) as the supplier requested. For total cell count, nucleus was counterstained with DAPI (Beyotime). The fluorescence microscope bought from Olympus (Tokyo, Japan) was applied for capturing the images and evaluating the apoptotic cells. TUNEL positive cells were calculated in three randomly selected different fields.
Microarray analysis
HL60 cells were transfected with miR-4260 inhibitor or control for 48 h. Total RNA was labeled with a low-input QuickAmp labeling kit (Agilent Technologies, Santa Clara, CA, USA) and subsequently hybridized via Agilent gene-expression hybridization kit (Agilent Technologies). Microarray expression data were analyzed with GeneSpring software (Agilent Technologies). Various expressed lncRNAs (P < .05 and fold change >2.0) were screened out.
Bioinformatics analysis
According to GEPIA (http://gepia.cancer-pku.cn/index.html), the expression profiling of LINC01301, PLAC4, APCDD1 L-AS1, LINC01128 and LINC01398 in AML tissues and normal tissues was acquired. The binding site of miR-4260 in LINC01128 sequences was obtained from Starbase 3.0 (http://starbase.sysu.edu.cn/). NR3C2 was identified as the potential target gene of miR-4260 via miRDB (http://mirdb.org/).
Luciferase reporter assay
LINC01128 or NR3C2 fragment containing the wild-type or mutant sequence of miR-4260 was synthesized and subcloned into the pmirGLO luciferase reporter vector (Promega, Madison, WI, USA) to construct the reporter vector LINC01128-WT/Mut and NR3C2-WT/Mut. 293 T cells were co-transfected with reporter vector LINC01128-WT/Mut or NR3C2-WT/Mut and miR-4260 mimic or NC mimic via Lipofectamine 2000. Forty-eight hour later, relative luciferase activity was analyzed by the dual-luciferase reporter assay system (Promega).
RNA immunoprecipitation (RIP) assay
A Magna RIP™ RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was applied for carrying out RIP assay following the relevant requirements. HL60 or KG-1a cells were reaped and lysed in complete radioimmunoprecipitation assay buffer (RIPA; Thermo Fisher Scientific) supplemented with RNase inhibitor (Thermo Fisher Scientific) and a protease inhibitor cocktail (Sigma-Aldrich). Cell extracts were then maintained with RIP buffer containing magnetic beads (Invitrogen) conjugated with an antibody against Ago2 (Millipore) or IgG (Millipore). Proteinase K (Absin, Shanghai, China) was used to digest proteins. Obtained RNA was subjected to RT-qPCR.
Western blot analysis
RIPA lysis buffer was utilized for extracting total protein from transfected HL60 or KG-1a cells. Equal amounts of total protein were separated to incubate with primary antibody against NR3C2 (1/250, ABIN396956, Antibodies-online, Beijing, China), and then with secondary antibodies. GAPDH was used for qualifying band intensities.
Statistical analysis
Results were imported into Prism 6.0 (GraphPad Software, La Jolla, CA, USA) and presented as mean ± standard deviation (SD). At least three independent experiments were required. Student’s t-test or ANOVA was performed to analyze statistical significance. A value of P < .05 denoted statistical significance.
Results
MiR-4260 was upregulated in AML, and knocking down miR-4260 prevented proliferation and facilitated apoptosis of AML cells
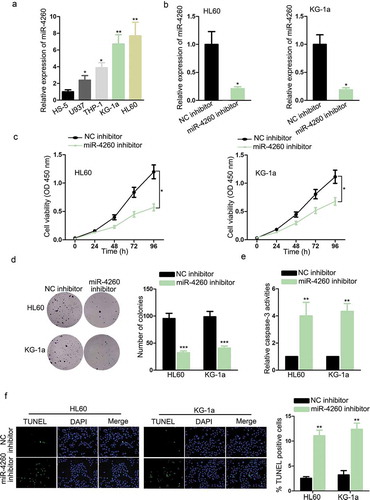
In the beginning, the implication of miR-4260 in AML was explored by examining its expression in cell lines. As a result, miR-4260 was conspicuously upregulated in AML cell lines versus normal cell line, with HL60 and KG-1a expressing the highest miR-4260 level (). Thereafter, we detected the functional role of miR-4260 in AML. HL60 and KG-1a cells transfected with miR-4260 inhibitor were confirmed to present the obvious downregulation of miR-4260 (). The absence of miR-4260 significantly reduced the proliferation of two AML cell lines (). Besides, trypan blue exclusion assay indicated that cell proliferation was inhibited in miR-4260 inhibitor-transfected cells (Figure S1A). The caspase-3 activity was evaluated to examine the apoptosis level of AML cells. MiR-4260 silence caused an increase in caspase-3 activity of both AML cells (). Additionally, TUNEL-stained AML cells were detected to increase upon the inhibition of miR-4260 (). Hence, it can be summarized that miR-4260 was upregulated in AML, and miR-4260 deficiency prevented proliferation and facilitated apoptosis of AML cells.
LINC01128 could sponge miR-4260 in AML
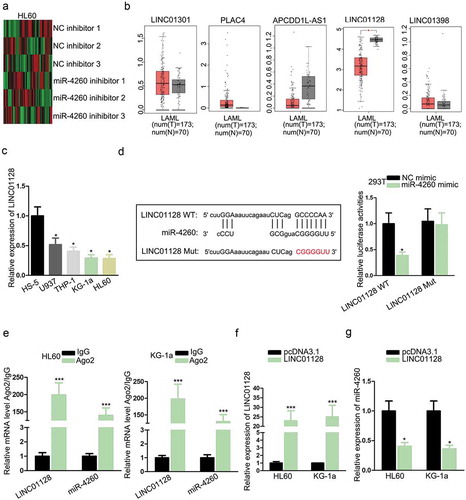
It has been reported that miRNAs could interact with and formed a negative feedback loop with lncRNAs in cancers and diseases.Citation20,Citation21 Therefore, we tried to identify the potent lncRNAs interacting with miR-4260 in AML. By searching lncBase (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r = lncbasev2%2 Findex-predicted), we found 379 candidate lncRNAs potentially combined with miR-4260. We detected the expression of these lncRNAs under the inhibition of miR-4260 in HL60 cells. We found that 34 lncRNAs were significantly upregulated while 28 were downregulated, and the 5 most upregulated lncRNAs were LINC01301, PLAC4, APCDD1L-AS1, LINC01128, and LINC01398 (). To further narrow the selection, we analyzed the expressions of 5 lncRNAs in AML samples through GEPIA (http://gepia.cancer-pku.cn/). Consequently, only LINC01128 exhibited significant downregulation in AML samples versus normal samples (). Hence, we put our focus on LINC01128. RT-qPCR results verified the low expression of LINC01128 in AML cell lines (). To detect the interplay between LINC01128 and miR-4260, we obtained the binding sites from lncBase and mutated the binding sites for luciferase reporter assay (). Overexpression of miR-4260 reduced the luciferase activity of LINC01128 WT reporter rather than LINC01128 Mut reporter (). Also, LINC01128 and miR-4260 were both abundant in the precipitates of Ago2 as shown by RIP analysis (). Additionally, we detected the influence of LINC01128 on miR-4260 expression. The overexpression of LINC01128 in HL60 and KG-1a cells were confirmed b by RT-qPCR (). The expression of miR-4260 dropped in response to the overexpression of LINC01128 (). In the collection, we revealed that LINC01128 acted as a sponge of miR-4260 in AML.
LINC01128 upregulated NR3C2 by targeting miR-4260 in AML
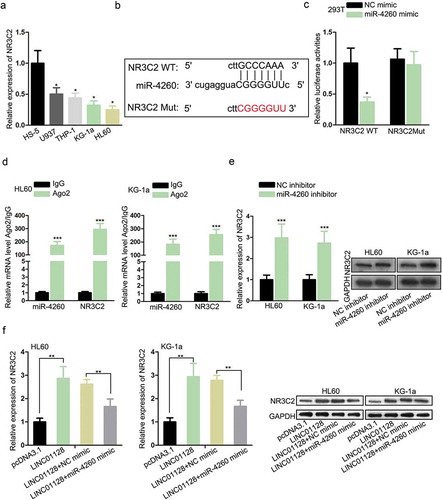
As axiomatically known, lncRNAs could competitively interact with miRNAs to save downstream genes from mRNA degradation or translation inhibition,Citation22,Citation23 including in AML.Citation15,Citation24 Thus, we exploited the downstream target for miR-4260. Through browsing miRDB (http://mirdb.org/), we discovered that NR3C2 was potently targeted by miR-4260. NR3C2 has been indicated by a number of works to perform as an anti-tumor gene in cancers,Citation17,Citation18 and its downregulation in AML has been reported by a previous study.Citation19 Hence, we speculated that miR-4260 could regulate NR3C2 in AML. RT-qPCR analysis confirmed that NR3C2 was downregulated in AML cell lines (). The miR-4260 sites on NR3C2 were identified through miRDB and the mutant sites were designed and presented in . Luciferase activity of NR3C2 WT reporter, rather than NR3C2 Mut reporter, was attenuated as a result of miR-4260 overexpression (). RIP assay demonstrated that NR3C2 mRNA and miR-4260 expressions were enriched by the immunoprecipitated products by Ago2 antibody (). Besides, the downregulation of miR-4260 resulted in an increased level of NR3C2 mRNA and protein in AML cells (). Furtherly, we observed that mRNA and protein levels of NR3C2 were induced by LINC01128 overexpression in AML cells, and such a result was reversed by miR-4260 mimic (). Altogether, the results suggested that LINC01128 upregulated NR3C2 by targeting miR-4260 in AML.
Role of LINC01128/miR-4260/NR3C2 axis in AML
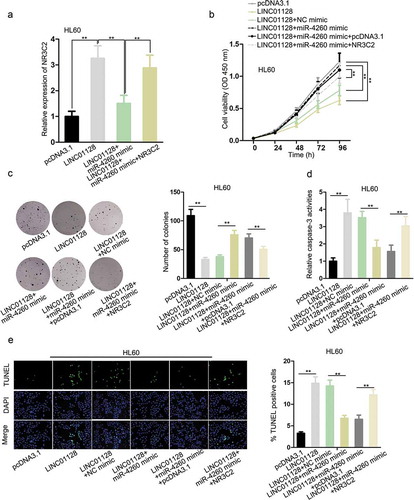
To detect the role of LINC01128/miR-4260/NR3C2 axis in AML, we designed rescue assays. RT-qPCR data demonstrated that high expression of NR3C2 in HL60 cells triggered by LINC01128 overexpression was reduced by miR-4260 mimic, and was further recovered by the transfection of pcRNA3.1/NR3C2 (). The inhibited proliferation of HL60 cells transfected with pcDNA3.1/LINC01128 was recovered by miR-4260 upregulation, and transfection of NR3C2 reversed such result (). Then, trypan blue exclusion assay indicated that the restrained cell proliferation triggered by LINC01128 overexpression could be compensated by enforced expression of miR-4260, and this compensation was reversed by additional upregulation of NR3C2 (Figure S1B). Besides, the apoptosis of HL60 cells induced by LINC01128 was retarded by miR-4260, and recovered by NR3C2 overexpression (). In sum, it was indicated that LINC01128 inhibited the proliferation and improved apoptosis of AML cells through regulating miR-4260/NR3C2 axis.
Discussion
As widely known, genetic abnormalities are implicated in AML pathogenesis through disrupting the growth differentiation, and metastasis of normal hematopoietic progenitor cells.Citation25,Citation26 MiRNAs are increasingly identified as essential participants in AML by exerting anti-tumor or oncogenic behaviors. For example, miR-628 inhibited AML proliferation through repressing IGF-1 R expressionCitation27; miR-21 was induced by STAT5 and promoted AML progression by targeting the tumor-suppressor gene PDCD4.Citation28 A previous study has stated that miR-4260 was upregulated in CRC and its overexpression facilitated CRC cell proliferation and prevented apoptosis,Citation9 indicating that miR-4260 could serve as a carcinogenic miRNA in human cancers. Present study firstly revealed the high expression of miR-4260 in AML cell lines and demonstrated that miR-4260 knockdown retarded proliferation and increased apoptosis in AML cells. These findings suggested that miR-4260 positively regulated AML progression in vitro.
Formerly, an ocean of studies has shown the interplay and reciprocal repression between lncRNAs and miRNAs in human cancers and diseases.Citation20,Citation21 Herein, our study firstly predicted through lncBase and confirmed that miR-4260 interacted with LINC01128. We found that LINC01128 was among the top 5 upregulated lncRNAs responding to miR-4260 silence in AML cells and LINC01128 demonstrated remarkable downregulation in AML samples according to bioinformatics analysis. These findings indicated that LINC01128 could sequester miR-4260 and participated in AML. Expectedly, we confirmed the low LINC01128 level in AML cell lines and showed that LINC01128 could inhibit miR-4260 in AML cells.
Moreover, accumulating works have delineated the function of lncRNAs mediating ceRNA networks in AML, whereby lncRNAs prevented miRNAs from resulting in the post-transcriptional silence of target genes.Citation15,Citation24 The current study firstly found through miRDB that NR3C2 was targeted by miR-4260. Formerly, multiple studies indicated that NR3C2 was a tumor-suppressor gene in cancer progression, which could negatively inhibit proliferation, migration, invasion, and EMT of cancer cells.Citation17,Citation18 A study also showed that the NR3C2 level was reduced in AML samples.Citation19 Accordingly, the present study validated that NR3C2 expression was suppressed in AML cell lines, and that miR-4260 combined with NR3C2 mRNA to repress its expression. In addition, we confirmed that LINC01128 upregulated NR3C2 expression through sequestering miR-4260. The rescue experiments indicated that LINC01128 attenuated the proliferation and encouraged the apoptosis of AML cells through miR-4260/NR3C2 axis.
In a word, our study firstly uncovered that LINC01128 resisted acute myeloid leukemia through regulating miR-4260/NR3C2, supplying new targets to push forward the treatment advance of AML.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (1.2 MB)Acknowledgments
Thank you for all involved in this study.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Somintara S, Leardkamolkarn V, Suttiarporn P, Mahatheeranont S. Anti-tumor and immune enhancing activities of rice bran gramisterol on acute myelogenous leukemia. PLoS One. 2016;11:e0146869–e. doi:10.1371/journal.pone.0146869.
- Bi L, Sun L, Jin Z, Zhang S, Shen Z. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol Lett. 2018;15:5611–5619. doi:10.3892/ol.2018.8050.
- Li S, Mason CE, Melnick A. Genetic and epigenetic heterogeneity in acute myeloid leukemia. Curr Opin Genet Dev. 2016;36:100–106. doi:10.1016/j.gde.2016.03.011.
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi:10.1038/nrc989.
- Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–e58. doi:10.1016/S1470-2045(12)70073-6.
- Monroig PDC, Chen L, Zhang S, Calin GA. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev. 2015;81:104–116. doi:10.1016/j.addr.2014.09.002.
- Albano F, Anelli L, Zagaria A, Coccaro N, Casieri P, Minervini A, Specchia G. SETBP1 and miR_4319 dysregulation in primary myelofibrosis progression to acute myeloid leukemia. J Hematol Oncol. 2012;5(1):48. doi:10.1186/1756-8722-5-48.
- Liu L, Ren W, Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell Physiol Biochem. 2017;41:1981–1992. doi:10.1159/000475277.
- Xiao J, Lv D, Zhou J, Bei Y, Chen T, Hu M, Zhou Q, Fu S, Huang Q. Therapeutic Inhibition of miR-4260 suppresses colorectal cancer via targeting MCC and SMAD4. Theranostics. 2017;7(7):1901–1913. doi:10.7150/thno.19168.
- Morlando M, Ballarino M, Fatica A. Long non-coding RNAs: new players in hematopoiesis and leukemia. Front Med (Lausanne). 2015;2:23. doi:10.3389/fmed.2015.00061.
- Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi:10.1038/bjc.2013.233.
- Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–376. doi:10.1016/j.semcdb.2011.01.001.
- Zhang Y, Huang Z, Sheng F, Yin Z. MYC upregulated LINC00319 promotes human acute myeloid leukemia (AML) cells growth through stabilizing SIRT6. Biochem Biophys Res Commun. 2018;509(1):314–321.
- Zhang T-J, Zhou J-D, Zhang W, Lin J, Ma J-C, Wen X-M, Yuan Q, Li -X-X, Xu Z-J, Qian J, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10(1):47. doi:10.1186/s13148-018-0486-z.
- Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39:330–336.
- Horisberger JD, Rossier BC. Aldosterone regulation of gene transcription leading to control of ion transport. Hypertension. 1992;19:221–227. doi:10.1161/01.HYP.19.3.221.
- Zhang Z, Che X, Yang N, Bai Z, Wu Y, Zhao L, Pei H. miR-135b-5p promotes migration, invasion and EMT of pancreatic cancer cells by targeting NR3C2. Biomed Pharmacother. 2017;96:S0753332217353143. doi:10.1016/j.biopha.2017.11.074.
- Zhao Z, Zhang M, Duan X, Deng T, Qiu H, Zeng G. Low NR3C2 levels correlate with aggressive features and poor prognosis in non-distant metastatic clear-cell renal cell carcinoma. J Cell Physiol. 2018;233(10):6825–6838. doi:10.1002/jcp.v233.10.
- Long MD, Campbell MJ. Pan-cancer analyses of the nuclear receptor superfamily. Nucl Receptor Res. 2015;2:101182. doi:10.11131/2015/101182.
- Tan J, Qiu K, Li M, Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589:3175–3181. doi:10.1016/j.febslet.2015.08.020.
- Tian W, Du Y, Ma Y, Gu L, Zhou J, Deng D. MALAT1-miR663a negative feedback loop in colon cancer cell functions through direct miRNA-lncRNA binding. Cell Death Dis. 2018;9:857. doi:10.1038/s41419-018-0925-y.
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi:10.1016/j.cell.2011.07.014.
- Zhan Y, Chen Z, Li Y, He A, He S, Gong Y, Li X, Zhou L. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37(1):273. doi:10.1186/s13046-018-0921-1.
- Yin X, Huang S, Zhu R, Fan F, Sun C, Hu Y. Identification of long non-coding RNA competing interactions and biological pathways associated with prognosis in pediatric and adolescent cytogenetically normal acute myeloid leukemia. Cancer Cell Int. 2018;18:122. doi:10.1186/s12935-018-0621-0.
- Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, Habdank M, Späth D, Morgan M, Benner A, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. doi:10.1056/NEJMoa074306.
- Woods BA, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Immunol Rev. 2015;263:22–35. doi:10.1111/imr.12246.
- Chen L, Jiang X, Chen H, Han Q, Liu C, Sun M. microRNA-628 inhibits the proliferation of acute myeloid leukemia cells by directly targeting IGF-1R. Onco Targets Ther. 2019;12:907–919. doi:10.2147/OTT.S192137.
- Espadinha AS, Prouzet-Mauléon V, Claverol S, Lagarde V, Bonneu M, Mahon FX, Cardinaud B. A tyrosine kinase-STAT5-miR21-PDCD4 regulatory axis in chronic and acute myeloid leukemia cells. Oncotarget. 2017;8:76174–76188. doi:10.18632/oncotarget.19192.