ABSTRACT
Background
Deemed as a member of malignant tumors, hepatocellular carcinoma (HCC) has been characterized as a lethal disease with high morbidity and mortality. It has been widely accepted that long noncoding RNAs (lncRNAs) play a big part in the complicated biologic processes of cancer.
Aim of the study
The purpose of the study is to figure out the role and molecular regulation mechanism of ST8SIA6-AS1 in HCC.
Methods
The role of ST8SIA6-AS1 in HCC was validated by RT-qPCR, colony formation, ki-67 detection, TUNEL, JC-1 detection, wound healing and transwell-invasion assays, furthermore, the binding ability between ST8SIA6-AS1/HOXB6 and miR-5195-3p were confirmed by RNA pull down and luciferase reporter assays. Besides, the regulatory mechanism of ST8SIA6-AS1 to HOXB6/miR-5195-3p was measured by RT-qPCR and western blot assays.
Results
We measured that ST8SIA6-AS1 was highly expressed in HCC cell lines. Then knockdown of it suppressed cell proliferation, migration and migration but activated cell apoptosis in HCC. Furthermore, ST8SIA6-AS1 could bind with miR-5195-3p and negatively regulated its expression in HCC. Subsequently, it confirmed that HOXB6 was target gene of miR-5195-3p and positively modulated by ST8SIA6-AS1 in HCC. Finally, we verified that miR-5195-3p deficiency or HOXB6 upregulation countervailed the repressing effects of ST8SIA6-AS1 depletion on HCC progression.
Conclusions
To sum up, ST8SIA6-AS1 promotes HCC progression by absorbing miR-5195-3p to regulate HOXB6, which might provide some worthy suggestions to research the development process of HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fourth main cause of cancer-related death across the world, capturing public attention.Citation1 Major risk factors of HCC include Hepatitis B virus (HBV), Hepatitis C virus (HCV) infections and alcohol intake.Citation2 The development of HCC is featured with accelerated cell growth, enhanced cell metastasis and inhibited cell apoptosis.Citation3 Although quite a few treatments have been introduced, such as radiotherapy, chemotherapy and surgical resection,Citation4 the overall prognosis of HCC is still unsatisfactory.Citation1 Hence, further exploring regulatory mechanisms of HCC occurrence would provide new insights for improving prognosis and finding novel treatment agents.
Long non-coding RNAs (lncRNAs), consisting of over 200 nucleotides in length, is a typical class of non-coding RNAs (ncRNAs), lacking protein-coding ability.Citation5,Citation6 Previous researches have elucidated that lncRNAs exert critical function in cancer occurrence and development. For example, lncRNA SNHG1 acts as an oncogene in non-small cell lung cancer via suppressing miR-101-3p and activating Wnt/β-catenin pathway.Citation7 LncRNA CCAT1 modulates miR-148b to weaken radiosensitivity of breast cancer cells.Citation8 Besides, the significance of lncRNAs in HCC has also been validated by extensive researches. For instance, lncRNA AWPPH interacts with YBX1 to play its oncogenic role in HCC.Citation9 LncRNA MIAT sponges miR-214 to promote HCC development.Citation10 LncRNA GHET1 represses KLF2 expression to accelerate cell growth in HCC.Citation11 More importantly, it validated that lncRNA ST8SIA6-AS1 contributed to cell proliferation, migration and invasion in breast cancer.Citation12,Citation13 In the recent researches, ST8SIA6-AS1 (also named as APAL) expression was upregulated in diverse human cancers, related to unsatisfactory clinical outcome of patients. ST8SIA6-AS1 downregulation resulted in mitotic catastrophe and accelerated apoptosis of breast, lung and pancreatic cancer cells. Upregulating ST8SIA6-AS1 facilitated cancer cell cycle progression, promoting proliferation and inhibiting chemotherapy-induced apoptosis.Citation14 Nevertheless, whether ST8SIA6-AS1 took part in the progression of HCC remained obscure and was worthy of further investigation.
This paper attempted to obtain a thorough understanding of the biological role and molecular mechanisms of ST8SIA6-AS1 in HCC, and our findings might contribute to provide some novel insights for HCC researches.
Materials and methods
Cell culture
The cancer cells (SNU-398, SK-Hep-1, SNU-182 and Hep3B) and the normal liver cells (THLE-3) were brought from Cell Biology of the Chinese Academy of Sciences (Shanghai, China). And DMEM medium was utilized to deposit the cells in the humid environment at 37°C. Additionally, the cells had already been supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin for cultivating with 5% CO2.
Cell transfection
Cells at approximately 70–80% cell fusion were reaped for 48 h of plasmid transfection by employing the Lipofectamine 2000 (Invitrogen). The shRNAs specific to ST8SIA6-AS1 or HOXB6 (sh-ST8SIA6-AS1#1/2 or sh-HOXB6#1/#2), negative control (NC; sh-NC), the pcDNA3.1/ST8SIA6-AS1 or pcDNA3.1/HOXB6 and NC (pcDNA3.1), were synthesized at the RiboBio (Guangzhou, China). The miR-5195-3p mimics and NC mimics, miR-5195-3p inhibitor and NC inhibitor, were all simultaneously procured from RiboBio. The transfection was completed in 48 hours later, and cancer cells were gathered for further experiments.
Real-time quantitative polymerase chain reaction (RT-qPCR) assay
On the basis of the protocols of suppliers, TRIzol reagent (Invitrogen) was adopted to extract the total RAN of Hep 3B, SNU-398 or THLE-3 cells. Then, RNA was subjected to reversely transcription to cDNA by the use of M-MLV Reverse Transcriptase. SYBR® Premix Ex Taq™ II was adopted to conduct the RT-qPCR on System of Applied Biosystems 7500 Real-time PCR. GAPDH or U6 was served as endogenous control.
Colony formation assay
A six-well plate was used to seed the transfected Hep 3B, SNU-398 or THLE-3 cells, which then were put in a culture medium containing 10% FBS. After 24 h incubation, the culture medium was replaced. The methanol and 0.1% crystal violet were separately utilized to fix and stain the cells after 14-day cultivation. At last, the cell colony formation capacity was evaluated by counting the number of stained colonies.
Fluorescence in situ hybridization (FISH) analysis
With regard to the FISH assay, Biosearch Technologies devise the probes that were conjugated with fluorescence. And the probes were incubated with the cell samples, which underwent non-denaturation treatment before. Then, antibodies were utilized to conduct the double FISH and the cell nuclei were counterstained by Hoechst. Finally, confocal microscopy was utilized to observe the cell staining.
Transwell-invasion assay
With regard of the experiment of invasion, Hep 3B, SNU-398 or THLE-3 cells were cultivated in the serum-free medium and deposited within the upper chamber containing matrigel. And 10% FBS was added into the lower chamber. After the 48 h cultivation, the cotton swabs were used to wipe off those cells which failed to cross over the membrane. Finally, crystal violet was utilized to stain the cells in the lower chamber.
Immunofluorescence
After 24 h cultivation, 4% paraformaldehyde and 0.2% Triton X-100 were utilized to fix and permeabilize the cells. Then cells were blocked by the use of 5% bovine serum albumin (BSA) for half an hour. Afterward, Ki-67 was adopted to label the cell samples for two hours at 37°C. Then the cells were cultivated with IgG which was coupling with Cy3 for half an hour at RT. Then DAPI was utilized to stain the nucleus. Finally, Olympus IX-71 and Olympus DP2-BSW software were adopted to observe and analyze the cells, separately.
Terminal-deoxynucleoitidyl transferase mediated nick end labeling (TUNEL) assay
In accordance with the user guide, a Colorimetric TUNEL Apoptosis Assay Kit was used to test cell apoptosis. After pretreated by CDMC, cells were fixed by 4% fresh paraformaldehyde for half an hour. Then they were cultivated with 0.1% Triton™ X-100 for 2 minutes at 4°C after rinsed by PBS. Cells were washed by H2O2 in methanol, and then rinsed by PBS for three times. Afterward, TdT was performed with Bio-16-dUTP for at least 1 hour incubation at RT. DAPI was utilized to stain cells for ten minutes. Finally, TUNEL positive cells were calculated.
Wound healing assay
6-well plates were adopted to cultivate the Hep 3B, SNU-398 or THLE-3 cells at 37°C. When cells were almost fusing together, the pipette tip was used to make a scratch on cells. Subsequently, the previous medium was replaced with serum-free medium. At 24 hours post cultivation, the width of wound was calculated.
JC-1 staining
JC-1 working solution was utilized to cultivate the Hep 3B, SNU-398 or THLE-3 cells in the dark for twenty minutes at 37°C and fluorescence microscopy was adopted to observe the cells. PBS was used to wash the fluorescence labeled cells, which were analyzed by an EnSpire Reader. GraphPad Prism 5 statistical software was applied to process the data and images. In normal mitochondria, JC-1 aggregated to form a polymer in the mitochondrial matrix, and the polymer emitted strong red fluorescence; unhealthy mitochondria could only exist in the cytoplasm as monomers due to the decrease or loss of membrane potential to produce green fluorescence.
Animal studies
5 week-old female BALB/c nude mice (Tianjin First Central Hospital) were used in this experiment. Mice were randomly divided into two groups (5 mice each group), including sh-NC and sh-ST8SIA6-AS1#1 groups. Mice were sacrificed 28 days’ post injection. The tumor volume was calculated using the formula “tumor volume = length × width × width/2”. Tumor weight was also measured. The animal experiments were approved by the Ethics Review Committee of Tianjin First Central Hospital.
Immunohistochemical staining (IHC)
The paraffin sections were prepared for hematoxylin and eosin (H&E) staining and IHC staining. The sections were dewaxed with xylene, and then hydrated with fan ethanol solution, and EDTA was added for antigen recovery. Subsequently, normal goat serum was used to block the sections for half an hour to eliminate nonspecific binding. Sections were cultivated with anti-ki-67 and anti-PCNA. Similarly, the sections were cultivated with biotin-labeled secondary antibodies for half an hour at room temperature and then stained by diaminobenzidine (DAB). Finally, the sections were counterstained with hematoxylin.
Subcellular fractionation
The part of cytoplasm and nuclear were extracted respectively by Cytoplasmic and Nuclear RNA Purification Kit. Then the RT-qPCR assay was adopted to test the relative expression of ST8SIA6-AS1, GAPDH and U6 were served as endogenous controls in cytoplasm or nuclear, respectively.
Luciferase reporter assay
The predicted binding sites of miR-5195-3p with ST8SIA6-AS1 and HOXB6-3ʹUTR were obtained from starBase. The binding and mutant sequences were respectively cloned into pmirGLO Dual-luciferase vectors (Gene Pharma, Shanghai, China). Hep 3B and SNU-398 cells were seeded in 96-well plates and transfected with the wild-type pmirGLO-ST8SIA6-AS1/HOXB6-3ʹUTR reporter plasmid or the mutated type and miR-5195-3p mimics or NC mimics with Lipofectamine 2000. At 48 hours post the cultivation, dual luciferase assay system was used to estimate luciferase activity, which was recorded as the ratio of firefly luciferase activity to renilla luciferase activity.
Western blot analysis
Total proteins were extracted by utilizing RIPA buffer reagent (Thermo Fisher Scientific). The concentration of the lysate was measured by a BCA kit (Thermo Fisher Scientific). Proteins were separated using 10% SDS-PAGE gels and transferred into PVDF membranes. After blocked by the 5% milk without fat, the membranes were cultivated with the primary antibodies for 15 hours in 4 °C and then cultured with secondary antibodies at room temperature for 3 hours. Western ECL Substrate Kit was adopted to detect the protein bands and ImageJ version 1.48 was utilized to analyze the densitometry. GAPDH was served as endogenous control.
RNA pull-down assay
T7 RNA polymerase and RNeasy Plus Mini Kit (Qiagen) were adopted to transcribe and purify ST8SIA6-AS1 RNAs in vitro. And RNase-free DNase I (Qiagen) was utilized to treat ST8SIA6-AS1 RNAs which were then biotin-labeled by using Biotin RNA Labeling Mix. Hep 3B and SNU-398 cell lysates were utilized to cultivate the mixture of biotinylated RNAs. The tubes of binding reaction were supplemented with magnetic beads for cultivating at 37 C. After the beads were rinsed, proteins were estimated by the conduction of western blot.
Statistical analysis
The collected data was analyzed by the utilization of SPSS 18.0 software. Student’s t-test or ANOVA was used to compare different groups. Means ± SD was adopted to indicate all data. P-value <0.05 was considered as statistically significant.
Results
ST8SIA6-AS1 promotes HCC development in vitro and in vivo
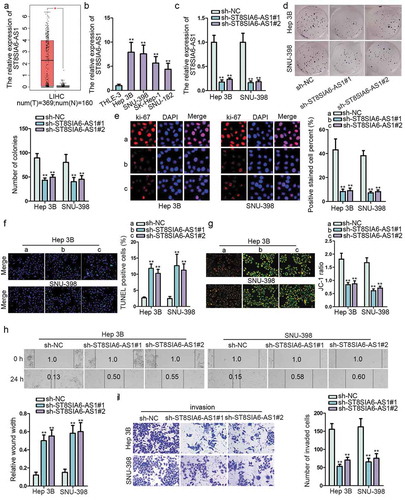
To explore the function of ST8SIA6-AS1 in HCC development, first of all, the expression of ST8SIA6-AS1 was found to be notably upregulated in LIHC (liver hepatocellular carcinoma) tissues through searching GEPIA public database online (). RT-qPCR analysis delineated that ST8SIA6-AS1 expression was evidently upregulated in HCC cell lines (Hep 3B, SNU-398, SK-Hep-1 and SNU-182) compared with normal liver epithelial cell lines (THLE-3). Besides, Hep 3B and SNU-398 cells contained the highest level of ST8SIA6-AS1 (). Then, ST8SIA6-AS1 was knocked down by transfecting sh-ST8SIA6-AS1#1/#2 into Hep 3B and SNU-398 cells, sh-NC as normal control. The results indicated that ST8SIA6-AS1 expression was markedly decreased (). Subsequently, functional experiments were performed. Colony formation and Ki-67 detection assays examined that ST8SIA6-AS1 deficiency inhibited the proliferation of HCC cells (,e). Meanwhile, TUNEL and JC-1 staining assays illustrated that knockdown of ST8SIA6-AS1 encouraged HCC cell apoptosis (,g). Moreover, ST8SIA6-AS1 depletion suppressed cell migration in HCC, which was elucidated by wound healing assay (). Besides, transwell assay validated that ST8SIA6-AS1 knockdown repressed cell invasion (). To further confirm the tumor facilitator role of ST8SIA6-AS1, gain-of-function experiments were conducted in THLE-3 cells. ST8SIA6-AS1 expression was increased in pcDNA3.1/ST8SIA6-AS1 transfected THLE-3 cells (Figure S1A). Then ST8SIA6-AS1 upregulation contributed to cell proliferation, migration and invasion, but obstructed cell apoptosis (Figure S1B-1G). Additionally, in vivo assays were carried out. ST8SIA6-AS1 knockdown inhibited tumor growth, including tumor volume and weight (Figure S2A-S2C). Results of H&E staining and IHC staining assays certified that ST8SIA6-AS1 depletion also decreased the tumor metastasis nodes as well as the expression of ki-67 and PCNA (two tumor cell proliferation factors) in tissue sections obtained from mice (Figure S2D). Overall, ST8SIA6-AS1 is highly expressed in HCC cells and it promotes HCC development both in vitro and in vivo.
ST8SIA6-AS1 sponges miR-5195-3p in HCC
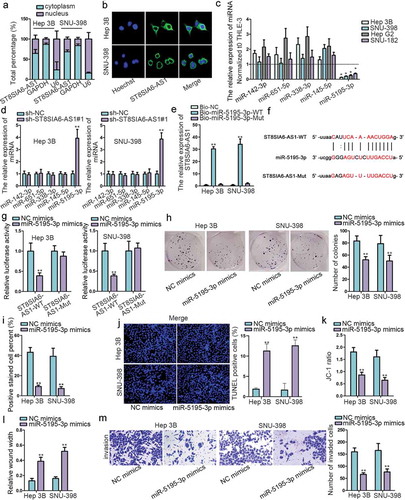
To investigate the regulatory mechanism of ST8SIA6-AS1 in HCC cells, subcellular fractionation and FISH assays detected that the expression of ST8SIA6-AS1 mainly distributed in cytoplasm, indicating the post-transcription regulation possibility of ST8SIA6-AS1 (,b), which was further confirmed by bioinformatics tool (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) (Figure S3A). Afterward, starBase was utilized to find out five possible downstream miRNAs of ST8SIA6-AS1 (miR-142-3p, miR-651-5p, miR-338-3p, miR-145-5p and miR-5195-3p) (Figure S3B). Then, RT-qPCR assay suggested that miR-5195-3p expression was downregulated the most in HCC cells and most regulated by sh-ST8SIA6-AS1 (,d). Therefore, miR-5195-3p was chosen as the focus in following researches. RNA pull-down assay tested that ST8SIA6-AS1 was enriched in Bio-miR-5195-3p-WT group but not in Bio-NC group or Bio-miR-5195-3p-Mut group (). As shown in , the binding site between miR-5195-3p and ST8SIA6-AS1 was predicted by starBase. Then luciferase reporter assay found that the luciferase activity of ST8SIA6-AS1-WT was significantly reduced by miR-5195-3p mimics compared with NC group while that of ST8SIA6-AS1-Mut presented no obvious change (). Similarly, several functional assays were employed to comprehend the role of miR-5195-3p in HCC. According to the data, on one hand, miR-5195-3p overexpression suppressed cell proliferation (,i and S3C), but activated cell apoptosis (,k and S3D). Furthermore, HCC cell migration and invasion were suppressed by miR-5195-3p upregulation (, S3E and 2M). On the other hand, miR-5195-3p inhibitor was also used in functional assays. The knockdown efficiency of miR-5195-3p was certified in THLE-3 cells firstly (Figure S4A). When silencing miR-5195-3p, cell proliferation, migration and invasion were induced, cell apoptosis was repressed (Figure S4B-S4G). In a word, ST8SIA6-AS1 sponges miR-5195-3p, which serves as a tumor inhibitor in HCC.
ST8SIA6-AS1 regulates HOXB6 expression by competitively binding with miR-5195-3p in HCC
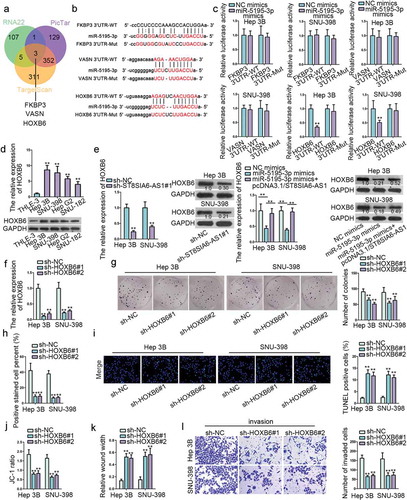
Through searching starBase, three messenger RNAs (mRNAs) including FKBP3, VASN and HOXB6 were found to have binding sites for miR-5195-3p (,b). Subsequently, luciferase reporter assay verified whether above binding sites were effective, and it found that miR-5195-3p mimics only had decreasing effects on the luciferase activity of HOXB6 3ʹUTR-WT (). Accordingly, RT-qPCR and western blot detected that the RNA and protein expressions of HOXB6 was remarkably upregulated in HCC cells compared with that in normal liver epithelial cells (). As displayed in , both ST8SIA6-AS1 knockdown and miR-5195-3p mimics reduced the mRNA and protein levels of HOXB6, yet the suppressive role of miR-5195-3p mimics was offset by ST8SIA6-AS1 overexpression. Similarly, the role of HOXB6 in HCC was explored. Firstly, the knockdown efficiency of HOXB6 was confirmed (). Then we observed that HOXB6 downregulation hindered cell proliferation (,h and S5A) and promoted cell apoptosis (,j and S5B), cell migration ( and S5 C) and invasion () were also inhibited by HOXB6 deficiency. Besides, several rescue experiments were performed. HOXB6 overexpression neutralized the inhibitory influence of miR-5195-3p mimics in cell proliferation, migration and invasion as well as the facilitating influence of that in cell apoptosis (Figure S6A-6F). Taken together, ST8SIA6-AS1 competitively binds with miR-5195-3p to upregulate expression of HOXB6, which promotes the development of HCC.
ST8SIA6-AS1 promotes HCC progression via regulating miR-5195-3p/HOXB6 axis
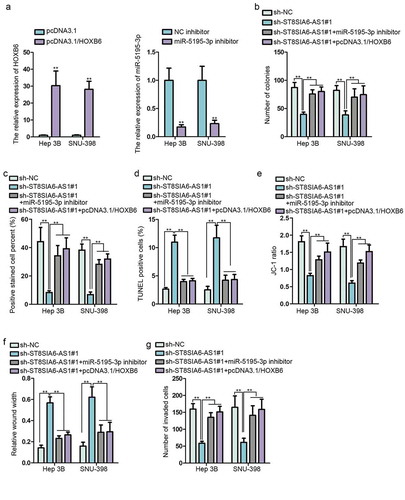
To further confirm the regulatory mechanism of ST8SIA6-AS1/miR-5195-3p/HOXB6 network in HCC, it was of great necessity to conduct the following assays. The results of RT-qPCR assays displayed that HOXB6/miR-5195-3p expression was obviously increased/decreased by pcDNA3.1/HOXB6/miR-5195-3p inhibitor (). Colony formation assay showed that miR-5195-3p knockdown or HOXB6 upregulation reversed ST8SIA6-AS1 depletion-mediated function on the cell proliferation (,c). Besides, miR-5195-3p inhibition or HOXB6 overexpression offset the ST8SIA6-AS1 suppression-mediated function on cell apoptosis (,e). Moreover, miR-5195-3p deficiency or HOXB6 augmentation countervailed the obstructing function of ST8SIA6-AS1 silencing on cell migration and invasion (,g). In summary, ST8SIA6-AS1 promotes HCC progression via regulating miR-5195-3p/HOXB6 axis.
Discussion
Numerous attempts have been dedicated to uncover the role of lncRNAs in cancer development. It is convinced that lncRNAs function as either tumor suppressers or promoters in the malignant processes of cancers, such as non-small cell lung cancer, breast cancer and HCC.Citation7,Citation8,Citation10 Our study revealed that ST8SIA6-AS1 was notably upregulated in LIHC tissues and HCC cell lines via public database and RT-qPCR analysis, respectively. ST8SIA6-AS1 silencing inhibited cell proliferation, migration and invasion as well as promoted cell apoptosis. Tumor growth was also suppressed by ST8SIA6-AS1 depletion, implying that ST8SIA6-AS1 is an oncogenic gene in HCC.
LncRNAs have been reported to competitively sponge microRNAs (miRNAs), thus weakening the regulatory effects of those miRNAs on downstream mRNAs.Citation15,Citation16 MiRNAs are another fraction of non-coding RNAs, with a length of 20–24 nucleotides, participating in the regulation of cancer development.Citation17,Citation18 As an illustration, lncRNA-UCA1 functions as a sponge of miR-204 to regulate Sox4 expression, thus promoting the occurrence of esophageal cancer.Citation19 LncRNA RSU1P2 acts as an oncogene via sponging let-7a in cervical cancer.Citation20 The tumor inhibitor role of miR-5195-3p has been confirmed in several cancers, like miR-5195-3p inhibits cell proliferation and invasion in bladder cancer through targeting oncogene KLF5.Citation21 MiR-5195-3p suppresses the expression of TGFβR1, TGFβR2, SMAD3 and SMAD4 to repress HCT116 cell activity.Citation22 And our work demonstrated that miR-5195-3p could bind with ST8SIA6-AS1 and its expression was considerably downregulated in HCC cells. In addition, through gain-of-function assays, miR-5195-3p overexpression repressed cell proliferation, migration and invasion in HCC. Through loss-of-function assays, miR-5195-3p inhibitor stimulated THLE-3 cell proliferation, migration and invasion, further proving tumor-suppressor role of miR-5195-3p.
Previous studies have manifested that miRNAs are able to affect cancer development through modulating their downstream mRNAs.Citation23,Citation24 For instance, miR-25 inhibits osteosarcoma via targeting SOX4.Citation25 MiR-377 deficiency promotes oral squamous cell carcinoma through modulating HDAC9.Citation26 As an mRNA, HOXB6 has been proofed to serve as an oncogenic gene in some diseases, such as barrett esophagus,Citation27 acute myeloid leukemia.Citation28 In current study, it was confirmed that HOXB6 was overexpressed in HCC cells and could bind with miR-5195-3p. HOXB6 expression was positively/negatively regulated by ST8SIA6-AS1/miR-5195-3p. ST8SIA6-AS1 could upregulate HOXB6 expression via sequestering miR-5195-3p. Additionally, HOXB6 knockdown inhibited the progression of HCC, including cell proliferation, migration and invasion. HOXB6 upregulation counteracted suppressive effects of miR-5195-3p overexpression on HCC cell proliferation, migration and invasion. Finally, we found that miR-5195-3p inhibitor or HOXB6 upregulation rescued the inhibitory role of ST8SIA6-AS1 silencing in HCC, suggesting that ST8SIA6-AS1 could sponge miR-5195-3p to upregulate HOXB6, thus facilitating HCC development.
On the whole, ST8SIA6-AS1 promotes HCC by absorbing miR-5195-3p to upregulate HOXB6, which possibly offer a novel direction for studying HCC development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (6.8 MB)Acknowledgments
We appreciate the technical supports of laboratory members.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi:10.3322/caac.21442.
- Dimitroulis D, Damaskos C, Valsami S, Davakis S, Garmpis N, Spartalis E, Athanasiou A, Moris D, Sakellariou S, Kykalos S, Tsourouflis G, Garmpi A, Delladetsima I, Kontzoglou K, Kouraklis G, et al. From diagnosis to treatment of hepatocellular carcinoma: an epidemic problem for both developed and developing world. World J Gastroenterol. 2017;23:5282–5294. doi:10.3748/wjg.v23.i29.5282.
- Sun X, Zhuo XB, Hu YP, Zheng X, Zhao QJ. A novel matrine derivative WM622 inhibits hepatocellular carcinoma by inhibiting PI3K/AKT signaling pathways. Mol Cell Biochem. 2018;449:47–54. doi:10.1007/s11010-018-3341-9.
- Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. Report of incidence and mortality in China cancer registries, 2009. Chin J Cancer Res. 2013;25:10–21. doi:10.3978/j..1000-9604.2012.12.04.
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (New York, NY). 2007;316:1484–1488. doi:10.1126/science.1138341.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi:10.1158/0008-5472.CAN-16-2634.
- Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi:10.18632/oncotarget.14854.
- Lai Y, Chen Y, Lin Y, Ye L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol Int. 2018;42:227–236. doi:10.1002/cbin.10890.
- Zhao X, Liu Y, Yu S. Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochimica Et Biophysica Acta Mol Basis Dis. 2017;1863:1805–1816. doi:10.1016/j.bbadis.2017.04.014.
- Huang X, Gao Y, Qin J, Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am J Physiol Gastrointestinal Liver Physiol. 2018;314:G559–g65. doi:10.1152/ajpgi.00242.2017.
- Jin L, He Y, Tang S, Huang S. LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. J Cell Physiol. 2018;233:4726–4734. doi:10.1002/jcp.26257.
- Jeong G, Bae H, Jeong D, Ham J, Park S, Kim HW, Kang HS, Kim SJ. A Kelch domain-containing KLHDC7B and a long non-coding RNA ST8SIA6-AS1 act oppositely on breast cancer cell proliferation via the interferon signaling pathway. Sci Rep. 2018;8:12922. doi:10.1038/s41598-018-31306-8.
- Fang K, Hu C, Zhang X, Hou Y, Gao D, Guo Z, Li L. LncRNA ST8SIA6-AS1 promotes proliferation, migration and invasion in breast cancer through the p38 MAPK ssignalling pathway. Carcinogenesis. 2019. doi:10.1093/carcin/bgz197.
- Luo ML, Li J, Shen L, Chu J, Guo Q, Liang G, Wu W, Chen J, Chen R, Song E. The role of APAL/ST8SIA6-AS1 lncRNA in PLK1 activation and mitotic catastrophe of tumor cells. J Natl Cancer Inst. 2019. doi:10.1093/jnci/djz134.
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol (Clifton, NJ). 2016;1402:271–286.
- Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99:494–501. doi:10.1002/cpt.355.
- Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9. doi:10.1016/j.jbior.2014.09.013.
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi:10.1016/j.molmed.2014.06.005.
- Jiao C, Song Z, Chen J, Zhong J, Cai W, Tian S, Chen S, Yi Y, Xiao Y. lncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol Rep. 2016;36:2960–2966. doi:10.3892/or.2016.5121.
- Liu Q, Guo X, Que S, Yang X, Fan H, Liu M, Li X, Tang H. LncRNA RSU1P2 contributes to tumorigenesis by acting as a ceRNA against let-7a in cervical cancer cells. Oncotarget. 2017;8:43768–43781. doi:10.18632/oncotarget.10844.
- Jiang Z, Zhang Y, Cao R, Li L, Zhong K, Chen Q, Xiao J. miR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res. 2017;25:1081–1087. doi:10.3727/096504016X14831120463349.
- Jahangiri Moez M, Bjeije H, Soltani BM. Hsa-miR-5195-3P induces downregulation of TGFbetaR1, TGFbetaR2, SMAD3 and SMAD4 supporting its tumor suppressive activity in HCT116 cells. Int J Biochem Cell Biol. 2019;109:1–7. doi:10.1016/j.biocel.2019.01.001.
- Srivastava SK, Bhardwaj A, Leavesley SJ, Grizzle WE, Singh S, Singh AP. MicroRNAs as potential clinical biomarkers: emerging approaches for their detection. Biotechnic Histochem. 2013;88:373–387. doi:10.3109/10520295.2012.730153.
- D’Angelo B, Benedetti E, Cimini A, Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36:5571–5575. doi:10.21873/anticanres.11142.
- Chen B, Liu J, Qu J, Song Y, Li Y, Pan S. MicroRNA-25 suppresses proliferation, migration, and invasion of osteosarcoma by targeting SOX4. Tumour Biol. 2017;39:1010428317703841. doi:10.1177/1010428317703841.
- Rastogi B, Kumar A, Raut SK, Panda NK, Rattan V, Joshi N, Khullar M. Downregulation of miR-377 promotes oral squamous cell carcinoma growth and migration by targeting HDAC9. Cancer Invest. 2017;35:152–162. doi:10.1080/07357907.2017.1286669.
- Di Pietro M, Lao-Sirieix P, Boyle S, Cassidy A, Castillo D, Saadi A, Eskeland R, Fitzgerald RC. Evidence for a functional role of epigenetically regulated midcluster HOXB genes in the development of Barrett esophagus. Proc Natl Acad Sci USA. 2012;109:9077–9082. doi:10.1073/pnas.1116933109.
- Fischbach NA, Rozenfeld S, Shen W, Fong S, Chrobak D, Ginzinger D, Kogan SC, Radhakrishnan A, Le Beau MM, Largman C, et al. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 2005;105:1456–1466. doi:10.1182/blood-2004-04-1583.