ABSTRACT
Patients with urothelial carcinoma (UC) of the bladder have a high risk of death in China. However, a lack of comprehensive molecular profiling in Chinese Han population hinders the development of targeted therapies for bladder cancer. In our present study, we collected fresh bladder tumors from low-grade (T1, N0, M0, G1) non-muscle invasive bladder cancer (NMIBC) patients (n = 16) and high-grade (T2-4, N0, M0, Gx) muscle-invasive bladder cancer (MIBC) patients (n = 16) with their paired normal bladder tissues, and subjected the total genomic DNAs to targeted next-generation sequencing (NGS) for 94 cancer-associated genes. NGS results showed that 30.9% of detected genes (29/94) was mutated in 32 urothelial carcinoma bladder tissues. Furthermore, our results and ICGC database showed that FGFR3, KMT2D, TP53, KDM6A, and ARID1A were the most frequently mutated genes in UC patients. Of note, NMIBC and MIBC displayed distinguishable genomic alterations. FGFR3, KMT2D, AKT1, ARID1A, and STAG2 were the most frequently mutated genes in NMIBC patients, whereas mutations of TP53, CREBBP, FGFR3, KDM6A, KMT2D, and ARID1A were frequently detected in MIBC. Intriguingly, gene ontology and clustering analysis revealed that these frequently mutated genes were highly enriched in signaling pathways responsible for cancer development. Taken together, the mutation frequency of genes associated with UC development in NMIBC and MIBC was screened out in Chinese Han population and elucidation of the related mechanisms provides theoretical basis and technical support for the development of early diagnosis and therapeutic strategies in UC.
Introduction
Bladder cancer (BCa) is one of the common malignancies worldwide with global prevalence at approximately 1 million each year,Citation1 which ranked the ninth in the world with sixth in men and tenth in the middle age of women.Citation2,Citation3 In China, the overall incidence of bladder cancer was about 8.05/10 million in 2015 and continued to increase in recent years.Citation4 Epidemiological studies have shown that genetic factors,Citation5,Citation6 environmental factors such as active smoking and passive smoking,Citation7,Citation8 occupational exposure in chemical and petrochemical industry,Citation9 air pollution,Citation10 food and drink,Citation11 use of medicines,Citation12 and sexual distinctionCitation13 can all affect the development and progression of BCa. In particular, tobacco smoking and gender difference are two important risk factors for BCa incidence.Citation7,Citation8 The pathogenesis of BCa involves a series of aberrant biological processes such as rapid DNA replication, increased cell proliferation, suppressed cell apoptosis, tumor cell invasion and metastasis, immune escape, and angiogenesis.Citation14,Citation15 Molecular biological analysis also revealed that alterations in oncogenes or tumor suppressor genes led to the development of BCa.Citation16
Typically, most BCa (70-80%) are superficial tumors, also called non muscle invasive BCa (NMIBC), with Bacillus of Calmette-Guerin (BCG) or chemotherapy treatment.Citation17,Citation18 Nevertheless, half of them can relapse and 5% to 20% of NMIBC will eventually become muscle-invasive BCa (MIBC in short) despite of treatments.Citation19,Citation20 On the contrary, 50% of MIBC patients with systemic chemotherapy or radical cystectomy eventually die due to metastases within 2 years of diagnosis.Citation21 Currently, no effective strategies have been applied to cure MIBC, suggesting an emergent development of novel targeted therapies to cure MIBC is needed.
Next generation sequencing (NGS) allows researchers to study biological functions at a massively broad level. NGS has been applied to decipher genomic alterations in BCa by numerous groups.Citation22–Citation26 Indeed, the active forms of FGFR3 and HRAS due to genomic mutation have been identified in the early stage of BCa, serving as biomarkers for NMIBC.Citation27–Citation30 Additionally, TP53 mutation and loss of Rb were associated with MIBC incidence.Citation6,Citation31-Citation33 These facts indicated that a better understanding of the genomic landscape of BCa was essential to improve its therapeutic strategies. Despite of its significance, the molecular profile of BCa at the genomic level in Chinese Han population, however, hasn’t been investigated yet. Here, we performed targeted NGS to test the mutation frequency of 94 well-known cancer-associated genes in 32 bladder cancer tissues. Our aim was to explore the potential biomarkers for BCa diagnosis and prognosis in order to provide novel targets for the treatment of BCa.
Our results showed that 30.9% of detected genes were mutated in our study cohort. MIBC and NMIBC displayed a unique mutation signature. Those unique-mutated genes may serve as potential biomarkers or therapeutic targets for BCa treatment.
Materials and methods
Clinical samples collection
Fresh clinical BCa tissues were collected according to the ethical guidelines of the Declaration of Helsinki (1975) and the Institutional Medical Ethics Committee of Xiamen University. Pathological diagnosis of BCa was verified by at least two qualified pathologists. A total of 32 fresh BCa tissues and their corresponding pairs of normal tissues were collected from low stage and low grade (T1, N0, M0, G1) of non-muscle invasive bladder cancer (NMIBC) patients and high stage and high grade (T2-4, N0, M0, G2-G3) of muscle-invasive samples (MIBC) patients. Sixteen pairs of human NMIBC and MIBC specimens were collected from the Department of Urology, The First Affiliated Hospital of Xiamen University from January 2016 to March 2017.
Genomic DNA extraction and target sequencing
Genomic DNA was extracted from RNA Later-preserved bladder cancer samples using DNeasy Blood & Tissue Kit (QIAGEN, Germany, Cat No.69506). The detection of genomic DNA quality was performed on 1% agarose gel, after that the DNA concentration was measured using a Qubit® DNA Assay Kit in Qubit® 2.0 Fluorometer (Invitrogen, USA) according to the manufacturer’s instruction.
To get the target gene regions, we designed probes on the website of Agilent (http://www.agilent.com/) for the 94 genes according to the design description. Initially, DNA fragmentation was performed by hydrodynamic shearing machine (Covaris, Massachusetts, USA) to generate 180–280 bp fragments. Extracted DNA was then subjected to ligation-mediated PCR (LM-PCR), followed by the product purification and hybridization with the probe for enrichment. All LM-PCR products (non-captured or captured) were used to estimate the magnitude of enrichment by real-time PCR. The captured library was subjected to cluster generation in an Illumina flow cell. Then, sequencing was performed to produce 150 bp paired-end reads by using an Illumina Hiseq platform. Finally, to ensure each sample meets the desired average fold coverage, the captured libraries were sequenced independently.
Sequence data quality control
The shortreads (Raw data) were transformed from the original fluorescence image files obtained from HiSeq platform by base calling. They were recorded in FASTQ format, including sequence data and their corresponding quality scores. Reads containing adapter contamination and low-quality/unrecognizable nucleotides were excluded through quality control. Then, downstream bioinformatics analyses were performed on clean data. At the same time, percentage of reads with average quality >20 and with average quality >30, sequencing error rate, the total reads number, and GC content distribution were calculated.
Reads mapping and somatic genetic alteration detection
Original mapping results were obtained and stored in BAM format by mapping valid sequencing reads to the human reference genome (UCSC hg19) using Burrows-Wheeler Aligner (BWA) software. After that, BAM files were sorted with SAM tools and Picard (http://broadinstitute.github.io/picard/). The resulting BAM files were obtained and used for calculating the sequence coverage and depth after duplicate marking, local realignment, and base quality recalibration.
MuTect and Strelka were used to call somatic single nucleotide variations (SNVs), small insertions, and deletions (InDels) from paired tumor-normal samples. In addition, polymorphisms of somatic SNVs and InDels referenced in the 1000 Genomes Project or Exome Aggregation Consortium (ExAC) with a minor allele frequency over 1% were removed to default filters. Finally, ANNOVAR was used for functional annotation of genetic variants.Citation34
Control-FREEC was used to identify somatic copy number variations (CNVs) and GISTIC algorithm was used to infer recurrently amplified or deleted genomic regions. Based on the frequency and amplitude of amplification or deletion affecting each gene, G-scores were calculated for genomic and gene-coding regions. Amplification or deletion region with a G-score > 0.1 and a corresponding p-value less than 0.05, based on a permutation-derived null distribution, was defined as a significant CNV region.
Results
Patient characteristics
Among these 32 identified BCa patients, 5 patients were female and 27 were male, with age of 33 y to 82 y (). Pathologically, 16 patients were classified as NMIBC and others were MIBC. The maximum diameter of mass from 27 patients (84.4%) was less than 5 cm while only 5 patients have their mass larger than 5 cm in diameter. Fifty percent of patients (n = 16) were in TNM stage I and 8 patients (25%) were in stage II, while only 3 and 5 patients had stage III and IV disease (9.375% and 15.625%), respectively. Sixteen (50%) patients were G1 degree and 16 (50%) had G2-G3 degree.
Table 1. Clinical information of BCa patients in our study.
Genomic characterization of NMIBC and MIBC cases
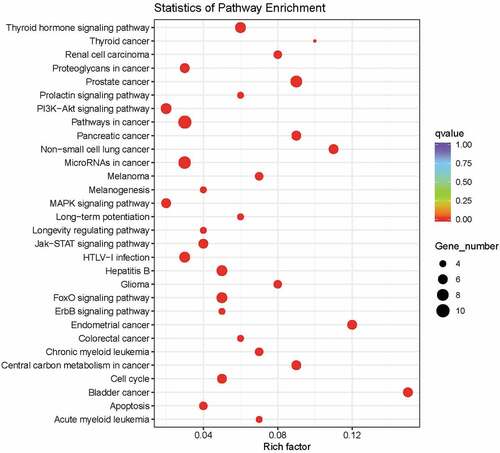
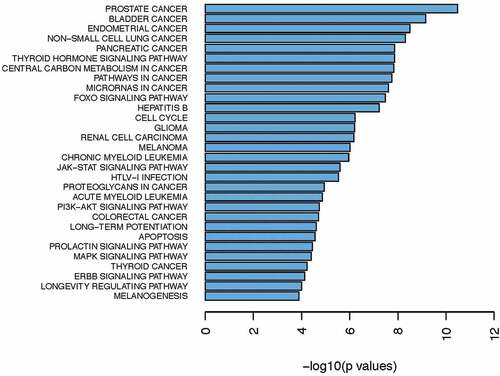
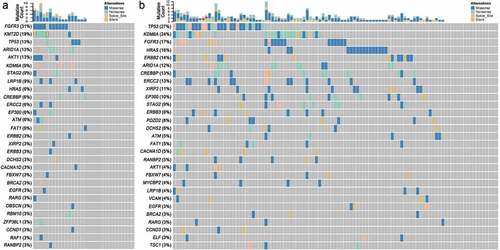
To characterize the landscape of genomic mutations in Chinese Han population, we collected 32 BCa tissues (the detailed clinical information is shown in ) and their paired normal bladder controls, then targeted next generation sequencing was applied to these samples for 94 well defined UC-associated genes (Table S1). The average sequencing depth for the Normal and tumor samples from 32 UBC patients analyzed by NGS was 474.5×, with 99.98% of the reference human genome covered as shown in . In the combined discovery and validation cohorts, we identified an average of 5595.4 point mutations and 806.3 inDels per sample. According to the NGS results, we noticed that 53.1% of UC patients (17/32) had different degree of gene mutation (). In addition, 30.9% of detected genes (29/94, & Table S2) were mutated in 32 urothelial carcinoma bladder tissues. Of note, FGFR3 (31.3%), KMT2D (18.8%), TP53 (12.5%), ARID1A (12.5%), AKT1 (12.5%), KDM6A (9.4%), STAG2 (9.4%), and LRP1B (9.4%) were the most frequently mutated genes in UC patients ( & ). We also analysis the sequencing data of 103 China bladder cancer samples download from ICGC database and found that TP53 (27.2%), KDM6A (24.3%), FGFR3 (16.5%), HRAS (15.5%), ERBB2 (13.6%), CREBBP (12.6%), ERCC2 (12.6%), ARID1A (11.7%) and XIRP2 (10.7%) were the top-mutated genes ( & Table S3). Consistent with our results, we all found coding genes of histone modification-related genes (KMT2D, KDM6A, EP300, and CREBBP), FGFR3, TP53, ARID1A, AKT1, STAG2, LRP1B, HRAS2, ERBB2, ERCC2, and FAT1 were frequently mutated genes in UC patients (). However, different from their results, FGFR3 is the most frequently mutated gene in our results other than TP53 in their results (). This may be due to sample capacity, different regions, diets and living habits, however, it needs more samples to further verify. Previous studies showed that the FGFR3 mutations occured predominantly in Urothelial cell carcinoma, whereas TP53 mutations were found in over 50% of human cancers.Citation35,Citation36 Data from The Cancer Genome Atlas (TCGA) Research Network showed that TP53 (49%), MLL2 (49%), ARID1A (49%), KDM6A (49%), PIK3CA (49%), EP300 (49%) and CDKN1A (49%) were the most frequently mutated genes.Citation37 These results are consistent with our results and complement each other, elucidate the molecular mutation basis for BCa development.
Table 2. Summary of gene mutation frequency in 32 BCa tissues.
Figure 1. The genomic landscape of bladder cancer. (a) Targeted NGS analysis of the mutated genes in 32 bladder cancer samples. (b) ICGC data showing the mutated genes in 103 bladder cancer samples of Chinese Han population.
Furthermore, subgroup analysis showed that FGFR3 (50.0%, 8/16), KMT2D (31.3%, 5/16), AKT1 (25.0%, 4/16), ARID1A (12.5%, 2/16) and STAG2 (18.8%, 3/16) were the most frequently mutated genes in NMIBC patients ( & ). FGFR3, one member of fibroblast growth factor receptor family, mediates cell proliferation in the response to FGF stimulation. The activation of FGFR3 due to gene amplification or gene mutation triggers numerous signaling pathways which are involved in the carcinogenesis of UC.Citation38,Citation39 Consistent with a previous report documenting that FGFR3 mutation (49%) was frequently detected in NMIBC,Citation26 our sequencing result also confirmed this finding. Another prevalently mutated gene was KMT2D (50%). As a histone methyltransferase, KMT2D mediates epigenetic modification by methylating lysine-4 position of histone 3,Citation40 the mutation of which may lead to aberrant epigenetic profile occurred in NMIBC. Therefore, our sequencing result not only strengthens the prevalence of FGFR3 and KMT2D in NMIBC but also directs further scientific investigations.
Figure 2. Twenty-nine mutated genes in NMIBC and MIBC tissues assayed by Targeted NGS. (a) Gene mutation frequency in NMIBC. (b) Gene mutation frequency in MIBC. Abbreviations: NMIBC, non-muscle invasive bladder cancer; MIBC, muscle-invasive bladder cancer.
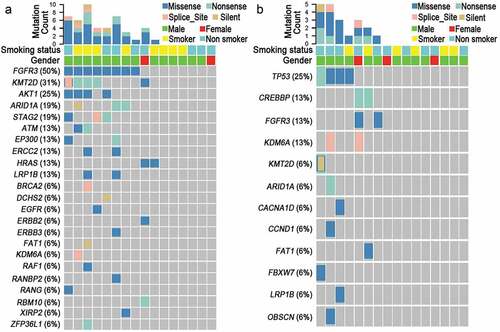
In addition, NSG results also showed that TP53 (25.0%, 4/16), CREBBP (12.5%, 2/16) and FGFR3 (12.5%, 2/16) were the most frequently mutated genes in MIBC patients ( & ). Of note, TP53, CREBBP, CACNA1D, and CCND1 were only frequently mutated in MIBC but not in NMIBC patients ( & ). We have followed up the operation patients and found there are 25% (8/32) of patients had relapse or metastasis (Data not shown). Among the genes with high mutation frequency, 75% (3/4) of patients with TP53 mutation, 66.7% (2/3) of patients with LRP1B or ARID1A and 33.3% (2/6) of patients with KMT2D mutation had relapse and metastasis, suggesting their mutations were correlated with recurrence, metastasis, or associated with other tumors such as gastric cancer and ovarian cancer.
Among these mutated genes, CACNA1D (Calcium Voltage-Gated Channel Subunit Alpha1 D), which is located in cell membrane and mediates calcium homeostasis, is highly associated with neuron diseases.Citation41,Citation42 However, the detailed function of CACNA1D in the development of MIBC is still elusive. We speculate that the CACNA1D mutation disrupts calcium homeostasis and alters calcium-related signaling. In comparison with other reports, CACNA1D seems to have a higher mutation rate in Chinese Han population, suggesting the geographic disparity exists in the carcinogenesis of BCa. Interestingly, another mutated gene is CREBBP (CREB binding protein), which complexes with phosphorylated CREB and enhances the transcription of cAMP-responsive genes. CREBBP also functions to regulate gene expression via acetylating histone and allowing chromatin to a relaxed state.Citation43,Citation44 Accordingly, an other group also demonstrated that CREBBP was commonly mutated in high grade BCa (13%).Citation24 Taking together, scientists have compelling rationale to develop these mutated genes as potential biomarkers or therapeutic targets for MIBC treatment.
Signaling pathway alterations
To investigate which signaling pathways these mutated genes are correlated with, we performed gene ontology and clustering analysis by using KEGG and Reactome program. Data revealed that these frequently mutated genes were tightly associated with signaling pathways which were implicated into cancer development, such as PI3 K/AKT, MAPK, Jak-STAT, FOXO and ERBB signaling pathways ( & ). Previous studies have reported that dysfunction of these signaling pathways drives cancer progression and targeting them with their specific inhibitors slows down tumor growth in various cancer types.Citation45,Citation46 Collectively, our NGS result suggests that activation of oncogenic signaling pathways (PI3 K/AKT, MAPK, Jak-STAT, FOXO and ERBB) in BCa development may partially result from genomic alterations.
Discussion
BCa is one of the leading causes of cancer deaths worldwide and continues to threaten human health. Although current treatments show promising therapeutic benefits, BCa can recur and advance to a high stage with limited curable options. Therefore, understanding of the molecular profiling of BCa not only provides rationale to develop targeted therapies but also helps identify potential diagnostic or prognostic biomarkers. Although genomic alterations in BCa have been comprehensively investigated by analyzing TCGA database or performing next negation sequencing by other groups,Citation24,Citation37 however, geographic disparity in BCa led us to screen gene mutations in Chinese Han population. Our results showed that 29/94 genes were mutated in 32 bladder cancer tissues, indicating a higher mutation burden in Chinese BCa patients. According to our analysis, a differential profile of mutated genes existed between NMIBC and MIBC, which may serve as the rationale to develop promising biomarkers to distinguish these two types of BCa. However, further investigation is required to strengthen our results given the relatively small sample size of our study.
Previous investigations into genomic alteration in Chinese Han population have identified some mutated genes. For instance, Cai et al. found that three genes (STAG2, ESPL1, TACC3), were involved in sister chromatid cohesion and segregation (SCCS) process and-, displayed a mutation in bladder cancer with transitional cell carcinoma.Citation47 Wu S. et al. performed whole-genome sequencing in 65 urothelial bladder carcinomas (UBCs) and targeted sequencing in an additional 196 UBCs. Besides FGFR3, TP53, and PIK3CA, they also found ZFP36L1, HRAS, and KDM6A were frequently mutated in UBCs. Notably, they find recurrent ADGRG6 enhancer mutations and FRS2 duplications which are associated with higher protein expression in the tumor and poor prognosis.Citation48 In our study, we also identified a mutated STAG2 and ZFP36L1 in BCa patients. This consistent data suggest that dysfunctional SCCS may be implicated into BCa carcinogenesis. Cai et al. also demonstrated that some chromatin remodeling genes (UTX, MLL-MLL3, EP300, NCOR1, and ARID1A) were frequently mutated in transitional cell carcinoma of BCa.Citation49 Our result revealed that three genes (KMT2D, EP300 and ARID1A) involved in chromatin remodeling were genetically changed in UC patients, implying that aberration in chromatin remodeling is a hallmark of BCa patients. Novel genetic alterations in BCa have continuously been identified in Chinese Han population,Citation50 however, none of these genes was genetically changed on the basis of our sequencing result, which may be explained by the fact that evident difference on genetic background exists in BCa patients.
Cystoscopy is the primary diagnostic tool for BCa patients.Citation51 Nevertheless, it is an invasive detection method because it needs urologists to take biopsy from patients. Therefore, a noninvasive diagnostic tool is emergently required to replace cystoscopy. Circulating tumor cells (CTCs) are tumor cells that move to the bloodstream, detection of biomarkers in CTCs represents a promising noninvasive tool in numerous cancers.Citation52,Citation53 Here, a higher mutation frequency of FGFR3 (50.0%), KMT2D (31.3%), and AKT1 (25.0%) was observed in NMIBC but not in MIBC, indicating these three mutation events occur in the early stage of urothelial carcinogenesis. Therefore, it may be practical to detect the mutated form of FGFR3, KMT2D, and AKT1 in CTCs for the diagnosis of early stage of BCa.
In addition, we found that several mutants (TP53, CREBBP, CACNA1D, CCND1, FAT1, FBXW7, and OBSCN) were only detected in MIBC but not in NMIBC. As to MIBC patients, few therapeutic options were effectively utilized to cure them. Consequently, investigating the biological functions of these mutated genes may provide us identify novel targets for MIBC. Actually, studies have also shown that mutation of TP53 has been identified to be associated with MIBC.Citation31 Here, in addition to TP53, we identified more mutated genes specific to MIBC. Our NGS results and ICGC data analysis also showed that FGFR3 and TP53 are the two most frequently mutated genes (). Interestingly, patients with FGFR3 mutation generally do not share with TP53 mutation (). So we can divide bladder cancer patients into FGFR3-mutation or TP53-mutation types. These mutated genes may give us a variety of directions to elucidate the underlying mechanisms responsible for MIBC development.
For the first time, we showed that CACNA1D has a high mutation rate in MIBC, suggesting the calcium pathway may play important roles in BCa progression of in Chinese Han population. Actually, calcium ATPase inhibitors have been pre-clinically used for cancers such as prostate cancer and breast cancer.Citation54,Citation55 However, these inhibitors have been limitedly tested in BCa. These results provide insights into potential effects of ATPase inhibitors in MIBC treatment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Competing interests
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Supplemental Material
Download Zip (28 KB)Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi:10.3322/canjclin.57.1.43.
- McGuire S. World Cancer Report 2014. World Health Organization, International Agency for Research on Cancer, WHO Press: Geneva (Switzerland), Advances in nutrition 2016; 7:418-9; 2015
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi:10.1002/ijc.25516.
- Wu C, Zheng X, Li X, Fesler A, Hu W, Chen L, Xu B, Wang Q, Tong A, Burke S, et al. Reduction of gastric cancer proliferation and invasion by miR-15a mediated suppression of Bmi-1 translation. Oncotarget. 2016;7(12):14522–14536. doi:10.18632/oncotarget.7392.
- Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, Tardón A, Serra C, Carrato A, García-Closas R, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366(9486):649–659. doi:10.1016/S0140-6736(05)67137-1.
- LaRue H, Allard P, Simoneau M, Normand C, Pfister C, Moore L, Meyer F, Têtu B, Fradet Y. P53 point mutations in initial superficial bladder cancer occur only in tumors from current or recent cigarette smokers. Carcinogenesis. 2000;21:101–106. doi:10.1093/carcin/21.1.101.
- Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. Jama. 2011;306:737–745. doi:10.1001/jama.2011.1142.
- Boffetta P. Tobacco smoking and risk of bladder cancer. Scand J Urol Nephrol Suppl. 2008:45–54. doi:10.1080/03008880802283664
- Ugnat AM, Luo W, Semenciw R, Mao Y. Occupational exposure to chemical and petrochemical industries and bladder cancer risk in four western Canadian provinces. Chronic Dis Can. 2004;25:7–15.
- Benbrahim-Tallaa L, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Guha N, Loomis D, Straif K. Carcinogenicity of diesel-engine and gasoline-engine exhausts and some nitroarenes. Lancet Oncol. 2012;13(7):663–664. doi:10.1016/S1470-2045(12)70280-2.
- Djousse L, Schatzkin A, Chibnik LB, D’Agostino RB, Kreger BE, Ellison RC. Alcohol consumption and the risk of bladder cancer in the Framingham Heart Study. J Natl Cancer Inst. 2004;96:1397–1400. doi:10.1093/jnci/djh263.
- Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. doi:10.1016/j.bbrc.2008.07.154.
- Weibull CE, Eloranta S, Altman D, Johansson AL, Lambe M. Childbearing and the risk of bladder cancer: a nationwide population-based cohort study. Eur Urol. 2013;63:733–738. doi:10.1016/j.eururo.2013.01.005.
- Karkoulis PK, Stravopodis DJ, Margaritis LH, Voutsinas GE. 17-Allylamino-17-demethoxygeldanamycin induces downregulation of critical Hsp90 protein clients and results in cell cycle arrest and apoptosis of human urinary bladder cancer cells. BMC Cancer. 2010;10:481. doi:10.1186/1471-2407-10-481.
- Konstantakou EG, Voutsinas GE, Velentzas AD, Basogianni AS, Paronis E, Balafas E, Kostomitsopoulos N, Syrigos KN, Anastasiadou E, Stravopodis DJ. 3-BrPA eliminates human bladder cancer cells with highly oncogenic signatures via engagement of specific death programs and perturbation of multiple signaling and metabolic determinants. Mol Cancer. 2015;14:135. doi:10.1186/s12943-015-0399-9.
- Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, Matos T, Cordon-Cardo C. Molecular pathways of urothelial development and bladder tumorigenesis. Urol Oncol. 2010;28:401–408. doi:10.1016/j.urolonc.2009.04.019.
- Racioppi M, Di Gianfrancesco L, Ragonese M, Palermo G, Sacco E, Bassi P. The challenges of Bacillus of Calmette-Guerin (BCG) therapy for high risk non muscle invasive bladder cancer treatment in older patients. J Geriatr Oncol. 2018;9(5):507–512. doi:10.1016/j.jgo.2018.03.020.
- Dougherty DW, Gonsorcik VK, Harpster LE, Trussell JC, Drabick JJ. Superficial bladder cancer metastatic to the lungs: two case reports and review of the literature. Urology. 2009;73:210:e3–5.
- Chamie K, Litwin MS, Bassett JC, Daskivich TJ, Lai J, Hanley JM, Konety BR, Saigal CS. Recurrence of high-risk bladder cancer: a population-based analysis. Cancer. 2013;119(17):3219–3227. doi:10.1002/cncr.28147.
- van den Bosch S, Alfred Witjes J. Long-term cancer-specific survival in patients with high-risk, non-muscle-invasive bladder cancer and tumour progression: a systematic review. Eur Urol. 2011;60:493–500. doi:10.1016/j.eururo.2011.05.045.
- Milowsky MI, Rumble RB, Booth CM, Gilligan T, Eapen LJ, Hauke RJ, Boumansour P, Lee CT. Guideline on muscle-invasive and metastatic bladder cancer (European association of urology guideline): American society of clinical oncology clinical practice guideline endorsement. J Clin Oncol. 2016;34:1945–1952. doi:10.1200/JCO.2015.65.9797.
- Hovelson DH, Udager AM, McDaniel AS, Grivas P, Palmbos P, Tamura S, Lazo de la Vega L, Palapattu G, Veeneman B, El-Sawy L. Targeted DNA and RNA sequencing of paired urothelial and squamous bladder cancers reveals discordant genomic and transcriptomic events and unique therapeutic implications. Eur Urol. 2018;74(6):741–753. doi:10.1016/j.eururo.2018.06.047.
- Mendiratta P, Grivas P. Emerging biomarkers and targeted therapies in urothelial carcinoma. Ann Transl Med. 2018;6(12):250. doi:10.21037/atm.2018.05.49.
- Kim PH, Cha EK, Sfakianos JP, Iyer G, Zabor EC, Scott SN, Ostrovnaya I, Ramirez R, Sun A, Shah R, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67(2):198–201. doi:10.1016/j.eururo.2014.06.050.
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25–41. doi:10.1038/nrc3817.
- Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, Ostrovnaya I, Baez P, Li Q, Berger MF, et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol. 2017;72(6):952–959. doi:10.1016/j.eururo.2017.05.032.
- Czerniak B, Cohen GL, Etkind P, Deitch D, Simmons H, Herz F, Koss LG. Concurrent mutations of coding and regulatory sequences of the Ha-ras gene in urinary bladder carcinomas. Hum Pathol. 1992;23(11):1199–1204. doi:10.1016/0046-8177(92)90285-B.
- Billerey C, Chopin D, Aubriot-Lorton M-H, Ricol D, Gil Diez de Medina S, Van RB, Bralet M-P, Lefrere-Belda M-A, Lahaye J-B, Abbou CC. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001;158(6):1955–1959. doi:10.1016/S0002-9440(10)64665-2.
- van Rhijn BW, Lurkin I, Radvanyi F, Kirkels WJ, van der Kwast TH, Zwarthoff EC. The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate. Cancer Res. 2001;61:1265–1268.
- Knowles MA, Williamson M. Mutation of H-ras is infrequent in bladder cancer: confirmation by single-strand conformation polymorphism analysis, designed restriction fragment length polymorphisms, and direct sequencing. Cancer Res. 1993;53:133–139.
- Esrig D, Elmajian D, Groshen S, Freeman JA, Stein JP, Chen S-C, Nichols PW, Skinner DG, Jones PA, Cote RJ, et al. Accumulation of nuclear p53 and tumor progression in bladder cancer. N Engl J Med. 1994;331(19):1259–1264. doi:10.1056/NEJM199411103311903.
- Shariat SF, Tokunaga H, Zhou J, Kim J, Ayala GE, Benedict WF, Lerner SP. p53, p21, pRB, and p16 expression predict clinical outcome in cystectomy with bladder cancer. J Clin Oncol. 2004;22(6):1014–1024. doi:10.1200/JCO.2004.03.118.
- Schroeder JC, Conway K, Li Y, Mistry K, Bell DA, Taylor JA. p53 mutations in bladder cancer: evidence for exogenous versus endogenous risk factors. Cancer Res. 2003;63:7530–7538.
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi:10.1093/nar/gkq603.
- Sibley K, Stern P, Knowles MA. Frequency of fibroblast growth factor receptor 3 mutations in sporadic tumours. Oncogene. 2001;20:4416–4418. doi:10.1038/sj.onc.1204543.
- Karoui M, Hofmann-Radvanyi H, Zimmermann U, Couvelard A, Degott C, Faridoni-Laurens L, Ahomadegbe J-C, Gazzeri S, Brambilla E, Clerici T. No evidence of somatic FGFR3 mutation in various types of carcinoma. Oncogene. 2001;20(36):5059–5061. doi:10.1038/sj.onc.1204651.
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi:10.1038/nature12965.
- Pandith AA, Shah ZA, Siddiqi MA. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol Oncol. 2013;31:398–406. doi:10.1016/j.urolonc.2010.07.014.
- Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP, Radvanyi F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi:10.1038/12615.
- Lv S, Ji L, Chen B, Liu S, Lei C, Liu X, Qi X, Wang Y, Lai-Han Leung E, Wang H. Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene. 2018;37(10):1354–1368. doi:10.1038/s41388-017-0026-x.
- He K, An Z, Wang Q, Li T, Li Z, Chen J, Li W, Wang T, Ji J, Feng G, et al. CACNA1C, schizophrenia and major depressive disorder in the Han Chinese population. Br J Psychiatry. 2014;204(1):36–39. doi:10.1192/bjp.bp.113.126979.
- Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D, Jones I, Forty L, Jones L, Gordon-Smith K, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2013;18(6):708–712. doi:10.1038/mp.2012.67.
- Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, Meyer SN, Mo T, Basso K, Brindle PK, et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer Discov. 2017;7(3):322–337. doi:10.1158/2159-8290.CD-16-1417.
- Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18(11):6777–6783. doi:10.1128/MCB.18.11.6777.
- Martin GS. Cell signaling and cancer. Cancer Cell. 2003;4(3):167–174. doi:10.1016/S1535-6108(03)00216-2.
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi:10.1038/nature04869.
- Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, Dean M, Huang Y, Jia W, Zhou Q, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459–1463. doi:10.1038/ng.2798.
- Wu TO S, Xing N, Jiang L, Wan S, Wang C, Zhang X, Yang F, Huang Y. Zhiming Cai Whole-genome sequencing identifies ADGRG6 enhancer mutations and FRS2 duplications as angiogenesis-related drivers in bladder cancer. Nat Commun. 2019;10:720. doi:10.1038/s41467-019-08576-5.
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. doi:10.1038/ng.907.
- Pan H, Xu X, Wu D, Qiu Q, Zhou S, He X, Zhou Y, Qu P, Hou J, He J, et al. Novel somatic mutations identified by whole-exome sequencing in muscle-invasive transitional cell carcinoma of the bladder. Oncol Lett. 2016;11(2):1486–1492. doi:10.3892/ol.2016.4094.
- Zaak D, Ohlmann C, Stenzl A. Current and established diagnostic modalities for bladder cancer. Urologe A. 2018;57(6):657–664. doi:10.1007/s00120-018-0650-2.
- Lorente D, Olmos D, Mateo J, Dolling D, Bianchini D, Seed G, Flohr P, Crespo M, Figueiredo I, Miranda S, et al. Circulating tumour cell increase as a biomarker of disease progression in metastatic castration-resistant prostate cancer patients with low baseline CTC counts. Ann Oncol. 2018;29(7):1554–1560. doi:10.1093/annonc/mdy172.
- Hashimoto M, Tanaka F, Yoneda K, Takuwa T, Kuroda A, Matsumoto S, Okumura Y, Kondo N, Tsujimura T, Nakano T, et al. The clinical value of circulating tumour cells (CTCs) in patients undergoing pulmonary metastasectomy for metastatic colorectal cancer. J Thorac Dis. 2018;10(3):1569–1577. doi:10.21037/jtd.2018.03.05.
- Muscella A, Calabriso N, Vetrugno C, Fanizzi FP, De Pascali SA, Storelli C, Marsigliante S. The platinum (II) complex [Pt(O,O’-acac)(gamma-acac)(DMS)] alters the intracellular calcium homeostasis in MCF-7 breast cancer cells. Biochem Pharmacol. 2011;81:91–103. doi:10.1016/j.bcp.2010.09.012.
- Sehgal P, Szalai P, Olesen C, Praetorius HA, Nissen P, Christensen SB, Engedal N, Møller JV. Inhibition of the sarco/endoplasmic reticulum (ER) Ca 2+ -ATPase by thapsigargin analogs induces cell death via ER Ca 2+ depletion and the unfolded protein response. J Biol Chem. 2017;292(48):19656–19673. doi:10.1074/jbc.M117.796920.