ABSTRACT
Detection rate of de novo EGFR T790 M mutation was increased up to 80% through recent ultrasensitive detection methods. Here, we investigated the clinical significance and its usefulness of detecting de novo EGFR T790 M using ultrasensitive droplet-digital polymerase chain reaction (ddPCR) method.
In total, 102 cases diagnosed as lung adenocarcinoma with EGFR-tyrosine kinase inhibitor (TKI) sensitizing mutations (mEGFR) and had been treated with 1st ~ 2nd generation EGFR-TKI alone were enrolled for this study. De novo T790 M status was tested using the tissues at the initial diagnosis and positivity was defined as the ratio of T790 M/wild-type copies over 0.00294 by ddPCR.
Seventy patients (68.6%) harbored the de novo T790 M. De novo T790 M was more frequently detected in cases with EGFR L858 R mutation than those with EGFR exon 19 deletion (E19d) mutations (P = 0.024). Forty-three patients underwent rebiopsy due to disease progression. The cases who experienced progression due to acquired T790 M were more likely to have E19d at initial diagnosis and the presence of de novo T790 M and the ratio of T790 M/wild-type copies did not relate to the emergence of acquired T790 M. On the other hand, the cases with a longer duration of disease-control by EGFR-TKI had higher change to get acquired T790 M mutation (P-value = 0.040).
The presence of de novo T790 M has limitation in predicting disease progression by acquired T790 M, suggesting that identifying de novo T790 M through the ultrasensitive methods may not be necessary identifying patients who would be beneficial by 3rd-generation EGFR-TKI as the 1st line treatment.
1. Introduction
Lung cancer is the leading cause of cancer-related death worldwide.Citation1 According to an analysis of 32 cancer groups in 195 countries, there were 1.7 million deaths from tracheal, bronchus, and lung cancer in 2015. In addition, the incidence of these respiratory system-related cancers has increased by 29% between 2005 and 2015.Citation2 Despite the prevalence of lung cancer, the prognosis of this disease is still poor; during the localized, regional metastasis, and distant metastasis stages, the 5-year survival rates are approximately 55%, 28%, and 4%, respectively. Based on recent advancements in therapeutic approaches, the prognosis of patients with lung cancer is expected to improve owing to the development of molecular-targeted anticancer drugs inhibiting EGFR, ALK, and ROS1 and to the use of immunosuppressive drugs.Citation3
The discovery of EGFR-tyrosine kinase inhibitor (TKI)-sensitizing mutation (mEGFR) and the application of first-generation EGFR-TKIs have dramatically improved clinical outcomes in patients with lung adenocarcinoma.Citation4 However, EGFR-mutation positive lung adenocarcinomas often exhibit acquired drug resistance to EGFR-TKIs, resulting in treatment failure.Citation5 For example, the acquired T790 M mutation has been shown to cause treatment failure and is observed in approximately 50% of cases of treatment failure.Citation6 The main risk factors for acquired resistance to TKIs by T790 M expression include prolonged exposure to EGFR-TKIs and the presence of the EGFR E19del mutant.Citation7,Citation8 Although the de novo EGFR T790 M mutation is rarely detected in non-small cell lung cancer (NSCLC) using conventional genotyping methods, some previous studies have shown that this mutation is present in up to 80% of cases when using other sensitive methods.Citation9,Citation10
Accordingly, as the use of such ultrasensitive methods has increased, the presence of the de novo EGFR T790 M has been estimated to be the main cause of short progression-free survival (PFS) and failure of EGFR-TKI therapy.Citation10 These two hypotheses (i.e., that acquired T790 M is predominantly found in patients who have used TKIs for a long time and that acquired T790 M appears rapidly in patients with the de novo EGFR T790 M mutation) are mutually exclusive and contradictory with regard to the cause of treatment failure by the acquired T790 M mutation.
Further research is needed to clarify the reasons for treatment failure. Indeed, if patients with high rates of the de novo EGFR T790 M mutation in the tumor experience rapid recurrence due to the mutation, prolonged responses can be expected by using initial treatment with a third-generation EGFR-TKI, such as osimertinib. Moreover, if there is no relationship between high frequency of the de novo EGFR T790 M mutation and recurrence due to the mutation, first- and second-generation EGFR-TKIs should be used first rather than early positioning of third-generation EGFR mutant selective inhibitor (EMSI). Accordingly, sequential therapies using third-generation TKIs are desirable after confirming acquisition of the T790 M mutation by histological examination in patients showing lung cancer progression.
In this study, we aimed to confirm the existence of the de novo EGFR T790 M mutation by droplet digital PCR (ddPCR) and to establish a significant cutoff for de novo T790 M in the treatment of naïve lung adenocarcinoma. Additionally, we investigated the relationships among the de novo EGFR T790 M mutation and responses of first- and second-generation EGFR-TKIs with relapse-free survival. Finally, multivariate analysis was conducted to determine whether the de novo EGFR T790 M mutation was a risk factor for acquired T790 M.
2. Results
2.1. Determination of meaningful parameters
ddPCR yielded six variables, as follows: (1) T790 M copy number/20 μL, (2) ratio of allele frequency of T790 M to that of wild-type EGFR, (3) fractional abundance of T790 M mutation, (4) proportion of T790 M-positive droplets in total droplets, (5) proportion of T790 M-positive droplets in total droplets for the T790 M reaction, and (6) relative ratio of T790 M copy numbers in a 20 μL reaction to amount of total DNA per reaction. Although there were significant correlations between these six parameters, the correlations among T790 M copy number, T790 M copy number/wild-type copy number, and T790 M copy number/total DNA were relatively low. Therefore, we examined which of the other three parameters had significant clinical implications. We plotted the area under the curve (AUC) graph for the following three continuous variables with acquired T790 M mutation as the dependent variables. Although the values for the AUC were not significantly different between the above six variables, the AUC (0.566) was the highest for the ratio of T790 M mutation to wild-type allele frequency. Therefore, we used the ratio of T790 M mutation to wild-type allele frequency to obtain the best predicted cutoff value for acquired T790 M mutation using the method of Contal and O’Quigley. We obtained a cutoff value in ratio of T790 M mutation to wild-type allele frequency of 0.000294. Based on this value, 102 patients were classified into two groups; those with a score of less than 0.000294 were defined as the de novo T790 M (-) group (n = 30), and those with a score of greater than 0.000294 were defined as the de novo T790 M (+) group (n = 72).
2.2. Characteristics of patients at initial presentation
The mean age of the 102 patients was 63.2 ± 12.2 years, and the proportion of women was 53.9% (n = 55). The EGFR exon 19 deletion mutant was the most common (59.8%), followed by the 858 R (or L861Q) mutation (38.2%) and the S768I mutation (2.0%). All of these patients were prescribed EGFR-TKIs; 90 patients used gefitinib, nine patients used afatinib, and three patients used erlotinib. EGFR-TKIs were used in 80 cases as first-line agents, 19 cases as second-line agents, and three cases as third-line agents.
The initial presentation characteristics of patients were divided according to the presence or absence of the de novo EGFR T790 M mutation (). When the de novo EGFR T790 M was investigate to have a significant association with baseline clinical measures at initial presentation, demographic characteristics, such as age, sex, smoking status, and tumor size, were not significantly associated with the presence or absence of the de novo T790 M mutation. In contrast, when classified according to the subtype of EGFR-TKI-sensitizing mutations, the L858 R subgroup had a higher chance of having the de novo EGFR T790 M mutation than the other EGFR subtype groups (84.6%, p = .024). There was only one case of a de novo T790 M mutation observed by conventional EGFR testing.
Table 1. Baseline characteristics of the study population.
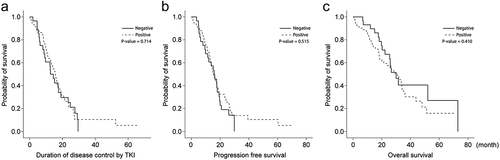
Evaluation of the relationships among the presence of the de novo T790 M mutation and TKI duration, PFS, and OS revealed that there were no statistically significant relationships among these parameters ().
Figure 1. The de novo T790 M mutation showed limited ability to predict the appearance of EGFR-TKI-resistant clones with T790 M (all patients, n = 102). (a) Duration of disease controlled by EGFR-TKIs. (b) Progression-free survival during EGFR-TKI treatment. (c) Overall survival of patients with EGFR-sensitizing mutations was compared according to the presence or absence of the de novo T790 M mutation.
2.3 Effects of the de novo EGFR T790 M mutation on treatment outcomes
In addition, we investigated whether the presence of the de novo EGFR T790 M mutation affected the course of treatment with EGFR-TKIs. First, examination of the differences in the duration of disease control in all patients revealed that the type of TKI did not significantly affect the duration of disease control (p = .384, by Kruskal-Wallis test). However, there were significant differences in PFS in the EGFR-TKI group according to first-, second-, and third-line therapies (p = .025, by Kruskal-Wallis test). These findings suggested that there were no differences in the EGFR-TKI type, but may be differences in positioning according to the type of EGFR-TKI. Therefore, additional analyzes were conducted in 80 patients using EGFR-TKIs as first-line therapy ().
Table 2. Duration of TKI use, progression-free survival, and overall survival in 80 patients using TKIs as first-line drugs.
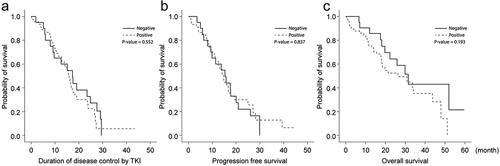
Of these, 22 were categorized into the T790 M (-) group, and 58 were categorized into the T790 M (+) group. Sixty-nine patients were administered gefitinib, nine were administered afatinib, and two were administered erlotinib. Other variables, including lung cancer stage at TKI start date, duration of TKI use, accepted droplets, and wild-type copy number, were not significantly different between the two groups. The duration of disease control by TKIs was slightly longer in the T790 M (-) group (15.3 versus 14.7 months, respectively), although this difference was not statistically significant (p = .837). Additionally, the OS was longer in the T790 M (-) group (31.5 versus 26.4 months, respectively; p = .193; ).
Figure 2. The de novo T790 M mutation showed limited ability to predict the appearance of EGFR-TKI-resistant clones with T790 M (80 patients using EGFR-TKIs as first-line treatments). (a) Duration of disease controlled by EGFR-TKIs. (b) Progression-free survival during EGFR-TKI treatment. (c) Overall survival in patients with EGFR-sensitizing mutations was compared according to the presence or absence of the de novo T790 M mutation.
In total, 69 patients who underwent EGFR-TKI therapy experienced disease progression, and 43 of these patients underwent rebiopsy. Rebiopsy was performed in 21 cases of primary lesion biopsy, 15 cases of metastatic lesion biopsy, and seven cases of liquid biopsy using pleural fluid.
To estimate the effects of the de novo T790 M mutation as a risk factor of resistance from acquired T790 M, we performed a multivariate analysis, including age and sex, on acquired T790 M mutation. Bifurcated logistic analysis was performed to identify the factors affecting acquired T790 M mutation (). Previous studies have shown that tumor size, EGFR subtype, and TKI use duration affected the acquired T790 M.Citation9,Citation11,Citation12 However, in this study, only the duration of TKI was significantly associated with acquired T790 M mutation. The presence of the de novo T790 M mutation tended to increase the risk of acquired T790 M mutation, although this result was not significant.
Table 3. Multivariate analysis of factors affecting acquired T790 M mutation by binary logic regression.
The de novo EGFR T790 M mutation was more likely to be observed in the L858 R subtype at initial diagnosis; however, in the rebiopsy sample, the acquired T790 M mutation was more likely to be found in the E19del subgroup (72.0% versus 50.0%, respectively; p = .141; ). In addition, patients with acquired T790 M mutation had a significantly longer duration of EGFR-TKI use than those without T790 M mutations (12.2 versus 17.9 months, respectively; p = .030). PFS was also significantly longer in the acquired T790 M group (p = .026). In summary, the presence of the de novo EGFR T790 M mutation did not contribute greatly to predicting the expression of acquired T790 M mutation.
Table 4. Duration of TKI use, progression-free survival, and overall survival according to acquired T790 M mutation.
3. Discussion
In this study, we detected de novo EGFR T790 M using ultrasensitive ddPCR and analyzed the relationships between de novo T790 M mutation and other clinical factors.
Until the early 2000 s, pathological and molecular classifications were not relevant to lung cancer treatment. Only categorization as small cell lung cancer or NSCLC was sufficient for decision-making regarding the chemotherapy regimen for patients with advanced lung cancer.Citation13 However, with the introduction of multiple-targeted agents, the pathological and molecular classification of lung cancer has become increasingly complex and important for devising a therapy strategy.Citation14–Citation16 Despite the large number of tests required, e.g., analysis of EGFR, ALK, and ROS1 expression/mutations, tissue samples obtained for lung cancer diagnosis have become smaller as biopsy approaches have become more noninvasive. Therefore, various techniques for analysis and characterization of smaller specimens have been developed. For example, ddPCR is a qualitative and quantitative targeted analysis approach with high sensitivity, enabling detection of de novo EGFR T790 M before TKI treatment.Citation17
The incidence of the de novo EGFR T790 M mutation depends on the diagnostic method used. For example, direct sequencing analyses showed that the incidence rates were 0.4–3.0% for all cases of NSCLC and 1.0–8.0% for cases of NSCLC with activating mutations.Citation12 With sensitive methods, such as mass spectrometry, laser microdissection, peptide nucleic acid clamping PCR, colony hybridization assays, and mutant-enriched PCR, the incidence of the de novo T790 M mutation increased from 31% to 80%.Citation13,Citation18,Citation19 Watanabe et al.Citation9 reported that the overall incidence of the EGFR T790 M mutation by ddPCR in patients who had not been treated with TKIs was 79.9% when analyzing surgically resected tumor tissue (frequency range: 0.009–26.9%). Similarly, in our study, the overall incidence of the de novo EGFR T790 M mutation was 73.5% (frequency range [percentage of EGFR T790 M mutation to wild-type allele frequency]: 0.013–6.5%).
Liu et al.Citation10 conducted a meta-analysis with 22 studies and showed that the presence of the de novo EGFR T790 M mutation was significantly related to worse PFS and OS than absence of the mutation. Additionally, longer PFS and post-progression survival was investigated in patients with acquired T790 M mutation in their study. On the other hands, retrospective studies have shown that acquired T790 M is likely to be found in patients who have used a short course of TKIs.Citation5,Citation10,Citation11 Moreover, the finding that TKI use in patients with the de novo T790 M mutation is related to short PFS was contradictory to the results of other retrospective studies showing that T790 M is likely to be expressed in patients with lung cancer who have been using TKIs for a long time. Additionally, in a study by Fujita et al., longer PFS was observed in the de novo EGFR T790 M group, although the difference was not significant,Citation18 potentially because of differences in the sample or detection method for de novo EGFR T790 M mutation. As in our study, the small portion of the de novo T790 M contained in the tumor cells was unlikely to significantly affect the clinical course associated with the use of TKIs. Even in the study by Fujita et al., when the T790 M group was divided into three groups based on the frequency of the mutation (0.0%, 0.0–0.5%, ≥ 0.5%), the time to treatment failure was significantly longer in the group with the highest frequency compared with those in the other two groups. Thus, large-scale, prospective studies are needed to further investigate the relationships between the de novo T790 M mutation and clinical course.
Liang et al.Citation20 conducted a pooled analysis of 25 studies on the EGFR T790 M mutation in 2018 and found that de novo T790 M was significantly more abundant in patients with L858 R mutation than in patients with E19del mutation. However, acquired T790 M was found more frequently in the E19del subgroup than in those with L858 R mutation. Similarly, in our study, although the frequency of de novo T790 M was higher in E19del mutation, the proportion of de novo EGFR T790 M mutation was more common in patients with L858 R or L861Q, and acquired T790 M was more common in patients with E19del than in patients with L858 R or L861Q. Thus, we further analyzed the EGFR subgroups according to the presence of acquired T790 M in patients who underwent rebiopsy (). Interestingly, in the E19del group, the proportion of acquired T790 M tended to be higher in the de novo T790 M-positive group (p = .097, Fisher’s exact test). However, in patients with L858 R or L861Q mutation, there were no significant relationships between de novo T790 M positivity and the acquired T790 M mutation (p = 1.000, Fisher’s exact test). Recently, Kim et al.Citation21 analyzed mutational characteristics in EGFR positive lung adenocarcinoma and reported a low mutation burden and high Ti/Tv ratio in the E19del subgroup compared with single nucleotide variations subgroup. Differences in these mutational characteristics between E19del and L858 R/L861Q may explain the differences in acquired T790 M according to the positivity of de novo T790 M.
Table 5. Alternations in acquired T790 M prevalence according to EGFR subtype and de novo T790 M mutation.
Although there were no significant differences in the duration of TKI use, PFS, and OS between the de novo EGFR T790 M-positive and -negative groups, there were significant differences in duration of TKI use and PFS according acquired T790 M. Additionally, patients with longer duration of TKI use showed higher acquired T790 M mutation positivity in this study. These findings are similar to those of previous studies.Citation7,Citation12,Citation22
4. Conclusion
In treatment naïve lung adenocarcinoma with mEGFR, de novo T790 M mutations were frequently observed when using ultrasensitive detection methods. Presence of the de novo EGFR T790 M mutation had limited ability to predict responses to first- or second-generation EGFR-TKIs and to predict disease progression by acquired T790 M. Therefore, detection of de novo T790 M through ultrasensitive methods have limited role in identifying patients who would be beneficial by 3rd generation EMSI as a 1st line treatment.
5. Materials and methods
5.1 Cell lines and clinical specimens
In total, 102 specimens from patients who visited Severance Hospital in Seoul, South Korea, from 2009 to 2017 were recruited from the institutional tissue bank. Detailed inclusion criteria for these cases were as follows: (1) pathologically proven lung adenocarcinoma, (2) confirmed EGFR status before EGFR-TKI treatment, (3) evaluation of response to EGFR-TKIs, (4) pathological or clinical stage IIIB–IV disease, (5) first-, second-, or third-line treatment with EGFR-TKIs, (6) sufficient formalin-fixed, paraffin-embedded (FFPE) tissue blocks for genomic DNA (gDNA) extraction, and (7) provided informed consent for the use of tissues. This study was approved by the Institutional Review Board of our institution (IRB #3-2017-5509). H1975 cells (harboring L858 R and T790 M mutations) and A549 cells (harboring wild-type EGFR) were used as positive controls for all relevant assays.
5.2. ddPCR
gDNA was extracted with a QIAamp DNA FFPE tissue kit, and the T790 M mutation was identified using a Bio-Rad ddPCR QX200 instrument (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions.
Using ddPCR, gDNA from FFPE samples was compartmentalized into 10,000–20,000 droplets containing 0 or more copies of target DNA. Allele frequencies were classified into two channels, i.e., wild-type and mutant, and these types were again divided according to the positivity and negativity of each type, resulting in four variables. When somatic mutations in cancer cells, which are present in low frequency among inflammatory cells and infiltrated tissues, are discovered, the most suitable method for determining the fraction of cancer cells with a specific mutation (e.g., the T790 M mutation) is still not clear. In this study, we simulated the cell line data to determine the detection threshold and to identify which parameters reflected the state of T790 M mutations in the tumor tissue as the ultimate dependent variable using ddPCR
5.3. Determination of detection threshold using cells with EGFR mutations
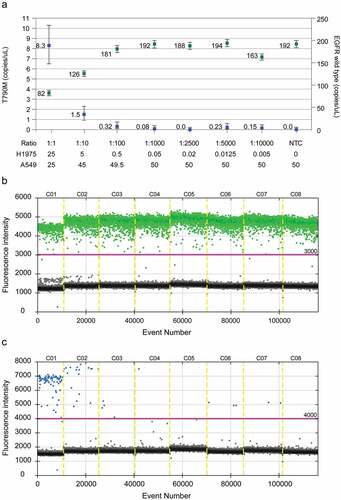
The detection threshold of the T790 M variant was set using various ratios of DNA mixtures from A549 cells, which contain the wild-type EGFR gene, and H1975 cells, which harbor both T790 M and L858 R mutations. When the detection limit was confirmed by serial dilution of the DNA from H1975 cells in a total of 50 ng pooled DNA from a wild-type DNA background, the amount of gDNA used less than 0.1 ng/reaction showed inconsistent results. Based on these results, we excluded samples from analysis if the amount of specimen used was less than 0.1 ng, as in cases of very small biopsies. After the detection threshold was defined, we also defined samples as “not detected” when the positive value was less than 10 copies/20 μL reaction ( and ). After performing ddPCR for tissue sample, fluorescence intensity of each well was manually inspected to determine the threshold, and once again validated through review.
Table 6. Concentration and ratio of the mixture of the control sample (EGFR wild-type DNA from A549 cells and T790 M mutant DNA from H1975 cells).
Figure 3. The detection threshold for the T790 M mutation was set using a mixture of EGFR wild-type DNA (from A549 cells) and T790 M mutant DNA (from H1975 cells). (a) Positivity was defined when the ratio of the T790 M mutant to the wild-type EGFR was 0.1% or more. (b) Fragmented template DNA from A549 cells was analyzed, and the threshold of fluorescence was set at 3,000. (c) Fragmented template DNA from H1975 cells was analyzed, and the threshold of fluorescence was set at 4,000.
5.4. Outcomes
Response Evaluation Criteria in Solid Tumors version 1.1 was used to assess clinical efficacy outcomes, including objective response rate, PFS, and overall survival (OS). PFS was defined as the duration between the first day of TKI treatment and the day of radiological evidence of disease progression. Clinical data (age, sex, smoking status, smoking amount, lung cancer TNM stage, duration of TKI use, and survival) were investigated in each patient. The data cutoff date for survival analysis was September 30, 2018.
5.5. Statistical analysis
Continuous variables were compared by t-tests, Kruskal-Wallis tests, or analysis of variance and presented as means ± standard deviations. Categorical variables were analyzed by Pearson’s chi-square tests or Fisher’s exact tests and presented as frequencies (n) and percentages (%). The method described by Contal and O’Quigley, which uses a log rank test statistic considering time, was conducted to determine the cutoff value of the ratio of T790 M to wild-type EGFR copy number for best predicting the recurrence of acquired T790 M.Citation23 A binary logistic regression model was used to identify the factors affecting pathologically acquired T790 M. All statistics were analyzed with SPSS Version 23.0 (SPSS, Chicago, IL, USA) or R Studio, Version 1.1.463 (RSTUDIO, MA, USA). We considered an adjusted p-value of less than 0.05 to be statistically significant.
Disclose of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19.
- Global Burden of Disease Cancer C, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, et al. 2017. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 3(4):524–548. doi:10.1001/jamaoncol.2016.5688.
- Boloker G, Wang C, Zhang J. Updated statistics of lung and bronchus cancer in United States (2018). J Thorac Dis. 2018;10:1158–1161. doi:10.21037/jtd.2018.03.15.
- Kuiper JL, Smit EF. Challenges in the management of EGFR-mutated non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors. Oncology. 2014;87:83–94. doi:10.1159/000362819.
- Gonzalez-Larriba JL, Lazaro-Quintela M, Cobo M, Domine M, Majem M, Garcia-Campelo R. Clinical management of epidermal growth factor receptor mutation-positive non-small cell lung cancer patients after progression on previous epidermal growth factor receptor tyrosine kinase inhibitors: the necessity of repeated molecular analysis. Transl Lung Cancer Res. 2017;6:S21–S34. doi:10.21037/tlcr.2017.10.03.
- Chang YS, Choi CM, Lee JC. Mechanisms of epidermal growth factor receptor tyrosine kinase inhibitor resistance and strategies to overcome resistance in lung adenocarcinoma. Tuberc Respir Dis (Seoul). 2016;79:248–256. doi:10.4046/trd.2016.79.4.248.
- Koo HJ, Kim MY, Park S, Lee HN, Kim HJ, Lee JC, Kim S-W, Lee DH, Choi C-M. Non-small cell lung cancer with resistance to EGFR-TKI therapy: CT characteristics of T790M mutation-positive cancer. Radiology. 2018;289:227–237. doi:10.1148/radiol.2018180070.
- Goag EK, Lee JM, Chung KS, Kim SY, Leem AY, Song JH, Jung JY, Park MS, Chang YS, Kim YS, et al. Usefulness of bronchoscopic rebiopsy of non-small cell lung cancer with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor. J Cancer. 2018;9(6):1113–1120. doi:10.7150/jca.21650.
- Watanabe M, Kawaguchi T, Isa S, Ando M, Tamiya A, Kubo A, Saka H, Takeo S, Adachi H, Tagawa T, et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res. 2015;21(15):3552–3560. doi:10.1158/1078-0432.CCR-14-2151.
- Liu Y, Sun L, Xiong ZC, Sun X, Zhang SL, Ma JT, Han CB. Meta-analysis of the impact of de novo and acquired EGFR T790M mutations on the prognosis of patients with non-small cell lung cancer receiving EGFR-TKIs. Onco Targets Ther. 2017;10:2267–2279. doi:10.2147/OTT.S133082.
- Kawamura T, Kenmotsu H, Omori S, Nakashima K, Wakuda K, Ono A, Naito T, Murakami H, Omae K, Mori K, et al. Clinical factors predicting detection of T790M mutation in rebiopsy for EGFR-mutant non-small-cell lung cancer. Clin Lung Cancer. 2018;19(2):e247–e52. doi:10.1016/j.cllc.2017.07.002.
- Huang YH, Hsu KH, Tseng JS, Chen KC, Hsu CH, Su KY, Chen JJW, Chen H-W, Yu S-L, Yang T-Y, et al. The association of acquired T790M mutation with clinical characteristics after resistance to first-line epidermal growth factor receptor tyrosine kinase inhibitor in lung adenocarcinoma. Cancer Res Treat. 2018;50:1294–1303. doi:10.4143/crt.2017.512.
- Bubendorf L, Lantuejoul S, de Langen AJ, Thunnissen E. Nonsmall cell lung carcinoma: diagnostic difficulties in small biopsies and cytological specimens: number 2 in the Series “pathology for the clinician” edited by Peter Dorfmuller and Alberto Cavazza. Eur Respir Rev. 2017;26(144):170007. doi:10.1183/16000617.0007-2017.
- Grigoriu B, Berghmans T, Meert AP. Management of EGFR mutated nonsmall cell lung carcinoma patients. Eur Respir J. 2015;45:1132–1141. doi:10.1183/09031936.00156614.
- Shaw AT, Kim D-W, Nakagawa K, Seto T, Crino L, Ahn M-J, De Pas T, Besse B, Solomon BJ, Blackhall F, et al. Crizotinib versus Chemotherapy in advanced ALK -positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. doi:10.1056/NEJMoa1214886.
- Bubendorf L, Buttner R, Al-Dayel F, Dietel M, Elmberger G, Kerr K, López-Ríos F, Marchetti A, Öz B, Pauwels P, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch. 2016;469(5):489–503. doi:10.1007/s00428-016-2000-3.
- Zhang R, Chen B, Tong X, Wang Y, Wang C, Jin J, Tian P, Li W. Diagnostic accuracy of droplet digital PCR for detection of EGFR T790M mutation in circulating tumor DNA. Cancer Manag Res. 2018;10:1209–1218. doi:10.2147/CMAR.S161382.
- Fujita Y, Suda K, Kimura H, Matsumoto K, Arao T, Nagai T, Saijo N, Yatabe Y, Mitsudomi T, Nishio K, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol. 2012;7(11):1640–1644. doi:10.1097/JTO.0b013e3182653d7f.
- Cabanero M, Sangha R, Sheffield BS, Sukhai M, Pakkal M, Kamel-Reid S, Karsan A, Ionescu D, Juergens RA, Butts C, et al. Management of EGFR-mutated non-small-cell lung cancer: practical implications from a clinical and pathology perspective. Curr Oncol. 2017;24(2):111–119. doi:10.3747/co.24.3524.
- Liang H, Pan Z, Wang W, Guo C, Chen D, Zhang J, Zhang Y, Tang S, He J, Liang W, et al. The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: a literature-based pooled analysis. J Thorac Dis. 2018;10(4):2311–2320. doi:10.21037/jtd.2018.03.150.
- Kim EY, Kim A, Lee G, Lee H, Chang YS. Different mutational characteristics of the subsets of EGFR-tyrosine kinase inhibitor sensitizing mutation-positive lung adenocarcinoma. BMC Cancer. 2018;18:1221. doi:10.1186/s12885-018-5116-9.
- Kogure Y, Shigematsu F, Oki M, Saka H. T790M correlates with longer progression-free survival in non-small cell lung carcinomas harboring EGFR mutations. In Vivo. 2018;32:1199–1204. doi:10.21873/invivo.11364.
- Contal C, O’Quigley J. An application of changepoint methods in studying the effect of age on survival in breast cancer. Comput Stat Data An. 1999;30:253–270. doi:10.1016/S0167-9473(98)00096-6.
