ABSTRACT
Glioma is the most prevalent brain malignancy with high mortality. In recent decades, the regulatory role of long noncoding RNAs (lncRNAs) has been unmasked in glioma. In this study, we focused on the function and mechanism of LINC00641 in glioma. First of all, we found that LINC00641 was expressed at a low level in glioma cell lines. Importantly, overexpression of LINC00641 prevented cell proliferation but enhanced cell apoptosis. Meanwhile, NRGN, a previously-reported downregulated mRNA in GBM, was disclosed as a tumor suppressor in glioma cells. Besides, we verified that NRGN could be positively regulated by LINC00641 in glioma cells. Moreover, the cellular distribution of LINC00641 was identified to be cytoplasmic. Therefore, bioinformatics analysis and mechanism experiments were carried out and we determined that miR-4262 was the shared miRNA between LINC00641 and NRGN. In contrast to LINC00641 and NRGN, miR-4262 was dramatically upregulated in glioma cells. Furthermore, we confirmed that LINC00641 acted as a ceRNA in glioma cells via absorbing miR-4262 to upregulate NRGN. More importantly, silenced NRGN countervailed the repression on glioma cell proliferation caused by LINC00641 upregulation. Collectively, our findings unveiled that LINC00641 serves as a tumor inhibitor in glioma by targeting miR-4262/NRGN axis, providing a new potential therapeutic target for glioma patients.
KEYWORDS:
Introduction
Glioma is the most prevalent brain malignancy that accounts for more than 50% of all the cerebral tumors.Citation1 On the basis of different histological characteristics, gliomas can be classified into five different forms including astrocytomas, glioblastomas, oligodendrogliomas, mixed oligoastrocytomas and pilocytic astrocytoma.Citation2 Although great progress has been made to explore glioma progressionCitation3,Citation4 and therapeutic methods,Citation5–Citation7 the clinical prognosis of glioma patients, especially those who are with glioblastoma, remains unsatisfactory.Citation8 Hence, it is urgent to figure out effective molecular targets for glioma.
Long non-coding RNAs (lncRNAs) are a large class of transcripts (>200 nucleotides) belonging to non-coding RNA and have nearly no capacity to encode proteins.Citation9 In recent years, dysregulated lncRNAs have been uncovered to participate in the pathogenesis and progression of various human cancers, including glioma.Citation8,Citation9 For example, lncRNA OIP5-AS1 targets miR-367-3p/CEBPA axis and modulates the biological processes in glioma.Citation10 LncRNA ECONEXIN plays an oncogenic role in gliomagenesis.Citation11 LncRNA CRNDE facilitates the glioma progression through regulating miR-384/PIWIL4/STAT3 signaling.Citation12 Recent years, long intergenic non-protein coding RNA 641 (LINC00641) has been recognized as a prognostic predictor and tumor suppressor in bladder cancer.Citation13 LINC00641 was also downregulated in GBM and it could predict clinical outcomes of GBM patients.Citation14 Nevertheless, the specific role of LINC00641 in glioma cells still needs further studying.
The aim of our work was to study the expression and precise role of LINC00641 in glioma cells. In addition, the potential mechanism that LINC00641 exerted functions in glioma cells was probed in this study.
Materials and methods
Cell culture
Normal human astrocytes (NHAs), human embryonic kidney cell (HEK-293 T) and human glioblastoma cells (T98 G, LN18, U251, LN229 and U87) used in this research were all purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin antibiotics (Beyotime, Shanghai, China). The medium was changed every three days. Cell culture was conducted at 37°C under a humidified atmosphere of 5% CO2.
Cell transfection
When cell confluence reached 50–80%, T98 G and LN18 cells were seeded in 96-well plates. The transfection of cells was conducted as the specification of Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA). Design and synthesis of miR-4262 mimics and miR-NC in this study were accomplished by Genepharma (Shanghai, China). To enhance the expression of LINC00641 or NRGN, the pcDNA3.1 vector targeting LINC00641 or NRGN and the empty vector as control were bought from Genechem (Shanghai, China) and transfected into T98 G and LN18 cells. The small interfering RNA against NRGN (si-NRGN#1 and si-NRGN#2) and its corresponding control (si-NC) were constructed simultaneously by Genechem for silencing NRGN expression. Transfection was ceased after 48 h. The sequences for all transfection plasmids were listed as following: si-NC: 5ʹ-ACUUCCCCAUUUUAGACAU-3ʹ; si-NRGN#1: 5ʹ-ACGACGACAUUCUAGACAU-3ʹ; si-NRGN#2 5ʹ-GGACGACGACAUUCUAGAC-3ʹ; miR-NC: 5ʹ-GAAAUUUUGACUACCUG-3ʹ; miR-4262 mimics: 5ʹ-GACAUUCAGACUACCUG-3ʹ.
Quantitative real-time PCR
TRIzol reagent (Invitrogen) was used to extract total RNAs which were then reverse-transcribed into cDNA via reverse transcriptase kit (Takara, Tokyo, Japan). Next, qRT-PCR was carried out by SYBR Premix Ex Taq™ II (TaKaRa) to amplify the cDNA. The conditions of amplification were shown as below: at 95°C for 30 s, then 40 cycles of amplification at 95°C for 5 s, at 59°C for 30 s afterward, finally at 72°C for 30 s. Relative gene expression was measured by the 2−ΔΔCt method. LINC00641 and NRGN expressions were normalized to GAPDH, while miR-4262 expression was normalized to U6.
Cell viability assay
T98 G and LN18 cells were harvested after transfection and then planted in 96-well plates with 2000 cells each well. After 24, 48, 72 and 96 h, Cell Counting Kit-8 solution (CCK-8; Dojindo, Tokyo, Japan) without FBS was added to per well. A microplate reader was employed to examine absorbance at 450 nm wavelength.
EdU staining
In this study, EdU staining with Click-iT Alexa Fluor 488 Imaging Kit (Invitrogen) was performed to assess the proliferative ability. After transfection, T98 G and LN18 cells were initially rinsed twice with 3% bovine serum albumin (BSA; Sigma-Aldrich Company, Burlington, Massachusett, USA), and then treated with 150 μl EdU-Click reaction-mix at 20°C for 30 min, followed by washing with 3% BSA. Cell nuclei were stained with DAPI in the dark.
TUNEL staining
T98 G and LN18 cells, after transfection for 48 h, were rinsed with PBS (Sigma-Aldrich). Cells were then maintained in 4% PFA (Sigma-Aldrich) at room temperature for 30 min. TUNEL assay kit (Roche, Mannheim, Germany) was applied to incubate the cells in line with the specification of the manufacturer. The apoptotic cells were observed via a fluorescence microscope (Olympus, Tokyo, Japan). The green regions represented the apoptotic cells, while the blue regions represented the cell nuclei.
Flow cytometry analysis
Apoptosis rate of cells was evaluated using flow cytometry analysis according to a previous report.Citation150.25% trypsin was used to digest the transfected cells. Then, cells were washed with PBS twice. To prepare a total of 1 × 106 cells/ml suspension, cells were added with 100 µl binding buffer next. Followed by addition of Annexin V-FITC and PI, cells were incubated at room temperature for 5 min without light, and then examined by FACSVerse flow cytometry system.
Western blot analysis
RIPA lysis buffer (Solarbio, Beijing, China) was applied to extract protein which was subsequently quantified by BCA protein assay (Beyotime). Protein was separated on 8–15% SDS-PAGE gels. After that, protein was transferred to PVDF membranes (Millipore, Bedford, MA, USA). 5% nonfat milk was utilized to block the membranes in TBST at room temperature for 1 h and then treated with the primary antibodies, including cleaved caspase-3 (ab214430; Abcam, Cambridge, England), total caspase-3 (ab184787; Abcam), cleaved caspase-9 (#20750; Cell Signaling Technology, Danvers, MA, USA), total caspase-9 (ab219590; Abcam), NRGN (ab217672; Abcam) and GAPDH (ab8245; Abcam). GAPDH was taken as an endogenous control. Afterward, TBST was used to rinse the membranes for three times to incubate with secondary antibodies. The protein bands were monitored by the ECL system (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
RNA isolation of nuclear and cytoplasmic fractions
The PARIS Kit was utilized to evaluate the subcellular localization of LINC00641 based on the protocol provided by supplier. T98 G and LN18 cells were reaped and cultivated in cell fractionation buffer and centrifuged. The cell supernatant was collected as cytoplasmic fraction. The remaining lysates were rinsed in cell fractionation buffer, centrifuged and subjected to cell disruption buffer. Cell nuclei were extracted and acquired. U6 and GAPDH were detected in isolated RNAs as control for nuclear RNA and cytoplasm RNA, respectively.
Luciferase reporter assay
The wild-type and mutant sequences of miR-4262 in 3ʹUTR of NRGN (NRGN-WT and NRGN-Mut; Genepharma) were separately cloned into the pmirGLO luciferase reporter vector. HEK-293 T cells were co-transfected with NRGN-WT or NRGN-Mut and miR-4262 mimics or miR-4262 mimics+pcDNA3.1/LINC00641 or miR-NC. Simultaneously, HEK-293 T cells were co-transfected with the reporter plasmids of LINC00641-WT or LINC00641-Mut (Genepharma) and miR-4262 mimics or miR-NC. Transfection was carried out using Lipofectamine 2000 as previously stated. 48 hours later, the dual-luciferase reporter assay system (Promega, Madison, WI, USA) was employed to evaluate the relative luciferase activity.
RNA pull down assay
T98 G and LN18 cells were treated with the biotinylated miR-4262-WT or miR-4262-Mut or NC. Then, streptavidin-coated magnetic beads (Ambion, Austin, TX, USA) were added in the cell lysate. When the biotin-coupled RNA complex was pulled down, analysis of the enrichment of LINC00641 and NRGN was accordingly performed via qRT-PCR.
Ago2-RIP assay
Ago2-RIP assay was carried out to detect the combination of miR-4262 and LINC00641 or NRGN. Imprint RNA immunoprecipitation kit (Sigma-Aldrich) was applied to perform Ago2-RIP assay in accordance with the protocol. T98 G or LN18 cells were lysed in RIP lysis buffer (Solarbio). Then, cells were incubated in media supplemented with proteinase K (Absin, Shanghai, China) and anti-Ago2 (Millipore, Bedford, MA, USA) or anti-IgG (Millipore) overnight at 4°C. Afterward, immunoprecipitated RNA was purified and qRT-PCR was performed.
Bioinformatics analysis
The binding sequence of LINC00641 or NRGN in miR-4262 was predicted from StarBase v3.0 (http://starbase.sysu.edu.cn/). Using TCGA dataset (http://gepia.cancer-pku.cn/index.html), LINC00641 and NRGN were discovered to be lowly-expressed in glioma tissues in comparison with human normal tissues. Besides, LINC00641 was mainly enriched in cytoplasm through online LncLocator prediction tool (http://www.csbio.sjtu.edu.cn/cgi-bin/lncLocator.py).
Statistical analysis
Independent experiment was conducted for no less than three times. Statistical analyzes was made by SPSS 23.0 (SPSS Inc, Chicago, IL, USA). Results in this research were shown as mean ± standard deviation (SD). The Student’s t-test and one-way analysis of variance (ANOVA) were two methods for statistical analyses. The threshold of significant difference was p-value < 0.05.
Results
Overexpression of LINC00641 suppresses the proliferation of glioma cells
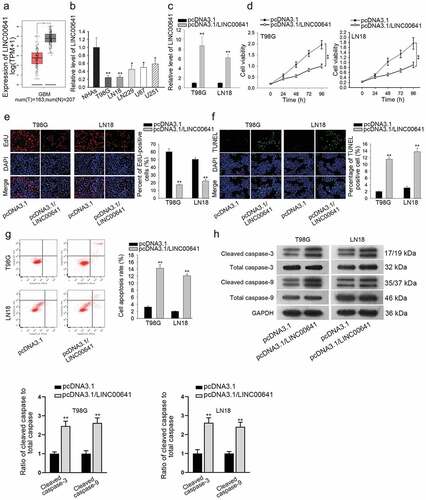
As predicted by TCGA, LINC00641 was markedly upregulated in GBM tissues when compared to the normal brain tissues (). Besides, the lower expression of LINC00641 was observed by qRT-PCR in glioma cell lines than the normal cell line NHAs (). Next, we explored the impact of LINC00641 on the malignant phenotypes of glioma cells through gain-of-function assays. As indicated in , the expression of LINC00641 was overtly enhanced in T98 G and LN18 cells transfected with pcDNA3.1/LINC00641. Consequently, upregulation of LINC00641 induced the distinct reduction of cell viability in T98 G and LN18 cells (). Consistently, the proliferation of T98 G and LN18 cells was also hindered in response to overexpression of LINC00641 through EdU assay (). Moreover, we found that the LINC00641 overexpression in glioma cells partially contributed to the promoted apoptosis (). Furthermore, cell apoptosis was confirmed by flow cytometry analysis and western blot analysis of apoptosis-related proteins. The enhanced apoptosis rate () and the increased protein level of cleaved caspase-3/9 were significantly observed in LINC00641-overexpressed cells (). Thus, we concluded that LINC00641 inhibits glioma cell proliferation and facilitates cell apoptosis.
Figure 1. LINC00641 was downregulated and acted as a tumor suppressor in glioma. (a) The expression of LINC00641 in GBM was obtained from TCGA. (b) Relative expression of LINC00641 in glioma cell lines and the normal NHAs was detected by qRT-PCR. (c) Transfection efficiency of pcDNA3.1/LINC00641 was evaluated using qRT-PCR. (d) CCK-8 assay was performed to assess cell viability upon LINC00641 overexpression. (e) The proliferation of T98 G and LN18 cells with or without LINC00641 overexpression was determined by EdU assay. (f) The apoptosis rate in above glioma cells was estimated through conducting TUNEL assay. (g) Flow cytometry analysis was implemented to evaluate apoptosis in T98 G and LN18 cells transfected with pcDNA3.1/LINC00641 or empty vector. (h) Apoptosis-related proteins were assessed in LINC00641-overexpressed cells by western blot analysis. *P < .05, **P < .01.
NRGN is an anti-tumor gene in the development of glioma
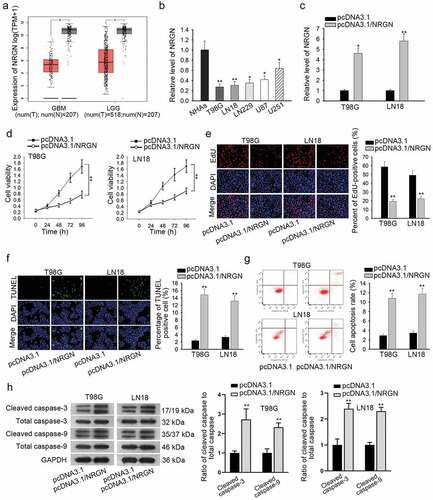
Subsequently, we attempted to explore the potential downstream effector that was involved in LINC00641-controlled glioma progression. NRGN, a gene that was proven to be downregulated in GBM.Citation16 Through TCGA database, NRGN expression was noticed at a lower level in both GBM and LGG (brain lower grade glioma) tissues compared to the normal ones (). Besides, qRT-PCR was carried out to verify that NRGN was downregulated in glioma cell lines (). Next, we investigated the functional role of NRGN in glioma cells through performing gain-of-function assays. The result of qRT-PCR indicated that NRGN was significantly overexpressed in T98 G and LN18 cells transfected with pcDNA3.1/NRGN (). Moreover, overexpression of NRGN weakened viability and impaired proliferative ability in both T98 G and LN18 cells ( and e). Conversely, the apoptosis rate was revealed to be obviously enhanced in NRGN-overexpressed glioma cells through TUNEL, flow cytometry and western blot analyses ( and h). Collectively, these findings suggested NRGN plays a tumor-suppressive role in the tumorigenesis of glioma.
Figure 2. NRGN was also an anti-tumor gene in glioma development. (a) TCGA revealed that NRGN was lowly-expressed in both GBM and LGG tissues. (b) qRT-PCR result presented NRGN expression level in indicated cells. (c) Overexpression efficiency of NRGN was assessed by qRT-PCR. (d and e) Cell viability and proliferation in T98 G and LN18 cells under NRGN upregulation were evaluated by CCK-8 and EdU assays, respectively. (f–g) TUNEL assay and flow cytometry analysis were carried out to determine the apoptosis of two glioma cells transfected with indicated plasmids. (h) Proteins associated with apoptosis were evaluated in NRGN-upregulated cells by western blot analysis. *P < .05, **P < .01.
Cytoplasmic LINC00641 is a positive regulator of NRGN in glioma
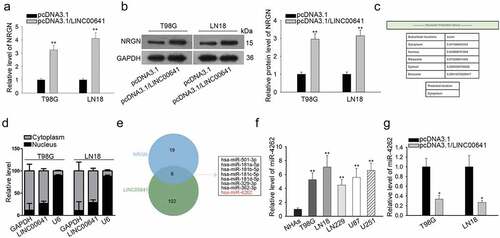
Further, we wondered whether there was an association between LINC00641 and NRGN in glioma. As a result, we uncovered that both mRNA and protein levels of NRGN were stimulated by overexpressed-LINC00641 in T98 G and LN18 cells ( and b). It indicated that NRGN was the downstream gene of LINC00641 in glioma cells. Considering that the functions of lncRNAs were dependent on its cellular distribution,Citation17 we examined the cellular localization of LINC00641 in glioma cells. According to lncLocator, LINC00641 was mainly located in the cytoplasm (). Besides, the cytoplasmic localization of LINC00641 in both T98 G and LN18 cells was also revealed by subcellular fractionation assay (). Recently, the implication of cytoplasmic lncRNAs in a competing endogenous RNA (ceRNA) network has been increasingly reported.Citation18 On this basis, we speculated that LINC00641 might affect NRGN expression through a ceRNA mechanism in glioma cells. As predicated by starBase v3.0, 8 miRNAs were found to bind with both LINC00641 and NRGN (). Among these 8 miRNAs, only miR-4262 has never been probed in glioma. Besides, the level of miR-4262 was apparently higher in glioma cell lines than that in the normal NHAs (). It was also shown that a notable decrease of miR-4262 level in T98 G and LN18 cells after overexpressing LINC00641 (). Taken together, LINC00641 may regulate NRGN expression through a ceRNA mechanism in glioma cells.
Figure 3. Cytoplasmic LINC00641 positively regulated NRGN in glioma cells. (a and b) The mRNA and protein expressions of NRGN in LINC00641-upregulated cells were assessed by qRT-PCR and western blot analyses, respectively. (c) The lncLocator (an online tool) predicted that LINC00641 mainly existed in the cytoplasm. (d) Subcellular fractionation plus qRT-PCR were used to confirm that LINC00641 located in the cytoplasm of T98 G and LN18 cells. (e) StarBase v3.0 suggested that there were 8 miRNAs that interacted with both LINC00641 and NRGN. (f) The level of miR-4262 in glioma cell lines and NHAs was tested using qRT-PCR. (g) qRT-PCR result was to display the expression of miR-4262 in LINC00641-overexpressed glioma cells. *P < .05, **P < .01.
LINC00641 is a ceRNA of NRGN in glioma by sponging miR-4262
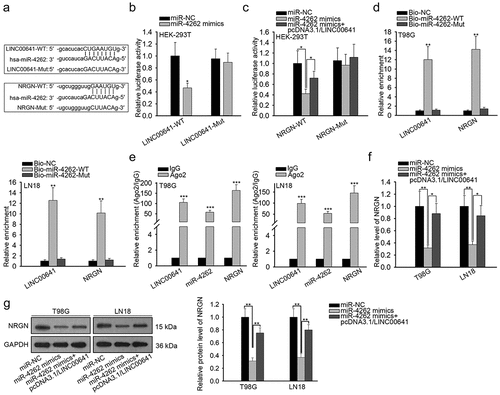
Thereafter, we focused on whether miR-4262 was involved in ceRNA mechanism, in which LINC00641 might modulate NRGN expression through miR-4262. In consequence, the interaction of miR-4262 with LINC00641 or NRGN was investigated. Firstly, the binding sequences of miR-4262 and LINC00641 or miR-4262 and NRGN were predicted by bioinformatics tools (). The luciferase reporter assay was conducted to verify that the luciferase activity of LINC00641-WT was sharply abated by miR-4262 mimics while that of LINC00641-Mut was not influenced by miR-4262 mimics (). Besides, the luciferase activity of NRGN-WT group but not NRGN-Mut group was markedly decreased by miR-4262 overexpression. However, overexpressed LINC00641 recovered the suppression of miR-4262 mimics on the luciferase activity of NRGN-WT (). Importantly, we validated that miR-4262 interacted with both LINC00641 and NRGN by RNA pull down assay and RIP assay ( and e). Furthermore, the decreased expression of NRGN at both mRNA and protein levels in cells transfected with miR-4262 mimics was later recovered by LINC00641 upregulation ( and g). These data revealed that LINC00641 enhances NRGN expression in glioma cells through absorbing miR-4262.
Figure 4. LINC00641 was a ceRNA of NRGN by absorbing miR-4262. (a) The sequences of miR-4262, LINC00641 (wild-type and mutated type) and NRGN (wild-type and mutated type) were represented. (b) The luciferase activity of LINC00641-WT/Mut with indicated transfections was proved by luciferase reporter assay. (c) Luciferase reporter assay was used to detect the luciferase activity of NRGN-WT/Mut in miR-NC, miR-4262 mimics and miR-4262 mimics+pcDNA3.1/LINC00641 groups, respectively. (d and e) The interaction of miR-4262 with LINC00641 and NRGN was validated through RNA pull down assay (d) and RIP assay (e). (f–g) The impact of miR-4262 overexpression and LINC00641 upregulation on NRGN expression was assessed using qRT-PCR (f) and western blot analysis (g). *P < .05, **P < .01, ***P < .001.
LINC00641 restrains glioma development through the NRGN-mediated pathway
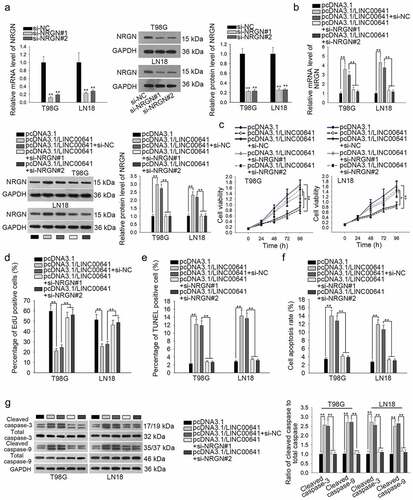
In depth, we wondered whether NRGN was implicated in LINC00641-suppressed glioma cells. Firstly, we silenced NRGN and both qRT-PCR and western blot analyses were utilized to confirm the interference efficiency of si-NRGN#1/2 in T98 G and LN18 cells (). The results showed that the mRNA and protein levels of NRGN were observably downregulated after silencing NRGN in glioma cells. Next, we confirmed that both the mRNA and protein levels of NRGN stimulated by LINC00641 overexpression were recovered by silenced NRGN in T98 G and LN18 cells (). Subsequently, rescue assays were conducted to examine the role of LINC00641/NRGN in the growth of two glioma cell line. As a consequence, the confined proliferation of LINC00641-upregulated T98 G and LN18 cells was revived by NRGN depletion through CCK-8 and EdU assays ( and d). Also, knockdown of NRGN offset the increased apoptosis rate caused by LINC00641 silencing in T98 G and LN18 cells through carrying out TUNEL assay, flow cytometry and western blot analyses (–g). Based on these findings, we concluded that LINC00641 dampens glioma cell proliferation through NRGN-dependent mechanism.
Figure 5. Inhibition of NRGN countervailed the suppressive effect of LINC00641 overexpression on the proliferation of glioma cells. (a) The mRNA and protein levels of NRGN in two glioma cell lines transfected with si-NRGN#1/2 were respectively evaluated using qRT-PCR and western blot analysis. si-NC group was taken as the negative control. (b) NRGN expression in LINC00641-overexpressed cells was evaluated after co-transfected with siRNAs targeting NRGN by qRT-PCR and western blot analysis. (c and d) The role of NRGN in LINC00641-mediated cell proliferation was evaluated by CCK-8 and EdU assays. (e–g) The influence of NRGN silence on the apoptosis of LINC00641-upregulated T98 G and LN18 cells was estimated via TUNEL assay, flow cytometry and western blot analyses. *P < .05, **P < .01.
Discussion
Amount of increasing evidence illustrates that dysregulated lncRNAs usually participate in the initiation and progression of diverse tumors,Citation19 including glioma.Citation11,Citation20 For instance, HOXA11-AS facilitates the glioma cell proliferation through modulating cell cycle.Citation21 CCND2-AS1 promotes glioma cell proliferation by regulating Wnt/β-catenin signaling.Citation22 CRNDE accelerates the growth of glioma cells via mTOR signaling.Citation23 In this study, we disclosed a novel lncRNA, LINC00641, as a tumor suppressor since it blocked cell proliferation but stimulated apoptosis in glioma cells, which was in accord with the previous findings that LINC00641 exerted an anti-cancer function in bladder cancer.Citation13
NRGN (neurogranin) is the human homolog of the neuron-specific rat RC3/neurogranin gene which exhibits an abnormal expression in brain. Recently, researchers have recognized the importance of NRGN in the pathogenesis of several diseases attacking the brain, such as Parkinson’s disease,Citation24 schizophreniaCitation25 and Alzheimer disease.Citation26 Previously, Yokota, T., et al. reported that NRGN was lowly-expressed in GBM tissues,Citation16 which was further confirmed by the data from TCGA. Nevertheless, the specific role of NRGN in glioma has not been identified before. In the present study, we elucidated that NRGN played an inhibitory role in the development of glioma for the first time.
In the past decades, lncRNAs have been widely reported as ceRNAs to regulate mRNA expression through competitively sequestering certain miRNA in various human cancers.Citation27 As an example, CRNDE contributes to glioma progression through targeting miR-384/PIWIL4/STAT3 signaling.Citation12 HOXD-AS1 sponges miR-130a to regulate glioma development by targeting E2 F8.Citation28 PVT1 aggravates glioma tumorigenesis by modulating miR-128-3p/GREM1 axis.Citation29 Herein, we certified that there was a competition between LINC00641 and NRGN to interact with miR-4262 in glioma cells. Here, we also confirmed that LINC00641 elicited its repressive function in glioma by upregulating NRGN via sponging miR-4262.
Collectively, our findings identified that LINC00641 plays an anti-tumorigenic role in glioma progression through releasing NRGN from miR-4262, which suggests a new therapeutic target for glioma patients.
Acknowledgments
We acknowledge the support of all individuals involved in our study.
References
- Morgan LL. The epidemiology of glioma in adults: a “state of the science” review. Neuro-oncology. 2015;17(4):623–624. doi:10.1093/neuonc/nou358.
- Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of gliomas. In: Raizer J, Parsa A, editors. Current understanding and treatment of gliomas. Cham, Switzerland: Springer International Publishing; 2015. p. 1–14.
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828.
- Reifenberger G, Wirsching H-G, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas — implications for classification and therapy. Nat Rev Clin Oncol. 2016;14:434. doi:10.1038/nrclinonc.2016.204.
- Fan Z, Zheng J, Xue Y, Liu X, Wang D, Yang C, Ma J, Liu L, Ruan X, Wang Z, et al. NR2C2-uORF targeting UCA1-miR-627-5p-NR2C2 feedback loop to regulate the malignant behaviors of glioma cells. Cell Death Dis. 2018;9:1165. doi:10.1038/s41419-018-1149-x.
- Lin X, Jiang T, Bai J, Li J, Wang T, Xiao J, Tian Y, Jin X, Shao T, Xu J, et al. Characterization of transcriptome transition associates long noncoding RNAs with glioma progression. Mol Ther Nucleic Acids. 2018;13:620–632. doi:10.1016/j.omtn.2018.10.009.
- Xu N, Liu B, Lian C, Doycheva DM, Fu Z, Liu Y, Zhou J, He Z, Yang Z, Huang Q, et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis. 2018;9:1139. doi:10.1038/s41419-018-1183-8.
- Linz U. Aspects on the survival of patients with glioblastoma and the origin and histology of oligodendrogliomas. Nat Rev Cancer. 2003;3:77–82. doi:10.1038/nrc1121-c1.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi:10.1038/nrc.2017.99.
- Liu X, Zheng J, Xue Y, Yu H, Gong W, Wang P, Li Z, Liu Y. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics. 2018;8:1084–1105. doi:10.7150/thno.21740.
- Deguchi S, Katsushima K, Hatanaka A, Shinjo K, Ohka F, Wakabayashi T, Zong H, Natsume A, Kondo Y. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene. 2017;36:4629. doi:10.1038/onc.2017.88.
- Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, Liu Y. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24:1199–1215. doi:10.1038/mt.2016.71.
- Li Z, Hong S, Liu Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun. 2018;503:1825–1829. doi:10.1016/j.bbrc.2018.07.120.
- Liang R, Zhi Y, Zheng G, Zhang B, Zhu H, Wang M. Analysis of long non-coding RNAs in glioblastoma for prognosis prediction using weighted gene co-expression network analysis, Cox regression, and L1-LASSO penalization. Onco Targets Ther. 2018;12:157–168. doi:10.2147/OTT.S171957.
- Hu Y, Ma Y, Liu J, Cai Y, Zhang M, Fang X. LINC01128 expedites cervical cancer progression by regulating miR-383-5p/SFN axis. BMC Cancer. 2019;19:1157.
- Yokota T, Kouno J, Adachi K, Takahashi H, Teramoto A, Matsumoto K, Sugisaki Y, Onda M, Tsunoda T. Identification of histological markers for malignant glioma by genome-wide expression analysis: dynein, α-PIX and sorcin. Acta Neuropathol. 2006;111:29–38. doi:10.1007/s00401-005-1085-6.
- Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9:e1471. doi:10.1002/wrna.1471.
- Ulitsky I. Interactions between short and long noncoding RNAs. FEBS Lett. 2018;2874–2883. doi:10.1002/1873-3468.13085.
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi:10.1158/2159-8290.CD-11-0209.
- Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, Chen Q, Wei C, Fu H, Xu T, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018;24:2002–2014. doi:10.1158/1078-0432.CCR-17-2376.
- Wang Q, Zhang J, Liu Y, Zhang W, Zhou J, Duan R, Pu P, Kang C, Han L. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016;373:251–259. doi:10.1016/j.canlet.2016.01.039.
- Zhang H, Wei D-L, Wan L, Yan S-F, Sun Y-H. Highly expressed lncRNA CCND2-AS1 promotes glioma cell proliferation through Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2017;482:1219–1225. doi:10.1016/j.bbrc.2016.12.016.
- Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi:10.1016/j.canlet.2015.03.027.
- Koob AO, Shaked GM, Bender A, Bisquertt A, Rockenstein E, Masliah E. Neurogranin binds α-synuclein in the human superior temporal cortex and interaction is decreased in parkinson’s disease. Brain Res. 2014;1591:102–110. doi:10.1016/j.brainres.2014.10.013.
- Ohi K, Hashimoto R, Yasuda Y, Fukumoto M, Yamamori H, Umeda-Yano S, Fujimoto M, Iwase M, Kazui H, Takeda M. Influence of the NRGN gene on intellectual ability in schizophrenia. J Hum Genet. 2013;58:700. doi:10.1038/jhg.2013.82.
- Kester MI, Teunissen CE, Crimmins DL, Herries EM, Ladenson JH, Scheltens P, van der Flier WM, Morris JC, Holtzman DM, Fagan AM. Neurogranin as a cerebrospinal fluid biomarker for synaptic loss in symptomatic alzheimer disease. JAMA Neurol. 2015;72:1275–1280. doi:10.1001/jamaneurol.2015.1867.
- Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8:30. doi:10.1186/s13045-015-0129-1.
- Chen Y, Zhao F, Cui D, Jiang R, Chen J, Huang Q, Shi J. HOXD-AS1/miR-130a sponge regulates glioma development by targeting E2F8. Int J Cancer. 2018;142:2313–2322. doi:10.1002/ijc.31262.
- Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15:1139–1157. doi:10.1007/s13311-018-0649-9.
