ABSTRACT
Background
Antibodies against epidermal growth factor receptor (EGFR), panitumumab, a fully human monoclonal antibody, and cetuximab, a human/mouse chimeric monoclonal antibody, have shown clinical efficacy in metastatic colorectal cancer (mCRC). In the phase 3 noninferiority ASPECCT (ClinicalTrials.gov, NCT01001377) study, panitumumab was demonstrated to be noninferior to cetuximab and provided a similar overall survival benefit for patients with chemotherapy-refractory wild-type KRAS exon 2 mCRC. However, some patients eventually develop resistance to anti-EGFR therapy. EGFR p.S492R mutation was previously identified as conferring resistance to cetuximab, but not to panitumumab.
Methods
This biomarker study analyzed plasma samples from ASPECCT collected at both baseline and posttreatment.
Results
No EGFR p.S492R mutations were identified at baseline; however, after treatment the EGFR p.S492R mutation was detected in 1% of patients treated with panitumumab versus 16% of those treated with cetuximab, supporting that, in a large population, this mutation is more likely to be induced by cetuximab than by panitumumab. There were, however, no significant differences in progression-free survival or overall survival between patients who were wild-type compared with those with the S492R mutation within the cetuximab arm or the overall population.
Conclusions
These results may support targeting treatment to small patient subgroups based on the presence of emerging EGFR mutations and provide a molecular rationale for rechallenging with a different anti-EGFR agent in patients who develop resistance. Prospective studies are needed to evaluate the efficacy of panitumumab in the EGFR p.S492R mutant population.
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwideCitation1 and is frequently associated with an overexpression of the epidermal growth factor receptor (EGFR).Citation2,Citation3 Panitumumab is a fully human monoclonal antibody that binds specifically to EGFR, competitively inhibiting ligand binding, and has demonstrated efficacy in the treatment of metastatic CRC (mCRC).Citation2 Cetuximab is a human/mouse chimeric monoclonal antibody that binds to the extracellular domain of human EGFR, also blocking ligand binding.Citation4 In CRC cell lines and patients treated with cetuximab, development of the S492R mutation in the EGFR cetuximab-binding epitope conferred resistance to cetuximab therapy. Although the S492R mutation blocked binding of cetuximab, panitumumab treatment was found to still be effective.Citation5,Citation6
The ASPECCT trial (A Study of Panitumumab Efficacy and Safety Compared to Cetuximab) was a phase 3 noninferiority study of panitumumab versus cetuximab monotherapy in patients with chemotherapy-refractory wild-type KRAS exon 2 mCRC and included 999 patients from 27 countries in North America, South America, Europe, Asia, Africa, and Australia.Citation7,Citation8 Patients who had previously received anti-EGFR therapy were excluded.Citation8 The primary endpoint was overall survival (OS), and key secondary endpoints included progression-free survival (PFS), objective response rate, and safety.Citation7,Citation8 In the intent-to-treat population, results indicated that panitumumab was noninferior to cetuximab for OS,Citation8 and the incidence of treatment-emergent adverse events (AEs) was similar in both treatment arms (~98% each).Citation8 Fatal serious AEs (SAEs) occurred in 6% and 10% of patients in the panitumumab and cetuximab arms, respectively.Citation8
Real-time monitoring of circulating cell-free DNA (cfDNA) has been used to detect mutations potentially conferring treatment resistance. It is estimated that up to 3.3% of tumor DNA may enter the blood daily,Citation9 with the fraction of circulating DNA that is tumor derived ranging between 0.01% and 93%.Citation10 Circulating cfDNA has a half-life ranging from 15 minutes to several hours and is cleared by the liver and kidney, thus making it a good biomarker for real-time emerging tumor mutation assessment.Citation9 Mutations detected in plasma show good agreement with tumor tissue mutations, particularly when samples are paired.Citation10–12
The objective of this analysis of data from the ASPECCT study was to determine whether the induced EGFR p.S492R mutation was specifically associated with resistance to cetuximab but not panitumumab in a large clinical trial population.
Results
In vitro receptor binding and activity
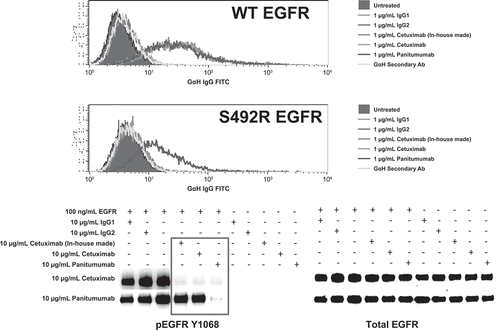
Both flow cytometry and immunoblotting data showed that panitumumab, but not cetuximab, bound to and inhibited the activation of S492R mutant EGFR ().
Mutational analyses
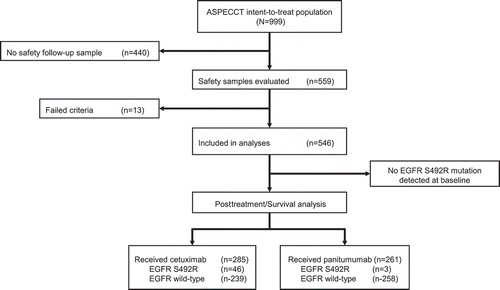
Of the 999 patients in the ASPECCT intent-to-treat population, safety follow-up samples, collected 4 weeks after the final dose was administered, were evaluated for 559 patients and were included in this analysis (). Of these, 13 samples were classed as failed based on the criteria of the analytical method. In the overall study, the median duration of treatment was 14 weeks in both arms. The ascertainment rates were similar between arms: 52.6% in the panitumumab arm (n = 261/496) and 56.7% in the cetuximab arm (n = 285/503).
The EGFR p.S492R mutation was detected in 1.1% of patients (n = 3/261) treated with panitumumab and 16.1% (n = 46/285) of those treated with cetuximab (P < .0001; ). Pretreatment plasma samples were subsequently analyzed from 48 mutation-positive samples and 51 randomly selected samples that were wild-type at safety follow-up. No mutations were observed at baseline for any samples analyzed.
Table 1. Frequency of EGFR p.S492R mutations in plasma samples for patients in the ASPECCT study
Survival analyses
Overall, PFS and OS analyses were evaluated in the 546 patients included in the mutational analyses; all patients had received at least one dose of cetuximab or panitumumab. Of these, 49 patients had tumors containing the S492R mutation. In total, 285 patients had received cetuximab, including 46 patients with the S492R mutation, and 261 patients had received panitumumab, including three patients with the S492R mutation.
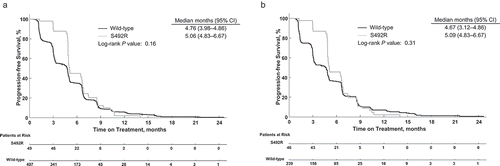
Overall, an initial separation in median PFS was observed; however, this difference was no longer apparent after 6 months (). Median PFS for all patients was 4.76 (95% CI, 3.98–4.86) months in the wild-type group and 5.06 (95% CI, 4.83–6.67) months in the S492R group (P = .16). For patients treated with cetuximab, median PFS was 4.67 (95% CI, 3.12–4.86) months in the wild-type group and 5.09 (95% CI, 4.83–6.67) months in the S492R group (P = .31; ). Data from patients treated with panitumumab were not included in PFS analyses because of the low number of patients with the S492R mutation.
Figure 3. Progression-free survival for patients with or without the EGFR p.S492R mutation for (a) all patients, (b) patients treated with cetuximab
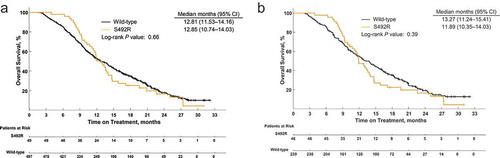
Similarly, an initial difference in median OS overall was observed; however, this difference was no longer apparent after 12 months (). Median OS for all patients was 12.81 (95% CI, 11.53–14.16) months in the wild-type group and 12.85 (95% CI, 10.74–14.03) months in the S492R group (P = .66). For patients treated with cetuximab, median OS was 13.27 (95% CI, 11.24–15.41) months in the wild-type group and 11.89 (95% CI, 10.35–14.03) months in the S492R mutant group (P = .39; ). Data from patients treated with panitumumab were also not included in OS analyses because of the low number of patients with the S492R mutation.
Discussion
The S492R mutation has potential implications for treatment because in preclinical models it selectively disrupts binding of cetuximab but not panitumumab, a finding that our results reinforce.Citation5,Citation6 In this analysis, no S492R mutations were identified in the baseline tumor samples analyzed, consistent with previous studies.Citation13 After treatment, the EGFR p.S492R mutation was detected more frequently in patients treated with cetuximab (16.1%) than in those treated with panitumumab (1.1%), although there were no significant differences in median OS and PFS between the wild-type and mutant groups.
Panitumumab and cetuximab have demonstrated clinical efficacy and improved OS in the first- or second-line treatment of mCRC leading to earlier use of anti-EGFR therapy in mCRCCitation14; however, a significant number of patients progress after two or more lines of therapy, thus raising the possibility of rechallenging with anti-EGFR therapy as a treatment option for this pretreated population, particularly in patients who had previously responded.Citation15–17
It has also been shown that the colorectal tumor genome can adapt dynamically in response to a treatment schedule; mutant clones of EGFR pathway genes such as KRAS have been shown to emerge during anti-EGFR treatment and then subsequently decline upon withdrawal of the anti-EGFR agent.Citation18 This change in the presence of EGFR pathway genes provides the molecular rationale for repeated use of anti-EGFR therapy in a rechallenge setting, with an understanding of markers of resistance allowing for more rational drug selection.
At present, consideration of rechallenge therapy seems to be largely based on initial response and the subsequent onset of secondary resistance. However, despite previous demonstrations of clinical benefit derived from rechallenge with anti-EGFR therapies in patients pretreated with the same protocol,Citation15,Citation18 as rechallenge becomes more relevant in the clinical setting, there is a need to identify additional indicators (e.g., biomarkers) to guide therapy options to achieve the optimal outcome.
This retrospective analysis of ASPECCT identified that 16.1% of patients in the cetuximab arm developed EGFR p.S492R mutations, compared with 1.1% in the panitumumab arm, indicating that the S492R mutation is more likely to be induced by cetuximab rather than panitumumab. The preclinical results shown here reinforce those previously reported, suggesting that patients with EGFR p.S492R mutant tumors may derive benefit from panitumumab versus cetuximab as rechallenge therapy or as a potential option for treatment after initial response and subsequent progression.
The optimal timing for testing of mutation status remains to be established and may depend on line of treatment (e.g., second line, before cetuximab rechallenge, or during therapy). Testing before rechallenge is important and should be considered where available in clinical practice. Furthermore, it has been shown that frequent monitoring of tumor heterogeneity (i.e., every 4 weeks) may help predict drug resistance depending on the expression levels of subclones that dominate resistance to particular therapies.Citation19 The advent of more sensitive assays for mutation detection, especially in the context of liquid biopsies, has allowed for tracking tumor clonal evolution. This will potentially provide a means by which one may predict how to best rechallenge a patient to achieve the most durable outcome.
Our analysis suggests that patients with EGFR p.S492R mutant tumors initially do well on anti-EGFR therapy, possibly due to the effect of treatment on EGFR wild-type cells. Over time cetuximab treatment becomes less effective, possibly as a result of the growth of the resistant S492R clone, which may be an early event preceding radiologic progression. This observation again supports the need for more frequent monitoring so that when a mutation first occurs, therapy can be modified or a switch to a different anti-EGFR agent can be considered.
Additional mutations in other EGFR pathway genes (KRAS, NRAS, BRAF, MAP2K1, PIK3CA, PTEN) and their downstream effectors have been associated with resistance to EGFR inhibitors in patients with mCRC.Citation20–24 Mutation profiles of EGFR pathway genes at both baseline and safety follow-up were reported in a previous analysis in patients from the panitumumab arm of the ASPECCT study. It was shown that baseline mutations in EGFR pathway genes were associated with shorter survival.Citation25 It is noted that cetuximab and panitumumab now include product labeling specifying that patients receiving treatment should have confirmed wild-type KRAS or RAS mCRC, respectively.Citation2,Citation4 Other EGFR mutations (e.g., R451C, S464L, G465R, K467T, and I491M) conferring resistance to cetuximab have been reported; of these panitumumab has been shown to prevent EGFR activation in the R451C and K467T mutants.Citation26 Further analysis of ASPECCT data may reveal the benefit of panitumumab in subgroups harboring these mutants.
Evidence from the Sym004-05 trial indicated that targeting anti-EGFR treatment according to the presence of mutations may be beneficial.Citation27 Using circulating tumor DNA as a guide, it was shown that there was no OS or PFS benefit with Sym004 (a mixture of two nonoverlapping anti-EGFR monoclonal antibodies) in unselected patients; however, OS was prolonged in a selected subgroup with “triple-negative” (i.e., RAS, BRAF, and EGFR extracellular domain wild-type) mCRC.Citation27 This serves as an additional example of the importance of understanding the molecular landscape at the time of commencing these new agents.
However, there are limitations to our analysis, and thus the survival results should be interpreted with caution and should not be used to guide clinical practice. First, the exact timing of EGFR p.S492R onset in our samples is unknown because the plasma samples were collected at only two time points (i.e., pretreatment and during safety follow-up). Therefore, the impact of this mutation on overall survival could not be accurately evaluated in the current retrospective analysis. Furthermore, the number of patients with the EGFR p.S492R mutation was very low in the panitumumab arm. As a result, no significant overall survival results could be derived for patients with wild-type tumors or for those with the EGFR p.S492R mutation in this treatment arm. However, another study found panitumumab to be effective after cetuximab resistance in a patient harboring the S492R mutation,Citation5 suggesting that switching to panitumumab may be a viable option. Tissue samples were not assessed in this study, which is a potential limitation; however, others have found a good agreement between mutations detected in plasma and those detected in tissue.10−12
Prospective studies are needed to evaluate the efficacy of panitumumab in the S492R mutant population. Overall our results suggest that there may be benefit targeting treatment to small patient subgroups based on the presence of emerging mutations. It remains unclear whether the S492R mutation is induced by treatment or if it occurs randomly; however, if it is the former, methods for predicting which patients are likely to develop the mutation may be beneficial.
Tumor sidedness was also not evaluated in this study as these data were not available. Furthermore, tissue analysis of extended RAS/BRAF mutations was not available for ASPECCT. A greater understanding of the impact of other mutations would be interesting and possibly clinically useful.
Conclusions
The EGFR p.S492R mutation was detected in 1.1% of chemotherapy refractory KRAS wild-type mCRC patients treated with panitumumab and in 16.1% of patients treated with cetuximab using a sensitive analysis technique (ddPCR), similar to results from prior publications. Mutations were not observed in pretreatment plasma samples, providing additional confidence in the low error rate of this assay and indicating that this resistance mutation probably arose as a result of predominantly cetuximab therapy. “Liquid biopsy” in combination with sensitive molecular testing may be feasible in patients with mCRC, with the potential to improve patient selection, detect disease progression and the emergence of resistance, and thereby guide therapeutic decisions. Rechallenge protocols could be developed using induced EGFR mutation analysis as a treatment guide. These protocols could also be useful for other mutations that may develop, such as additional cetuximab-induced mutations and newly identified panitumumab-induced mutations. It is possible that the different emerging mutations that arise may indicate subtle differences in the mechanism of action of the two monoclonal antibodies, possibly due to differences in their binding mechanisms.
Patients and methods
In vitro receptor binding and activity assays
In this analysis, the binding of panitumumab and cetuximab to wild-type EGFR or mutant EGFR p.S492R was analyzed in transduced Chinese hamster ovary cells by flow cytometry. The activity of panitumumab and cetuximab on wild-type EGFR and mutant EGFR p.S492R was assessed by immunoblotting.
Mutational analyses
Patient plasma samples from ASPECCT (ClinicalTrials.gov, NCT01001377) were collected before treatment and at the safety follow-up visit 4 weeks after the last dose of anti-EGFR therapy. Testing of the safety-follow-up samples was conducted without any knowledge of the treatment the patient had received, and the analysis plan was finalized before any data were transferred. Subsequently, the pretreatment samples for the mutation-positive patients and a randomly selected sample from patients expressing wild-type EGFR were analyzed. Mutation frequency in EGFR codon 492 was monitored in patient plasma samples using droplet digital polymerase chain reaction (ddPCR) on a BioRad Qx100 instrument. The ddPCR assay partitions a plasma sample into thousands of discrete amplification events, counting individual target molecules, thereby allowing the detection of rare mutations. The ddPCR assay has several advantages over the standard quantitative polymerase chain reaction (qPCR) assay, specifically improved precision, sensitivity, and reproducibility. The ddPCR can detect ≤1.1-fold differences compared with twofold differences for qPCR; endpoint analysis is less sensitive to inhibitors with ddPCR, and there is no PCR bias due to efficiency of the assays, there is no normalization to housekeeping genes required, and there are no standard curves.
The ddPCR assay was conducted using a basic TaqMan® assay (ThermoFisher Scientific, Waltham, MA) and a single primer set. A multiplex of four primers/probes was used to target three point mutations (i.e., nucleotide changes, C1476A [AGA], C1476G [AGG], and A1474C [CGC]) as well as wild-type EGFR [i.e., corresponding nucleotide, AGC]. The assay was designed to detect the presence of the EGFR p.S492R mutation in plasma using a sliding threshold for positivity at 0.2% at high DNA concentrations (i.e., >20,000 copies) and 1% at low DNA concentrations (i.e., 2000 copies). The thresholds were selected to ensure samples labeled mutant were truly mutant with a posterior probability of at least 99.0%. The false-positive rate was approximately 1 in 16,5000.
Survival analyses
PFS and OS were assessed for all patients with evaluable safety-follow-up samples. PFS and OS were also assessed in pre-specified patient subgroups (i.e., those with tumors expressing wild-type EGFR and those with tumors expressing the EGFR p.S492R mutation). Kaplan–Meier curves depicting cumulative probability of OS and PFS were developed.
Disclosure of potential conflicts of interest
T. Price has served as a consultant/advisory board member for Amgen Inc., Merck Serono, and Takeda. A. Ang and M. Boedigheimer are employees of and own stock in Amgen Inc. T. W. Kim has received research funding from Merck Serono, AstraZeneca, and Pfizer. J. Li has received speakers bureau honoraria from Eli Lilly. S. Cascinu has received research funding from Eisai and Celgene, consulting fees from Amgen Inc., and speakers bureau honoraria from Amgen Inc, and Servier. P. Ruff has received institutional research funding from Amgen Inc., and institutional speakers bureau honoraria from Amgen Inc., Merck Serono, and Sanofi. A. S. Suresh has nothing to disclose. A. Thomas has received consulting fees from Pierre Fabre and speakers bureau honoraria from Amgen Inc. S. Tjulandin owns stock in RosPharmTech and has received speakers bureau honoraria from AstraZeneca, Lilly, Merck Sharpe & Dohme, and Biocad. M. Peeters has received speakers bureau honoraria from and has served as a consultant/advisory board member for Amgen Inc.
Acknowledgments
The authors thank Lee Hohaia, PharmD, and Meghan Johnson, PhD (ICON, North Wales, PA, USA), whose work was funded by Amgen Inc., for medical writing assistance in the preparation of this manuscript.
Data sharing statement
There is a plan to share data. This may include de-identified individual patient data for variables necessary to address the specific research question in an approved data-sharing request; also related data dictionaries, study protocol, statistical analysis plan, informed consent form, and/or clinical study report. Data sharing requests relating to data in this manuscript will be considered after the publication date and 1) this product and indication (or other new use) have been granted marketing authorization in both the US and Europe, or 2) clinical development discontinues and the data will not be submitted to regulatory authorities. There is no end date for eligibility to submit a data sharing request for these data. Qualified researchers may submit a request containing the research objectives, the Amgen product(s) and Amgen study/studies in scope, endpoints/outcomes of interest, statistical analysis plan, data requirements, publication plan, and qualifications of the researcher(s). In general, Amgen does not grant external requests for individual patient data for the purpose of re-evaluating safety and efficacy issues already addressed in the product labeling. A committee of internal advisors reviews requests. If not approved, requests may be further arbitrated by a Data Sharing Independent Review Panel. Requests that pose a potential conflict of interest or an actual or potential competitive risk may be declined at Amgen’s sole discretion and without further arbitration. Upon approval, information necessary to address the research question will be provided under the terms of a data sharing agreement. This may include anonymized individual patient data and/or available supporting documents, containing fragments of analysis code where provided in analysis specifications. Further details are available at the following: https://wwwext.amgen.com/science/clinical-trials/clinical-data-transparency-practices/
Additional information
Funding
References
- International Agency for Research on Cancer, World Health Organization. Colorectal cancer. [Accessed 2018 Sept 17]. http://gco.iarc.fr/today/data/factsheets/cancers/10_8_9-Colorectum-fact-sheet.pdf
- Vectibix® (panitumumab). Full prescribing information. Thousand Oaks (CA): Amgen Inc. 2017
- Radinsky R, Risin S, Fan D, Dong Z, Bielenberg D, Bucana CD, Fidler IJ. Level and function of epidermal growth factor receptor predict the metastatic potential of human colon carcinoma cells. Clin Cancer Res. 1995;1(1):19–31.
- Erbitux® (cetuximab). Full prescribing information. Indianapolis (IN): Eli Lilly and Company. 2018
- Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, Salido M, Gallen M, Marsters S, Tsai SP, et al. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18(2):221–223. doi:10.1038/nm.2609.
- Bardelli A, Janne PA. The road to resistance: EGFR mutation and cetuximab. Nat Med. 2012;18(2):199–200. doi:10.1038/nm.2646.
- Kim TW, Peeters M, Thomas A, Gibbs P, Hool K, Zhang J, Ang AL, Bach BA, Price T. Impact of emergent circulating tumor DNA RAS mutation in panitumumab-treated chemoresistant metastatic colorectal cancer. Clin Cancer Res. 2018;24(22):5602–5609. doi:10.1158/1078-0432.CCR-17-3377.
- Price TJ, Peeters M, Kim TW, Li J, Cascinu S, Ruff P, Suresh AS, Thomas A, Tjulandin S, Zhang K, et al. Panitumumab versus cetuximab in patients with chemotherapy-refractory wild-type KRAS exon 2 metastatic colorectal cancer (ASPECCT): a randomised, multicentre, open-label, non-inferiority phase 3 study. Lancet Oncol. 2014;15(6):569–579. doi:10.1016/S1470-2045(14)70118-4.
- Wang R, Li X, Zhang H, Wang K, He J. Cell-free circulating tumor DNA analysis for breast cancer and its clinical utilization as a biomarker. Oncotarget. 2017;8(43):75742–75755. doi:10.18632/oncotarget.20608.
- Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi:10.1038/nrclinonc.2013.110.
- Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Fetting J, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18(12):3462–3469. doi:10.1158/1078-0432.CCR-11-2696.
- Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, Higashiyama M, Kodama K, Imamura F, Kato K. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17(24):7808–7815. doi:10.1158/1078-0432.CCR-11-1712.
- Esposito C, Rachiglio AM, La Porta ML, Sacco A, Roma C, Iannaccone A, Tatangelo F, Forgione L, Pasquale R, Barbaro A, et al. The S492R EGFR ectodomain mutation is never detected in KRAS wild-type colorectal carcinoma before exposure to EGFR monoclonal antibodies. Cancer Biol Ther. 2013;14(12):1143–1146. doi:10.4161/cbt.26340.
- Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–1422. doi:10.1093/annonc/mdw235.
- Santini D, Vincenzi B, Addeo R, Garufi C, Masi G, Scartozzi M, Mancuso A, Frezza AM, Venditti O, Imperatori M, et al. Cetuximab rechallenge in metastatic colorectal cancer patients: how to come away from acquired resistance? Ann Oncol. 2012;23(9):2313–2318. doi:10.1093/annonc/mdr623.
- Pietrantonio F, Perrone F, Biondani P, Maggi C, Lampis A, Bertan C, Venturini F, Tondulli L, Ferrari D, Ricci V, et al. Single agent panitumumab in KRAS wild-type metastatic colorectal cancer patients following cetuximab-based regimens: clinical outcome and biomarkers of efficacy. Cancer Biol Ther. 2013;14(12):1098–1103. doi:10.4161/cbt.26343.
- Liu X, George GC, Tsimberidou AM, Naing A, Wheler JJ, Kopetz S, Fu S, Piha-Paul SA, Eng C, Falchook GS, et al. Retreatment with anti-EGFR based therapies in metastatic colorectal cancer: impact of intervening time interval and prior anti-EGFR response. BMC Cancer. 2015;15(1):713. doi:10.1186/s12885-015-1701-3.
- Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795–801. doi:10.1038/nm.3870.
- Khan KH, Cunningham D, Werner B, Vlachogiannis G, Spiteri I, Heide T, Mateos JF, Vatsiou A, Lampis A, Damavandi MD, et al. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C phase II colorectal cancer clinical trial. Cancer Discov. 2018;8(10):1270–1285. doi:10.1158/2159-8290.CD-17-0891.
- Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023–1034. doi:10.1056/NEJMoa1305275.
- Van Cutsem E, Lenz H-J, Köhne C-H, Heinemann V, Tejpar S, Melezínek I, Beier F, Stroh C, Rougier P, van Krieken JH, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol. 2015;33(7):692–700. doi:10.1200/JCO.2014.59.4812.
- Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69(5):1851–1857. doi:10.1158/0008-5472.CAN-08-2466.
- van Brummelen EMJ, de Boer A, Beijnen JH, Schellens JHM. BRAF mutations as predictive biomarker for response to anti-EGFR monoclonal antibodies. Oncologist. 2017;22(7):864–872. doi:10.1634/theoncologist.2017-0031.
- Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, Godfrey JT, Nishimura K, Lynch KD, Mermel CH, et al. Clinical acquired resistance to RAF Inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 2015;5(4):358–367. doi:10.1158/2159-8290.CD-14-1518.
- Peeters M, Price T, Boedigheimer M, Kim TW, Ruff P, Gibbs P, Thomas A, Demonty G, Hool K, Ang A, et al. Evaluation of emergent mutations in circulating cell-free DNA and clinical outcomes in patients with metastatic colorectal cancer treated with panitumumab in the ASPECCT study. Clin Cancer Res. 2019;25(4):1216–1225. doi:10.1158/1078-0432.CCR-18-2072.
- Arena S, Bellosillo B, Siravegna G, Martinez A, Canadas I, Lazzari L, Ferruz N, Russo M, Misale S, Gonzalez I, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res. 2015;21(9):2157–2166. doi:10.1158/1078-0432.CCR-14-2821.
- Montagut C, Argilés G, Ciardiello F, Poulsen TT, Dienstmann R, Kragh M, Kopetz S, Lindsted T, Ding C, Vidal J, et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti-EGFR therapy and molecularly selected by circulating tumor DNA analyses: a phase 2 randomized clinical trial. JAMA Oncol. 2018;4(4):e175245. doi:10.1001/jamaoncol.2017.5245.