ABSTRACT
This study aimed to investigate the influence of miR-221-3p and O6-methylguanine-DNA methyltransferase (MGMT) interaction in human hepatocellular carcinoma (HCC), thereby revealing a novel molecular mechanism of hepatic carcinogenesis involving miR-221-3p and MGMT.
Fluorescence qPCR and immunoblot assays were performed to determine the expression of RNA and protein in HCC tissues and cell lines. We also employed the firefly and Renilla luciferase assay to verify the target relationship between miR-221-3p and MGMT mRNA. Assessments including the MTT assay, wound-healing assay, transwell assay, colony foci formation experiment, and flow cytometric experiment were carried out to determine the viability, migration, invasion, proliferation, cell cycle progression, and apoptosis of SMMC-7721 and BEL-7404 cell lines with the modulated expression of miR-221-3p and MGMT. Compared to healthy tissues and cell line HL7702, miR-221-3p was significantly upregulated but MGMT was significantly downregulated in carcinomas and cancerous cell lines. Forced miR-221-3p overexpression was found to enhance the proliferation, migration, invasion, and clonogenicity of cell lines, but it suppressed cell apoptosis. Findings also revealed that forced miR-221-3p overexpression had little effect on cell cycle progression. After MGMT was confirmed to be atarget gene of miR-221-3p, it was found that the forced upregulation of miR-221-3p downregulated MGMT mRNA and protein levels significantly. MiR-221-3p was identified as an HCC promoting factor, and it specifically inhibited the expression of the MGMT. Besides, the upregulation of miR-221-3p had apositive influence on HCC pathogenesis by inhibiting MGMT expression.
KEYWORDS:
Introduction
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer mortality globally. In 2018, about 781,631 individuals worldwide died of liver cancer.Citation1 HCC represents 75%-85% of primary liver cancer cases, and it can result from chronic infection, aflatoxin-contaminated foodstuffs, heavy alcohol intake, obesity, smoking, and type 2 diabetes.Citation2 While research suggests that early diagnosis and treatment can prolong the chances of survival of HCC patients,Citation3 the survival of advanced or metastatic HCC patients cannot be prolonged with conventional chemotherapy, which is the most commonly used therapy today. This limitation is, in part, because the mechanism that influences HCC is not yet clear. Besides, this mechanism is complicated in that it involves several genes and proteins that induce abnormal phenotypes, including cellular cycle, apoptosis, proliferation, cell migration, and invasion.Citation4,Citation5,Citation6 Researchers have intensified efforts to investigate genes and proteins associated with HCC, but they are yet to make substantive progress given that the specificity and sensitivity of genes and proteins as therapy targets are limited.Citation7,Citation8,Citation9,Citation10 Hence, it is crucial to identify novel HCC pathogenesis factors.
miRNAs belong to a non-coding small RNA family that includes miRNAs, snoRNAs, and exRNAs. They are single-stranded and 22 ~ 28 nucleotides in length and are responsible for interrupting the protein translation process by pairing up with 3ʹUTRs of mRNAs. In recent years, scientists have paid more attention to miRNAs’ involvement in cell proliferation, apoptosis, migration, cell cycle, invasion, and other biological processes.Citation11,Citation12,Citation13,Citation14,Citation15 Researchers have also observed that different miRNAs have different biological functions. To name a few, miR-105 played a negative role in the proliferation of HCC cells through the PI3K/AKT signaling pathway.Citation16 miR-221 was found to be overexpressed in hepatic carcinoma tissues compared with healthy hepatic tissues.Citation17 This RNA molecule could also improve the hepatocellular cancer cells’ growth ability, as indicated by the increased ratio of cells in the synthesis phase of the cell cycle.Citation18 In 2008, a study reported that miR-221-3p bound to the 3ʹUTRs of cyclin-dependent kinase inhibitor genes p27Kip1 and p57Kip2, thus enhancing cell proliferation in HCC.Citation18 Cyclin-dependent kinase inhibitor proteins was also discovered to inhibit cyclin-dependent kinases, thereby limiting cell cycle progression and acting as tumor suppressors. The DNA damage-inducible transcript 4 (DDIT4) is a gene that is ontologically related to protein binding. In 2010, Pineau et al.Citation19 found that DDIT4 was the target of miR-221-3p. Besides, OCitation6-methylguanine-DNA methyltransferase (MGMT) is a DNA repair enzyme that can suppress and eliminate guanine-alkyl groups influenced by alkylating agents, which are potent carcinogens.Citation20 In several experiments, MGMT promoter methylation has been found to inactivate MGMT, which is commonly observed in human neoplasms such as head and neck cancers, pulmonary cancers, and glioma.Citation21,Citation22 Nonetheless, only a few studies have reported the target relationship between miR-221-3p and MGMT.
In the current study, we carried out bioinformatics analysis and cellular experiments to determine whether miR-221-3p could directly bind to MGMT mRNA. To investigate the influence of the interaction of miR-221-3p and MGMT on HCC, we conducted several cytological and molecular biological experiments such as the RT-qPCR, western blot, MTT assay, and transwell assay. By carrying out these experiments, we are optimistic that our results may provide more insights into a new therapy target axis in HCC.
Materials and methods
Liver cancer samples and cell culture
Materials for this study were collected at the Guangxi Medical University Affiliated Tumor Hospital, and they included snap-frozen healthy hepatic tissues (n = 10), snap-frozen hepatic carcinomas (n = 20), and snap-frozen adjacent tissues (n = 20). All the tissues used in this study were based on the guidelines approved by the Ethics Committee. HL-7702 (cultured with RPMI 1640), Hep G2 (cultured with DMEM), BEL-7404 (cultured with RPMI 1640), Huh-7 (cultured with DMEM), SMMC-7721 (cultured with RPMI 1640) and 293 T (cultured with DMEM) cell lines were purchased from the Shanghai Institute of Life Sciences Cell Resource Center (China). The HL-7702 cell line was the human normal hepatocyte cell line, while HepG2, BEL-7404, Huh-7, and SMMC-7721 cell lines were the human hepatoma cell lines. 293 T cell line was used specifically for the dual luciferase assay. These cell lines were authenticated using short tandem repeat (STR) authentication, a service that was provided by Shanghai GENECHEM Co., Ltd (China). Ten STR loci were detected, and the matches were more than 80%. The phenotypes of SMMC-7721 and BEL-7404are showed in Supplementary Figure 1. All the cell culture media were complemented with 10% heat-inactivated fetal bovine serum and 1% antibiotic-antimycotic solution (Gibco). Before the experiments were performed, all the cells were kept at 37°C with 5% CO2 in an incubator.
Sample preparation and RNA isolation
Hepatic carcinoma, adjacent and normal hepatic tissues, were collected in sterile containers and immediately frozen in liquid nitrogen to avoid RNA degradation. The total RNAs were obtained from tissues and cells using TRIzol reagent (Tiangen Biotech, Beijing, China). 600–3,750 ng/µL total RNA was obtained from the 700 µL of tissue samples. The quality and quantity of the isolated total RNAs were evaluated using the spectrophotometric reading method.
qRT-PCR
Equal amounts of RNA were converted into cDNA, and RT-qPCR was performed using the Power SYBR Green PCR Master Mix. Afterward, U6 expression was used to normalize the expression of miR-221-3p,Citation23 and GAPDH expression was utilized to normalize the expression of MGMT. The primer sequences of miR-221-3p were F: 5ʹ-GGCATGAACCTGGCATACAA-3ʹ and R: 5ʹ-TTTCCAGGTAGCCTGAAACCC-3ʹ. The primer sequences of MGMT were F: 5ʹ-ATGAAACGCACCACACTGGA-3ʹ and R: 5ʹ-AATAGAGCAAGGGCAGCGTT-3ʹ. The comparative CT method (2−ΔΔCT) was used to calculate relative RNA expression.Citation24
Cell transfection
SMMC-7721 and BEL-7404 cell lines were chosen for all the following experiments except for the firefly luciferase reporter assay. The two cell lines were transfected with several molecules, including miR-221-3p mimic, miR-221-3p inhibitor, negative control RNA, and MGMT overexpression plasmids. After culturing the cells to 70% confluence, the transfection was performed. The cells in the control group were not transfected, however. To transfect the experimental cells, MGMT Human Tagged ORF Clone was purchased from OriGene (Cat#: RC229131, Shanghai, China). Apart from the overexpression plasmids, all the other molecules were designed and purchased from GENECHEM (Shanghai, China).
MTT assay
The selected BEL-7404 and SMMC-7721 cell lines were seeded in 96-well plates in RPMI 1640 with 10% FBS. After 12 h, both cell lines were washed with phosphate-buffered saline (PBS) twice and transfected with the molecules. The MTT reagent purchased from Sigma (USA) was later used to detect cell proliferation every day for five days. Next, 20 µL MTT reagents were incubated in each well for 14 min. Optical absorbance was then read at 490 nm.
Cell cycle and apoptosis assays
The cell cycle profiles were analyzed by flow cytometry, and the percentage of cells in different phases were calculated. BEL-7404 and SMMC-7721 cell lines in various groups were harvested and fixed with 80% ethanol. Then, the fixed cells were incubated with 0.1 mg/ml propidium iodide (PI) dye (eBiosciences, USA) and 0.1 mg/ml RNase A for 50 min at 37°C without light. The BD-LSRFortessa flow cytometer (BD Biosciences) was used to measure the cell cycle, and the data were analyzed using ModFit LT™ Trial and Reader software version 4.0 (USA).
Like in cell cycle flow cytometric assay, in cell apoptosis assay, the ethanol-fixed cells were resuspended in 1.25 μL of Annexin V-FITC and 10 μL of PI staining solutions (mixed in 500 μL 1× binding buffer, eBiosciences, USA), and kept in the dark for less than half an hour. Cells stained with FITCwere defined as early apoptotic cells, cells stained with both FITC and PI were defined as late apoptotic cells. Both early and late apoptotic cells were regarded as apoptotic cells.
Wound-healing assay
The transfected cells were seeded in 12-well plates at a density of 30,000 cells per well and then cultured under 5% CO2 at 37°C. At 90% confluence, the cell monolayers of each well were scraped to make cell-free areas with pipet tips. After washing them in PBS, the serum-free medium was used to culture cells under 5% CO2 at 37°C. The migration of cells into the scratch was observed at 24 h using an inverted microscope system (model IX83, Olympus, Japan). The wound distance was measured using Adobe Photoshop, a graphics editor. The migration rate was defined as the ratio of migration distance at 24 h to the wound width at 0 h.
Transwell invasion assay
The matrigel (BD Bioscience, USA) was diluted in RPMI-1640 media in a volume ratio of 1:3 before it was added to transwell inserts (BD Bioscience, USA) overnight for incubation at 37°C. The cell suspension in 100 μL serum-free medium was then added to the upper chambers mentioned above for 2-day incubation. The cells on the downside of the upper chambers were fixed in 4% paraformaldehyde for 30 min. After washing the cells with PBS and staining with 0.1% crystal violet for 20 min, the stained cells were counted at 200× magnification.
Colony foci formation assay
The transfected cells at the log phase were harvested and digested by mild trypsinization (Sigma, USA). Next, they were washed in PBS three times and seeded in a 6-well plate at a cell concentration of 400 ~ 1,000 cells per well. After 14 days of growth in 10% FBS RPMI 164 media, the colonies were fixed with 4% paraformaldehyde (Santa Cruz Biotechnology, USA) for 30 min, stained for 1 h using 1 mL/well crystal violet solution, and photographed using an inverted microscope (model IX83, Olympus, Japan). Then, the colony foci number was determined to assess the cells’ ability to form colonies.
Immunoblot assay
The transfected cells were cultured for 3 days and were analyzed with western blot in order to determine MGMT protein expression. Equal quantities (40 μg) of proteins extracted from BEL-7404 and SMMC-7721 cells were separated on 12% SDS gels by electrophoresis and then transferred onto nitrocellulose blotting membranes (Invitrogen, USA). After that, 5% nonfat milk was dissolved in Tris-buffered saline with 0.1%. Tween 20 was later used to block the blotting membranes for 1 h. Then, the blocked membranes were washed with TBST (Tris-Buffered Saline, 0.1% Tween® 20 Detergent) four times and incubated at 4°C for 20 h with the primary antibodies against MGMT (Cat#: ab39253, Abcam, UK) and GAPDH (Cat#: ab9484, Abcam, UK). The primary antibodies were subsequently washed off with TBST, and the membranes were incubated with Rabbit Anti-Mouse IgG secondary antibody (Cat#ab6728, Abcam, UK) for 90 min. Using TBST to wash PVDF membranes four times, the membranes were exposed according to the Pierce™ ECL Western Blotting Substrate protocol (Thermo Fischer, USA).
Immunohistochemistry
Hepatic carcinoma, adjacent and normal hepatic tissues were collected at biopsy, fixed in 4% formaldehyde, embedded in paraffin, and cut into 10 slices for immunohistochemical staining. The xylene-deparaffinized tissue slices were rehydrated in ethanol and incubated with 3% hydrogen peroxide to remove endogenous peroxidase. After washing them three times, the tissue slices were incubated overnight with primary antibodies against MGMT (Cat#: ab39253, Abcam, UK) at 4°C. The secondary antibody labeled by horseradish peroxidase (HRP) was incubated with the slices for another 20 min. Then, the tissue slices were stained with DAB-Peroxidase Substrate Solution (Cat#: 34002, Thermo Fischer, USA). The sections were later counterstained with Mayer’s hematoxylin solution (Sigma, USA). The whole tissue slices were then observed at ×100 magnification to identify a ‘hot spot.’ For further analysis, the field was shifted to ×400 magnification. Three pathologists who had no idea about the study results read the IHC pictures based on a 5-tier scoring system (see ).
Firefly luciferase reporter assay
RNA hybrid, microRNA, Pictar, and TargetScan were used to predict the targets of miR-221-3p. The sequence of human MGMT mRNA 3ʹ-UTR contains the complementary sequence motif of miR-221-3p, which was amplified with PCR. The specific primers of MGMT were designed as 5ʹ-ACCGTTTGCGACTTGGTACT-3ʹ (MGMT-3ʹUTR-F) and 5ʹ-GGTGAACGACTCTTGCTGGA-3ʹ (MGMT-3ʹUTR-R). The mutated human MGMT mRNA 3ʹ-UTR did not include the binding site of miR-221-3p, which was amplified using PCR. The primers of mutated human MGMT mRNA 3ʹ-UTR were designed as 5ʹ-CGCTGCTGTCTGATACTTCAT-3ʹ(MGMT-3ʹUTR-Mut-F) and 5ʹ-CATATGCAGCTAGCACGGTTTC-3ʹ(MGMT-3ʹUTR-Mut-R). Containing the firefly luciferase reporter, the GV272 vector (5010 bp in length) was purchased from GENECHEM (Shanghai, China). The promoter of the GV272 vector was increased from 48 bp to 250 bp (). The constructed GV272-MGMT or GV272-MGMT-Mut was transfected into the 293 T cell line. The cells were then seeded in 24-well plates and transfected with 0.1 μg of either GV272-MGMT or GV272-MGMT-Mut per well together with 0.01 μg of pRL-TK vectors (Promega, USA), which contained Renilla luciferase and 0.4 μg of the miR-221-3p mimics or negative control (NC) of mimics. The transfection was done using X-tremeGENE HP (Cat. No. 06366236001, Sigma, USA) in a final volume of 0.5 mL. After 1-day transfection, the Dual-Luciferase Reporter Assay System (Promega, USA) was employed to measure the firefly and Renilla luciferase intensities. Results were reported as firefly luciferase intensity/Renilla luciferase intensity, and each transfection was done in triplicate.
Statistical analysis
Data of this study were reported as mean±SD. Statistical differences between the blank or negative control group (in case of tissue examinations, the comparison was made to healthy tissue) and experimental groups were analyzed using a one-way analysis of variance method by SPSS software 19.0. P(probability)<0.05 was defined as statistically significant.
Results
Hepatocellular carcinomas and hepatocellular carcinoma cells showed upregulation of miR-221-3p
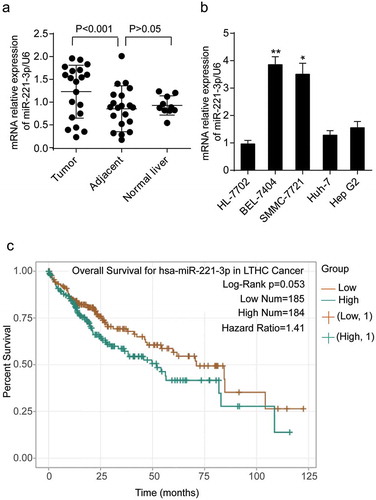
To reveal the expression of miR-221-3p in HCC, we detected the level of miR-221-3p in liver carcinomas (n = 20), adjacent-to-tumor tissues (n = 20), and healthy liver tissues (n = 10) and 5 cell lines (HL-7702, BEL-7404, SMMC-7721, Huh-7 and Hep G2) using real-time qPCR. As illustrated in , the expression level of miR-221-3p in tumors was 1.33-fold of that in healthy tissues. Compared with HL-7702 cell line (normal human hepatic cells), human hepatoma cells BEL-7404, SMMC-7721, Huh-7, and Hep G2 showed significant upregulation of miR-221-3p, especially SMMC-7721 and BEL-7404 cell lines (up to about 3.8- and 3.5-fold of healthy hepatic cells, respectively) (). BEL-7404 and SMMC-7721 cell lines were chosen for subsequent experiments because of this significant upregulation of miR-221-3p. Because we wondered whether the aberrant upregulation of miR-221-3p played a crucial role in HCC prognosis, we used the pan-cancer data analysis algorithm of StarBase (http://starbase.sysu.edu.cn/) to analyze the prognosis value of miR-221-3p in HCC. The result showed that HCC patients (n = 369) with a high level of miR-221-3p exhibited poorer prognosis than those with a low level of miR-221-3p, with a slight statistical significance (hazard ratio = 1.41, log-rank p = .053) ().
Figure 1. The expression of miR-221-3p in liver tissues and cell lines. (a) The expression of miR-221-3p was detected in the tumorous, adjacent, and normal liver tissues using RT-qPCR. (b) The expression of miR-221-3p was detected in HL-7702, BEL-7404, SMMC-7721, Huh-7, and Hep G2 cell lines using RT-qPCR. HL-7702 was the normal cell line. * p < .05 and ** p < .001, compared with HL-7702 cell line. (c) The overall survival results of HCC patients divided by miR-221-3p level. The survival curve comparing the patients with high (n = 184) and low (n = 185) expression of miR-221-3p in HCC was plotted using StarBase pan-cancer data. Data represented the mean ± SD of three independent experiments. Log-rank p = .053
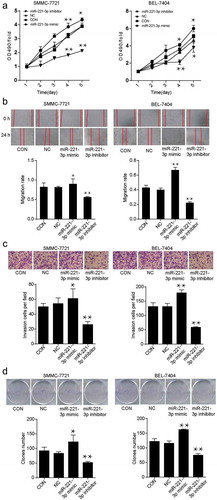
Overexpression of miR-221-3p induced cell viability, migration, and invasion
In the MTT assay, cell viability was detected in SMMC-7721 and BEL-7404 cell lines after transfecting with exogenous miR-221-3p mimic or miR-221-3p inhibitor for 5 days. The optical density (OD) values at 490 nm wavelength were illustrated. Compared with the negative control (NC) group, the miRNA mimic transfection caused a 1.17-and 1.11-fold increase in cell viability at day 4 and day 5, respectively. However, the miRNA inhibitor transfection caused a 1.81-and 1.85-fold decrease in cell viability at day 4 and day 5 in SMMC-7721 cells, respectively (). BEL-7404 cells displayed similar results as SMMC-7721 cells. Meanwhile, SMMC-7721 and BEL-7404 cells transfected with miR-221-3p mimic, miR-221-3p inhibitor, or negative control RNA were photographed at 0 h and 24 h after the scratch was leveraged to analyze the ability of migration. In SMMC-7721 and BRL-7404 cell lines, the migration rate of cells transfected with miR-221-3p mimic was enhanced by 1.1-and 1.73-fold, and the migration rate of cells transfected with miR-221-3p inhibitor was reduced by 69% and 59% compared with the NC group (). Moreover, the transwell invasion assay results demonstrated that the invasion ability was enhanced by 1.22-and 1.37-fold with the treatment of miR-221-3p mimic; however, it was reduced by 30% to 50% with the treatment of miR-221-3p inhibitor (). As for the colony foci formation assay, the miR-221-3p mimic-transfected cells formed more clones, whereas the miR-221-3p inhibitor-transfected cells resulted in fewer clones (). The statistical analysis results suggested that miR-221-3p was possibly hepatocellular carcinogenetic.
Figure 2. The effects of miR-221-3p on viability, migration, invasion, and proliferation in SMMC-7721 and BEL-7404 cell lines. (a) The cell viability was detected by MTT assay after transfection with miR-221-3p mimic, miR-221-3p inhibitor, negative control RNA for continuing five days. All the OD values were read at 490 nm. (b) Wound healing assay was used to assess the effect of miR-221-3p on migration rate. SMMC-7721 and BEL-7404 cells were transfected with miR-221-3p mimic, miR-221-3p inhibitor, and negative control RNA. Migration rate was defined as the ratio of migrating distance at 24 h to cell spacing at 0 h. (c) The ability of invasion was detected by transwell assay in SMMC-7721 and BEL-7404 cells in different groups. SMMC-7721 and BEL-7404 cells were transfected, as stated in Methods. The X-axis represents different groups depending on the transfection reagents, and the Y-axis represents the invasion cell number. (d) Colony foci formation results of transfected SMMC-7721 and BEL-7404 cells. The X-axis represents different groups, and the Y-axis represents the number of clones. All the bars in the figure represent mean ± SD from three independent experiments. * p < .05 and ** p < .001, compared with con (blank control) group
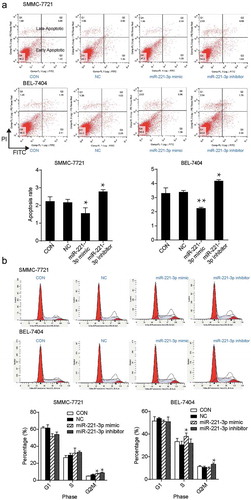
miR-221-3p inhibition induced cell apoptosis but had no effects on cell cycle in vitro
Both SMMC-7721 and BEL-7404 cells transfected with miR-221-3p inhibitor exhibited a 1.25-fold increase in the apoptosis rate when compared with the NC group (). In the SMMC-7721 cell line, the transfection of miR-221-3p mimic and inhibitor resulted in a higher ratio of cells in the G2/M phase (1.2-and 1.36 fold of the NC group, respectively), whereas no changes were observed for the percentage of cells in G1 phase and S phase compared with the NC group (). Similarly, the percentage of BEL-7404 cells transfected with miR-221-3p inhibitor was 1.26-fold of the NC group in the G2/M phase but not in G1 and S phase (). The data showed that miR-221-3p downregulation promoted the apoptosis of hepatic carcinoma cells. However, it did not influence HCC cell apoptosis by changing cell cycle progress.
Figure 3. The effects of miR-221-3p on apoptosis and cell cycle in SMMC-7721 and BEL-7404 cells. (a) Flow cytometric assay was conducted to detect cell apoptosis after the transfection of SMMC-7721 and BEL-7404 cells with miR-221-3p mimic, miR-221-3p inhibitor, and negative control RNA. (b) The cell cycle was measured using the flow cytometric assay. The SMMC-7721 and BEL-7404 cells were transfected with the same molecules as mentioned above. The bars come from three independent experiments. Values are mean ± SD. * p < .05 and ** p < .001, compared with CON (blank control) group
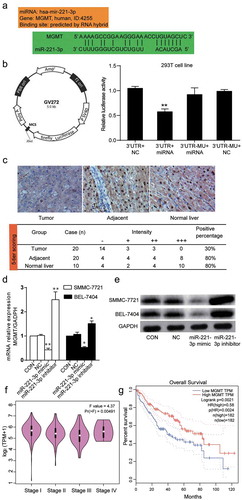
MGMT was the target of miR-221-3p and was downregulated in hepatic carcinoma
microRNA target prediction software applications such as RNA hybrid, microRNA, Pictar, and TargetScan were used to explore the targets of miR-221-3p. The software predicted that MGMT mRNA 3ʹ UTR contained a binding site for miR-221-3p (). To validate the binding relationship between MGMT mRNA 3ʹUTR and miR-221-3p, the wild-type or mutated 3ʹ UTR sequences of MGMT were respectively inserted into the firefly luciferase reporter plasmids. Afterward, the constructed vectors were transfected into 293 T cells together with miR-221-3p mimic or NC RNA to conduct a dual-luciferase reporter gene assay. As indicated in , the miR-221-3p overexpression reduced the luciferase activity by approximately 50% when it was co-transfected with wild-type MGMT mRNA 3ʹUTR. Nevertheless, it showed little effect in luciferase activity when co-transfected with the mutated 3ʹ UTR sequence of MGMT mRNA. Through immunohistochemical staining, the expression of MGMT protein in carcinomas was downregulated, as indicated by a lower average IHC score (). We then confirmed the transfection efficiency of miR-221-3p mimic and inhibitor on MGMT expression in hepatic carcinoma cells. As presented in , the result from RT-PCR showed that the transfection of miR-221-3p mimic into the SMMC-7721 and BEL-7404 cells led to significantly low levels of MGMT mRNA (42% and 50% decrease, respectively). The western blot assay produced a result that was similar to the RT-PCR assay: the transfection of miR-221-3p mimic into both cell lines decreased the level of MGMT protein (). By performing Gene Expression Profiling Interactive Analysis (GEPIA), we found that MGMT expression at stage II and III was lower than that at stage I of HCC (p = .00191, F-test was used). GEPIA acquires data from the TCGA open database (n = 377). MGMT expression was negatively correlated with staging; however, at stage IV, the level of MGMT expression was higher than that at stage II and III. According to the TNM system, at stage II and III, the tumor is larger, and it grows into nearby tissues or lymph nodes. At stage IV, the tumor reached other organs. We thought that MGMT might try to repair the DNA at stage IV to prevent the aggressive progression of HCC (). However, MGMT expression gradually increased at stage IV. We also found that a lower level of MGMT was an indicator of inferior prognosis results for HCC patients (). The data obtained from the dual luciferase assay, RT-PCR, immunohistochemical experiment, western blot, GEPIA analysis, and prognosis analysis proved that MGMT was not only the target of miR-221-3p but also a tumor suppressor gene of HCC.
Figure 4. MGMT was the target of miR-221-3p and it’s down-regulated in HCC tissues and cells. (a) The targeting relationship between miR-221-3p and MGMT mRNA was predicted through RNA hybrid, microRNA, Pictar, and TargetScan. The binding scheme was illustrated. (b) The information of GV272 vector that was used to build GV272-MGMT or GV272-MGMT-Mut vectors for transfection in 293 T cell line. The relative luciferase activity was measured using a dual luciferase reporter gene assay. The 293 T cells were co-transfected with pRL-TK vectors containing Renilla luciferase and 0.4 μg of the miR-221-3p mimic or negative control (NC). (c) The immunohistochemical staining was used to measure the expression of MGMT protein in the tumors, adjacent liver tissues, and normal liver tissues. The positive percentage equals the ratio of positive cases to total cases. -: negative; +: positive. (d) The expression of MGMT mRNA was detected by RT-qPCR in SMMC-7721 and BEL-7404 cells transfected with miR-221-3p mimic, miR-221-3p inhibitor, negative control RNA. The bars represent mean ± SD of mRNA relative expression from three independent qPCR experiments. * p < .05 and ** p < .001, compared with CON (blank control) group. (e) The expression of MGMT protein was detected using western blot. In SMMC-7721 and BEL-7404 cells after transfection with miR-221-3p mimic, miR-221-3p inhibitor, and negative control RNA. (f) The expressions of MGMT at different stages of HCC was plotted using GEPIA data (an online gene profiling analysis tool). (g) The overall survival for MGMT in HCC patients. The survival curve comparing the patients with high and low expression of MGMT in HCC was also plotted using GEPIA. Log-rank p = .0021
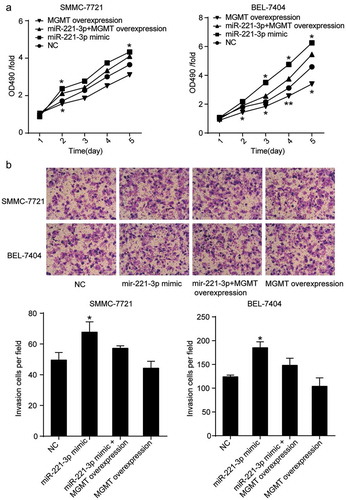
Overexpression of MGMT inhibited miR-221-3p-induced cell viability and migration
To further validate the effect of miR-221-3p on HCC by directly targeting MGMT, MTT and transwell assays were performed by co-transfecting miR-221-3p mimic and MGMT ORF clone in hepatic carcinoma cells. MTT experimental results revealed that forced MGMT overexpression attenuated the promoting effect of miR-221-3p on cell viability in the SMMC-7721 cell line (). A similar result was observed in BEL-7404 (). As for the effect of MGMT overexpression on cell migration, the transwell assay results showed that SMMC-7721 and BEL-7404 cell lines co-transfected with miR-221-3p mimic and MGMT ORF clone resulted in a 1.21-and 1.26-fold decrease in the number of invading cells per field, respectively, compared with the cells that were transfected with miR-221-3p mimic (). These data further proved that by targeting MGMT, miR-221-3p could promote the viability, migration, and invasion of hepatocellular carcinoma cells.
Figure 5. MGMT overexpression attenuated the effect of miR-221-3p on cell viability, and invasion in HCC cells. (a) MGMT overexpression was proved to inhibit HCC cell viability induced by miR-221-3p. Herein, the MTT assay was again employed to assess whether the forced overexpression of MGMT could influence miR-221-3p mediated increase of cell viability. (b) The MGMT overexpression restored the effect of miR-221-3p mimic on the invasion of SMMC-7721 and BEL-7404 cells. All the data represent mean ± SD from three independent experiments. Compared with the NC (negative control) group, * p < .05, and ** p < .001
Discussion
To confirm that the target gene of miR-221-3p plays a vital role in defining novel therapeutic targets, we verified its target relationship with its target gene MGMT and then inspected the potential mechanism involving miR-221-3p and MGMT in HCC. We also observed in this study that miR-221-3p was upregulated in human hepatic tumors and cell lines. As well as augmenting the viability, migration, and invasion of HCC cells, miR-221-3p inhibited cell apoptosis without affecting the cell cycle progression of hepatocellular carcinoma cells. According to the prediction of the bioinformatics software, MGMT mRNA 3ʹUTR contained a complementary site of miR-221-3p. In short, we found that miR-221-3p reduced the transcription and translation of MGMT genes to promote the viability, migration, and invasion of HCC cells.
A number of researchers have proved that miRNAs have a close relationship with cancer progression. Based on the evidence from the literature, miRNAs play essential roles in breast cancer, malignant glioma, esophageal cancer, and pancreatic cancer.Citation25,Citation26,Citation27,Citation28 Apart from promoting tumor development and the progression of tumor cells, miR-221 has been reported to inhibit cell apoptosis in HCC.Citation29,Citation30 miR-221-3p and miR-221-5p are the mature spliceosomes of miR-221, and miR-221-3p can be a more significant participant in various physiological processes because it is broadly conserved. Based on this fact, we selected miR-221-3p as the miRNA of interest to investigate its function in HCC cells. Forced miR-221-3p upregulation in our cell models revealed that miR-221-3p contributed to tumorigenesis in HCC. Our findings agree with the results of several previous reports, in which miR-221-3p has been demonstrated to be an onco-miRNA in liver carcinogenesis.Citation17,Citation31,Citation32,Citation33
What’s more, several studies have shown that miR-221-3p, as an oncogenic miRNA, promoted the clonogenicity and invasiveness of gastric carcinoma and cervical cancer cells significantly.Citation34,Citation35,Citation36 Also consistent with the research conducted by Yuan et al.Citation37 our results indicated that a positive impact on cell proliferation occurred when the miR-221-3p expression was upregulated in our experimental cell line models. As for apoptosis, some contradictory reports have been found. Dai et al.Citation38 for instance, used thapsigargin (1 mM) to induce the endoplasmic reticulum stress in HepG2 and SMMC-7721 cell lines. They found that miR-221 mimic enhanced ER stress-induced apoptosis. Gramantieri et al.,Citation18 on the other hand, discovered that miR-221 upregulation increased the cell ratio in S phase, thereby leading to the proliferation of HCC-derived cell line Hep3B. Compared with the studies conducted by Dai et al. and Gramantieri et al., a different influence of miR-221-3p on apoptosis or proliferation in hepatic carcinoma cells occurred. This might result from different cell lines or endoplasmic reticulum stress. We used flow cytometry not only to detect the apoptosis rate of SMMC-7721 and BEL-7404 cells but also to explore the positive or negative impact of miR-221-3p on apoptosis of hepatic carcinoma cells. Our result revealed that the cell apoptosis percentage reduced in cells associated with the upregulation of miR-221-3p. This revelation suggests that miR-221-3p suppressed HCC cell apoptosis. We also used flow cytometry to detect the cell cycle. Our findings indicated that miR-221-3p did not affect the HCC cell cycle even though a recent study reported that the downregulated miR-221 led to cell-cycle arrest in HCC.Citation39 Furthermore, it was reported that migration and invasion were enhanced in HCC cells that were exposed to miR-221 mimic.Citation40 Our data also support this finding. After performing wound-healing assay, transwell assay and colony formation assay, our data suggested that the upregulation of miR-221-3p significantly promoted cell migration, cell invasion, and colony formation in HCC.
Given that miRNAs have enormous effects on cancers by regulating their target genes, the study of miRNAs and their target genes has become an attractive area of research for scientists as it provides new possibilities for curing cancers. For example, upregulating miR-122 suppressed the expression of its target gene, ubiquitin-specific peptidase 53 (P53), to induce apoptosis in bile duct carcinoma cells.Citation41 In their research, Hu et al. found that miR-221 augmented osteosarcoma cell growth, migration, and invasion by limiting cyclin-dependent kinase inhibitor 1B (p27, Kip1).Citation42 In our study, we confirmed that the MGMT mRNA had a target relationship with miR-221-3p. We also found that MGMT mRNA and protein level was upregulated and accompanied by miR-221-3p suppression, but they were downregulated along with the upregulation of miR-221-3p. The regulation of miR-221-3p on MGMT mRNA significantly affected the migration, invasion, and colony foci formation of HCC cells (SMMC-7721 and BEL-7404 cell lines).
As promoter methylation has been found to serve as an essential mechanism for gene silencing, Gu et al. quantified the strong association between MGMT promoter methylation and non-small cell lung cancer-genesis using a meta-analysis protocol. Their analysis indicated that the MGMT gene was frequently silenced in non-small cell lung cancer tissues because promoter methylation resulted in the loss of function of MGMT. Eighteen studies carried out from 2001 to 2011 were included, and further prospective studies were carried out to confirm the association.Citation43 After analyzing the high methylation frequency and loss-of-function of MGMT in 82 gastric cancer tissues using western blotting, Yousuf et al. found that the loss of MGMT protein concomitantly with MGMT promoter hypermethylation was observed in 37 cases.Citation44 In the study performed by Zekri et al.,Citation45 the methylation of MGMT promoter was found with a significantly high methylation index in HCC tissues than in corresponding adjacent healthy tissues. These findings indicated that MGMT expression might be downregulated in HCC tissues. Our data also suggested that the expression level of MGMT protein in hepatic tumors was significantly lower than that in healthy liver tissues. A lower level of MGMT was a poor indicator of HCC prognosis. This result has been supported by the findings of Matsukura S et al., who showed that the downregulation of MGMT was a poor prognostic factor using a cox proportional-hazard regression model.Citation46
Nonetheless, the results of this research have been compromised by limitations such as lack of in vivo assays. To some extent, insufficient animal model experiments degraded the generalization of this research. According to GEPIA analysis, the expression of MGMT at stage II and III of HCC was lower than that of other stages of HCC. Nonetheless, the MGMT expression at stage IV was a little higher than that at stage III. It seemed that MGMT might be trying to recover the DNA at stage IV in order to prevent further HCC progression. Nevertheless, this needs further investigation.
Conclusion
Our study confirms the targeting relationship between MGMT mRNA and miR-221-3p in human HCC cells. In other words, this research demonstrates how the interaction between MGMT mRNA and miR-221-3p plays a crucial role in HCC cells. Based on our findings, miR-221-3p can promote the viability, migration, and invasion of HCC cells by suppressing MGMT transcription and translation. This interaction between miR-221-3p and MGMT mRNA can be regarded as a novel gene therapy target for HCC. In sum, our research not only explains human liver carcinogenesis but also provides insights into the prognosis biomarker for HCC.
Supplemental Material
Download Zip (9.6 MB)Disclosure statement
The authors declare that there is no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. PMID:30207593. doi:10.3322/caac.21492.
- Schottenfeld D, Fraumeni JF. 2006. Cancer epidemiology and prevention. Irvine (CA) (USA): Oxford University Press.
- Yang LY, Fang F, Ou DP, Wu W, Zeng ZJ, Wu F. Solitary large hepatocellular carcinoma: a specific subtype of hepatocellular carcinoma with good outcome after hepatic resection. Ann Surg. 2009;249:118–123. PMID:19106686. doi:10.1097/SLA.0b013e3181904988.
- Liu X, Liao W, Yuan Q, Ou Y, Huang J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget. 2015;6:34309–34320. PMID:26418879. doi:10.18632/oncotarget.5295.
- Karavias D, Maroulis I, Papadaki H, Gogos C, Kakkos S, Karavias D, Bravou V. Overexpression of CDT1 is a predictor of poor survival in patients with hepatocellular carcinoma. J Gastrointest Surg. 2016;20:568–579. PMID:26408331. doi:10.1007/s11605-015-2960-7.
- Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. PMID:20639898. doi:10.1038/onc.2010.236.
- Ezaka K, Kanda M, Sugimoto H, Shimizu D, Oya H, Nomoto S, Sueoka S, Tanaka Y, Takami H, Hashimoto R, et al. Reduced expression of adherens junctions associated protein 1 predicts recurrence of hepatocellular carcinoma after curative hepatectomy. Ann Surg Oncol. 2015;22(Suppl 3):S1499–507. PMID:26122373. doi:10.1245/s10434-015-4695-9.
- Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. PMID:26073087. doi:10.1038/onc.2015.223.
- Hu Z, Zhao W. Novel insights into the molecular mechanisms of alpha-fetoprotein expression and malignant phenotypes of hepatocellular carcinoma. Cell Mol Immunol. 2012;9:7–8. PMID:21860406. doi:10.1038/cmi.2011.30.
- Merle P, Trepo C. Molecular mechanisms underlying hepatocellular carcinoma. Viruses. 2009;1:852–872. PMID:21994573. doi:10.3390/v1030852.
- Cai K, Shen F, Cui JH, Yu Y, Pan HQ. Expression of miR-221 in colon cancer correlates with prognosis. Int J Clin Exp Med. 2015;8:2794–2798. PMID:25932237.
- Sun L, Hu J, Xiong W, Chen X, Li H, Jie S. MicroRNA expression profiles of circulating microvesicles in hepatocellular carcinoma. Acta Gastroenterol Belg. 2013;76:386–392. PMID:24592541.
- Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. PMID:23081718. doi:10.1002/hep.26095.
- Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297–303. PMID:22213236. doi:10.1002/mc.21864.
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. PMID:21125669. doi:10.1002/path.2806.
- Shen G, Rong X, Zhao J, Yang X, Li H, Jiang H, Zhou Q, Ji T, Huang S, Zhang J, et al. MicroRNA-105 suppresses cell proliferation and inhibits PI3K/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. 2014;35:2748–2755. PMID:25280563. doi:10.1093/carcin/bgu208.
- Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. PMID:23320393. doi:10.1186/1471-2407-13-21.
- Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. PMID:18521080. doi:10.1038/onc.2008.178.
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. PMID:20018759. doi:10.1073/pnas.0907904107.
- Li CC, Yu Z, Cui LH, Piao JM, Liu M. Role of P14 and MGMT gene methylation in hepatocellular carcinomas: a meta-analysis. Asian Pac J Cancer Prev. 2014;15:6591–6596. PMID:25169493. doi:10.7314/apjcp.2014.15.16.6591.
- Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, Schackert G, Kreth FW, Pietsch T, Loffler M, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129:659–670. PMID:21425258. doi:10.1002/ijc.26083.
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. PMID:10029064.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. PMID:18663219. doi:10.1073/pnas.0804549105.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. PMID:11846609. doi:10.1006/meth.2001.1262.
- Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee NPY, Law S, Xu LY, Li EM, Chan KW, et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986–4000. PMID:28288140. doi:10.1038/onc.2017.29.
- Kanno S, Nosho K, Ishigami K, Yamamoto I, Koide H, Kurihara H, Mitsuhashi K, Shitani M, Motoya M, Sasaki S, et al. MicroRNA-196b is an independent prognostic biomarker in patients with pancreatic cancer. Carcinogenesis. 2017;38:425–431. PMID:28186267. doi:10.1093/carcin/bgx013.
- Madhavan D, Peng C, Wallwiener M, Zucknick M, Nees J, Schott S, Rudolph A, Riethdorf S, Trumpp A, Pantel K, et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016;37:461–470. PMID:26785733. doi:10.1093/carcin/bgw008.
- Chang JH, Hwang YH, Lee DJ, Kim DH, Park JM, Wu HG, Kim IA. MicroRNA-203 modulates the radiation sensitivity of human malignant glioma cells. Int J Radiat Oncol Biol Phys. 2016;94:412–420. PMID:26678661. doi:10.1016/j.ijrobp.2015.10.001.
- Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15:5073–5081. PMID:19671867. doi:10.1158/1078-0432.CCR-09-0092.
- Igoucheva O, Alexeev V. MicroRNA-dependent regulation of cKit in cutaneous melanoma. Biochem Biophys Res Commun. 2009;379:790–794. PMID:19126397. doi:10.1016/j.bbrc.2008.12.152.
- de Conti A, Ortega JF, Tryndyak V, Dreval K, Moreno FS, Rusyn I, Beland FA, Pogribny IP. MicroRNA deregulation in nonalcoholic steatohepatitis-associated liver carcinogenesis. Oncotarget. 2017;8:88517–88528. PMID:29179453. doi:10.18632/oncotarget.19774.
- Lupini L, Bassi C, Ferracin M, Bartonicek N, D’Abundo L, Zagatti B, Callegari E, Musa G, Moshiri F, Gramantieri L, et al. miR-221 affects multiple cancer pathways by modulating the level of hundreds messenger RNAs. Front Genet. 2013;4:64. PMID:23630541. doi:10.3389/fgene.2013.00064.
- Fu X, Wang Q, Chen J, Huang X, Chen X, Cao L, Tan H, Li W, Zhang L, Bi J, et al. Clinical significance of miR-221 and its inverse correlation with p27Kip(1) in hepatocellular carcinoma. Mol Biol Rep. 2011;38:3029–3035. PMID:20146005. doi:10.1007/s11033-010-9969-5.
- Wei WF, Zhou CF, Wu XG, He LN, Wu LF, Chen XJ, Yan RM, Zhong M, Yu YH, Liang L, et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis. 2017;8:3220. PMID:29242498. doi:10.1038/s41419-017-0077-5.
- Shi J, Zhang Y, Jin N, Li Y, Wu S, Xu L. MicroRNA-221-3p plays an oncogenic role in gastric carcinoma by inhibiting PTEN expression. Oncol Res. 2017;25:523–536. PMID:27712596. doi:10.3727/096504016X14756282819385.
- Zhu J, Liu F, Wu Q, Liu X. MiR-221 increases osteosarcoma cell proliferation, invasion and migration partly through the downregulation of PTEN. Int J Mol Med. 2015;36:1377–1383. PMID:26397386. doi:10.3892/ijmm.2015.2352.
- Yuan Q, Loya K, Rani B, Mobus S, Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2013;57:299–310. PMID:22821679. doi:10.1002/hep.25984.
- Dai R, Li J, Liu Y, Yan D, Chen S, Duan C, Liu X, He T, Li H. miR-221/222 suppression protects against endoplasmic reticulum stress-induced apoptosis via p27(Kip1)- and MEK/ERK-mediated cell cycle regulation. Biol Chem. 2010;391:791–801. PMID:20624000. doi:10.1515/BC.2010.072.
- Xu Q, Li M, Yang M, Yang J, Xie J, Lu X, Wang F, Chen W. alpha-pinene regulates miR-221 and induces G2/M phase cell cycle arrest in human hepatocellular carcinoma cells. Biosci Rep. 2018;38. PMID:30473536. doi:10.1042/BSR20180980.
- Huang S, Zhou D, Li YX, Ming ZY, Li KZ, Wu GB, Chen C, Zhao YN. In vivo and in vitro effects of microRNA-221 on hepatocellular carcinoma development and progression through the JAK-STAT3 signaling pathway by targeting SOCS3. J Cell Physiol. 2019;234:3500–3514. PMID:30370582. doi:10.1002/jcp.26863.
- Wu C, Zhang J, Cao X, Yang Q, Xia D. Effect of Mir-122 on Human Cholangiocarcinoma Proliferation, Invasion, and Apoptosis Through P53 Expression. Med Sci Monit. 2016;22:2685–2690. PMID:27472451. doi:10.12659/msm.896404.
- Hu XH, Zhao ZX, Dai J, Geng DC, Xu YZ. MicroRNA-221 regulates osteosarcoma cell proliferation, apoptosis, migration, and invasion by targeting CDKN1B/p27. J Cell Biochem. 2019;120:4665–4674. PMID:30582227. doi:10.1002/jcb.27755.
- Gu C, Lu J, Cui T, Lu C, Shi H, Xu W, Yuan X, Yang X, Huang Y, Lu M. Association between MGMT promoter methylation and non-small cell lung cancer: a meta-analysis. PLoS One. 2013;8:e72633. PMID:24086261. doi:10.1371/journal.pone.0072633.
- Yousuf A, Bhat MY, Pandith AA, Afroze D, Khan NP, Alam K, Shah P, Shah MA, Mudassar S. MGMT gene silencing by promoter hypermethylation in gastric cancer in a high incidence area. Cell Oncol (Dordr). 2014;37:245–252. PMID:25008999. doi:10.1007/s13402-014-0179-3.
- Zekri AR, Bahnasy AA, Shoeab FE, Mohamed WS, El-Dahshan DH, Ali FT, Sabry GM, Dasgupta N, Daoud SS. Methylation of multiple genes in hepatitis C virus associated hepatocellular carcinoma. J Adv Res. 2014;5:27–40. PMID:25685469. doi:10.1016/j.jare.2012.11.002.
- Matsukura S, Miyazaki K, Yakushiji H, Ogawa A, Chen Y, Sekiguchi M. Combined loss of expression of O6-methylguanine-DNA methyltransferase and hMLH1 accelerates progression of hepatocellular carcinoma. J Surg Oncol. 2003;82:194–200. PMID:12619064. doi:10.1002/jso.10209.
