ABSTRACT
FGF13, a member of the FGF subfamily, has been found to be highly expressed in cancer cells such as prostate cancer, melanoma, glioma and multiple myeloma. However, the mechanism of FGF13 function during cancer cell proliferation remains to be unexplored, especially Non-small cell lung cancer (NSCLC). In this study, the cell proliferation effect of FGF13 on A549 cells was checked by CCK-8, clone formation, Ki67 immunofluorescence staining and Flow Cytometry assay. Localization of FGF13 within A549 cells was performed with confocal laser scanning microscope. The protein variations and interaction were measured by western blotting and co-immunoprecipitation analysis. It showed that FGF13 was mainly distributed in the cytoplasm and exhibited a high expression level in A549 cells. High expression of FGF13 activated AKT-GSK3 signaling pathway, and inhibited the activity of p21 and p27. Thus, FGF13 enhanced the process of transition from G1 to S phase and promoted A549 cells proliferation. Furthermore, the interaction between FGF13 and SHCBP1 was confirmed. Meanwhile, FGF13 and SHCBP1 had a cooperative effect to accelerate the cell cycle progression, especially the ability to promote cell proliferation is significantly enhanced via protein interaction. Hence, we conclude that FGF13 played a positive regulation role during A549 cells proliferation. FGF13 interacted with SHCBP1 to facilitate cell cycle progression, providing new insights into deep understanding of non-small cell lung cancer mechanisms of proliferation and regulation function of FGF13.
Introduction
Lung cancer has become the most serious malignant tumor globally because of its high invasive and metastatic features. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer cases.Citation1 NSCLC is further categorized into several major histological subtypes, such as adenocarcinoma, squamous cell carcinoma, and large-cell carcinoma.Citation2 Despite the effectiveness of surgical, radiotherapy and chemotherapy treatments have been applied, the treatment effect is not satisfactory, and the five-year survival rate is still only about 15%.Citation1 Moreover, the differential gene expression patterns observed in the different subtypes of lung cancers have a significant influence on the choice of chemotherapy drugs, suggesting the diversity and complexity of the mechanism of NSCLC.Citation3 Therefore, it is very important for the early diagnosis and treatment to fully understand the molecular mechanism of NSCLC development and explore potential targets for invasion and metastasis.
Fibroblast growth factors (FGFs) regulate cell differentiation and migration, which control a wide range of organisms development and regeneration processes.Citation4 Fibroblast growth factor homologues (FGF11, FGF12, FGF13 and FGF14, also known as FHF3, FHF1, FHF2 and FHF4) belong to the FGF family but differ from other FGF proteins due to the lack of signal peptides.Citation5 They cannot be secreted by cells, so they cannot activate the FGF receptor and can only act as an intracellular protein in a manner independent of FGFR.Citation6,Citation7 However, the FHFs are intracellular proteins that interact with various intracellular partners.Citation7,Citation8 Like all FHFs, FGF13 has features that distinguish it from other FGFs. Not only is it not secreted by cells, FGF13 cannot activate or antagonize any of the known FGF receptors (FGFRs).Citation7,Citation8 In humans, the FGF13 (FHF2) shows high expression level in brain and skeletal muscle of patients with Borjeson-Forssman-Lehmann syndrome (BFLS) l.Citation9,Citation10 Intracellular FGFs often colocalize with Nav channels in the initial segments of axons and nodes of Ranvier.Citation11,Citation12 Moreover, FGF13 has been confirmed as a microtubule-stabilizing protein that regulates neuronal polarization and migration.Citation13,Citation14 In addition, accumulating evidence has demonstrated that FGF13 is possibly involved in several cancers. A recent study found that FGF13 mRNA was significantly elevated in a subset of lung adenocarcinomas and might enable oncogene-driven cancer cells to avoid excessive accumulation of potentially toxic aberrant proteins, conferring a survival advantage.Citation15,Citation16 Indeed, high expression of FGF13 was associated with liver metastasis at diagnosis or at follow-up, even in patients who had complete primary tumor resection.Citation17 Likewise, FGF13 expression was significantly increased in prostate cancer (PCa) cells in both the cytoplasm and the nuclei, which was associated with biochemical recurrence following radical prostatectomy.Citation4 Even though, these observations suggest that FGF13 may be viewed as a cancer facilitator or enabler, and it is ill-defined which signaling pathways are involved in the proliferation of NSCLC.
The SHC binding and spindle associated 1 (SHCBP1) gene is a member of Src homolog and collagen homolog (Shc) family. Shc is a cell surface receptors adaptor protein, and can activate multiple growth factor receptor signaling pathways, such as insulin receptor (IR), insulin growth factor receptor (IGFR), epidermal growth factor receptor (EGFR) and fibroblast growth factor receptor (FGFR).Citation18,Citation19 Consequently, it is very important for the regulation of growth and development in mammalian cells. Recent studies demonstrated that SHCBP1 played critical roles in the regulation of cell proliferation, migration, adhesion, and cell cycle progression, especially in the carcinogenesis.Citation20 SHCBP1 was highly expressed in various cancers, such as leukemia, lymphoma,Citation21 hepatocellular carcinomaCitation22 gliomas,Citation23 synovial sarcomaCitation20 and breast cancer.Citation24 Meanwhile, loss of SHCBP1 function in these cancer cells leads to inhibition of cell proliferation and enhancing cell apoptosis.Citation22 These findings collectively showed that SHCBP1 played an active role in cancers and may be a potential diagnosis biomarker for most cancers. However, the cellular functions and underlying mechanisms of SHCBP1 action during the proliferation of NSCLC cells remain largely unknown. In the present study, we identify FGF13 as a proliferate regulator of NSCLC cells, and it acts by regulating AKT signaling. Mechanistically, FGF13 enhances cell proliferation by activating the phosphorylation of AKT and promoting the increase of GSK3α (Ser21) and β (Ser9) phosphorylation levels. Concomitantly, there is no mutual regulation between FGF13 and SHCBP1. Instead, FGF13 interacts with SHCBP1 which is more beneficial to NSCLC cells proliferation. Together, our data indicated that FGF13 provided temporal control of AKT signaling in the proliferation of NSCLC cells.
Materials and methods
Cell culture
Non-small cell lung cancer A549 cells were maintained at 37°C in a 5% CO2 humidified incubator and were grown in RPMI-1640 (GIBCO) plus 10% FBS (GIBCO) and 1% penicillin/streptomycin (MP). HEK293T and BEAS-2B cells were maintained at 37°C in a 5% CO2 humidified incubator and were grown in DMEM (GIBCO) plus 10% FBS (GIBCO) and 1% penicillin/streptomycin (MP).
Cell transfection
Togenerate A549 cells overexpressing FGF13, pcDNA3.1-FGF13 or empty vector was transfected in A549 cells with Superfect Plus reagent (QIAGEN), according to the manufacturer’s protocol. The cell lysates were harvested 48 h or 72 h post-transfection.
The sequences of the siRNA-FGF13 and siRNA-NC were obtained from GenePharma, Shanghai, China. A549 cells were seeded in 60 mm plates 24 h prior to transfection. siRNA-FGF13 or siRNA-NC was transfected using Lipofectamine 2000 (Invitrogen) with antibiotic-free medium. Five hours post-transfection, the cells were placed in complete medium. The cell lysates were harvested 48 h or 72 h after transfection.
Isolation of total RNA and qPCR
Cells were lysed and total RNA was extracted using TRIzol reagent (Takara) according to the manufacturer’s protocol and transcribed into cDNA using Reverse transcription Kit (Takara). Real-time PCR was performed using SYBR Premix EX Taq (Takara). The primers used in this study were listed in .
Table 1. Primers for real-time qPCR
Antibodies and western blotting
Antibodies directed against the following proteins were used: FGF13 (Abcam); SHCBP1 (Proteintech); Ki67 (Proteintech); CY3 (Boster); FITC (Boster). β-actin, p21, p27, cyclinE1, AKT, Phospho-AKT, gsk-3α/β, Phospho-gsk-3α, and Phospho-gsk-3β were purchased from Cell Signaling Technology (CST). Primary antibodies were detected by HRP-conjugated secondary antibodies to rabbit, mouse, and goat immunoglobulins. Immunoreactivity was determined with the ECL Prime Western Blotting Detection System (GE Healthcare).
Cell counting Kit-8 (CCK-8) cell viability assay
A549 cells transfected with siRNA-FGF13, siRNA-NC, pcDNA3.1 or pcDNA3.1-FGF13 were seeded in 96-well plates at a density of 5 × 103 per well and cultured for 0 day, 1 day, 2 days and 3 days. Cell viability was assessed by the Cell Counting Kit-8 (CCK-8, Dojindo) at day 0, 1, 2 and 3.
Clonogenic assay staining and analysis
The cells were seeded in 35 mm plate and incubated at 37°C for 2 weeks. For crystal violet staining, plates were washed once with PBS and were incubated in cold methanol for 5 min and then in crystal violet solution (0.4% in methanol) for 15 min. Plates were subsequently washed twice with double-distilled water, air-dried, and scanned using a Canon scanner.
Ki-67 immunofluorescence staining
A549 cells were seeded on Petri dish and transfected with siFGF13 or siNC, respectively. After 72 h, the cells were fixed and incubated with Ki-67 antibody (CST) for 1 h and then incubated with Alexa-488 at room temperature for 20 min. Cells were counterstained with DAPI to stain the cell nucleus.
Flow cytometry
For the cell cycle analysis, 5 × 106 cells per well were inoculated on a 6-well plate. At 48 h after transfection, the A549 cells were digested with trypsin (without Ethylenediamine Tetraacetic Acid (EDTA), centrifuged at 3,000 x g for 3 min at room temperature and washed with ice‑cold PBS twice at room temperature. Cells were fixed with 75% ethanol overnight at 4°C and subsequently dyed with propidium iodide for 30 min at 37°C in the dark. For each cell suspension, ≥10,000 events were analyzed using a BD flow cytometer (FACSAria) and FlowJo software (version 7.6.1) for data analysis.
Immunofluorescence staining
Briefly, cells were washed twice with PBS and fixed with 3% paraformaldehyde in PBS for 10 min. After fixation, cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min, followed by 15- to 30-min incubation with antibody solution (3% BSA in 0.2% Triton X-100 in PBS). Next, cells were incubated for 1–4 h with the indicated primary antibodies at 1:100–1:200 dilution and with an appropriate secondary antibody conjugated. Nuclei were counterstained with DAPI. Images were taken with a FV3000 confocal laser scanning microscope (Olympus, Japan).
Yeast two-hybrid screening
The yeast two-hybrid screening was performed using the yeast two-hybrid system (Takara Bio-Clontech) according to the manufacturer’s instructions. All reagents used in this screen were purchased from Takara Bio-Clontech, unless stated otherwise. The FGF13 coding sequence was subcloned into the EcoRI and BamHI sites of pGBKT7, and was used to transform Saccharomyces cerevisiae AH109. The Saccharomyces cerevisiae AH109 (pGBKT7-FGF13) was hybridized with the human lung cDNA library (Takara, 9506) according to the description of the Yeast protocol instructions. cells were grown at 30°C for 4 days on QDO/X/A-agar plates-a synthetic defined agar medium without tryptophan, leucine, histidine and adenine, and obtain positive clones for sequencing, and compared with non-redundant sequence databases using BLAST.
Co-immunoprecipitation
A549 cells were transfected with pcDNA3.1-Flag-FGF13 or pcDNA3.1-HA-SHCBP1 plasmid DNA, respectively. After 72 h, the cells were harvested and cell lysates were prepared with RIPA lysis buffer. For co-immunoprecipitation (Co-IP), total cell lysates were incubated overnight with rabbit anti-HA polyclonal antibody (CST) or mouse anti-Flag (CST) antibody and further incubated with protein A Sepharose for 4 h. The complex was washed three times with cell lysis buffer, and processed for further western blotting analysis.
Statistical analysis
The statistical analysis was performed using SPSS software, version 22 (SPSS Inc.). Differences were considered statistically significant when the p values were less than 0.05. Statistical significance was defined as *p < .05; **p< .01; ***p < .001. All experiments were performed three times independently.
Results
Loss of FGF13 expression delays A549 cells proliferation
Previous research has shown that the level of FGF13 mRNA is markedly increased in several cancer cell lines, and acts as a potential novel prognostic marker in prostate cancer.Citation4 However, the function of FGF13 in lung cancer remained unexplored. To determine how FGF13 affects proliferation of NSCLC, we used the human NSCLC cell line A549 to analyze subcellular localization of FGF13. Immunofluorescence staining showed FGF13 was mainly distributed in the cytoplasm and exhibited a high expression level, supporting the idea that FGF13 is a cytoplasmic protein (). Notably, the FGF13 expression levels of mRNA and protein were significantly elevated in A549 cells, relative to normal lung epithelial cells (BEAS-2B) (, c). Intriguingly, the expression level of FGF13 was gradually increased during the proliferation of A549 cells (, e). These data suggested that FGF13 up-regulation might benefit the cancer cells. For the reason, we silenced endogenous FGF13 expression in A549 cells by siRNA approach. After knock down FGF13 expression in A549 cells (, b), cell proliferation rate was measured by CCK8 assay. Remarkably, transient FGF13 knockdown weaken the rate of cell proliferation at 72 h compared to the control group (). Cell colony formation experiments further confirmed that transient FGF13 knockdown reduced the clonogenicity of A549 cells (). Next, we stained with Ki67 during A549 cell proliferation, a cellular marker of proliferation commonly used as a biomarker for estimating A549 cell outcome. The results showed that the number of Ki67 positive cells in the FGF13 knock down group was reduced by 56.3% compared with the control group (). To assess which phase of the cell cycle might be influenced by FGF13, cell cycle progression was analyzed by flow cytometry (FACS). The FACS analysis indicated a substantial increase of cells at G1 phase and a visibly decrease of cells at S phase upon FGF13 silencing as compared to control cells (), which implied that FGF13 knockdown could induce G1/S cell cycle arrest and retardation of A549 cell proliferation.
Figure 1. The expression levels of FGF13 were significantly elevated in A549 cells, which might be benefit the cancer cells. (a) The localization of FGF13 was analyzed by confocal laser scanning microscope in A549 cells, and siFGF13 as negative controls. (b) Relative mRNA expression and (c) protein levels of FGF13 were detected in BEAS-2B and A549 cells, respectively. (d) The mRNA expression level and (e) protein levels of FGF13 were checked during A549 cells proliferation. Results are presented as mean± S. E. M. n = 3. *P < .05, ** P < .01, *** P < .001
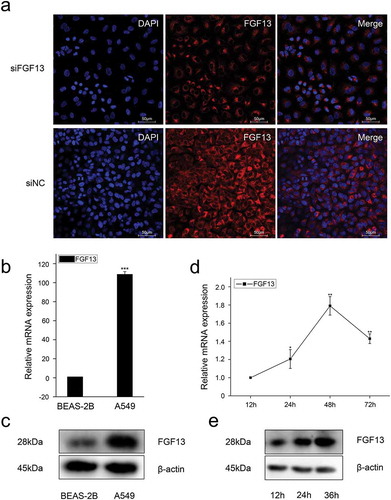
Figure 2. Knock-down of FGF13 inhibits A549 cells proliferation.A549 cells were transfected with siRNA-FGF13 or N.C siRNA, (a) the interference efficiency and relative mRNA expression levels of FGF13 were measured by real-time qPCR. (b) The protein expression level of FGF13 with the interference efficiency was detected by Western blotting. (c) Cell proliferation of A549 was measured by CCK8 assay at 0, 24, 48, 72 h post-transfection, respectively. (d) Cell colony formation was performed. (e) The number of A549 cells proliferation was checked by Ki67. (f) FGF13-silenced cells were harvested and samples were analyzed on the flow cytometer 72 h post-transfection. The cell numbers of G1 phase and S phase were counted in (f). Results are presented as mean± S. E. M. n = 3. *P < .05, ** P < .01
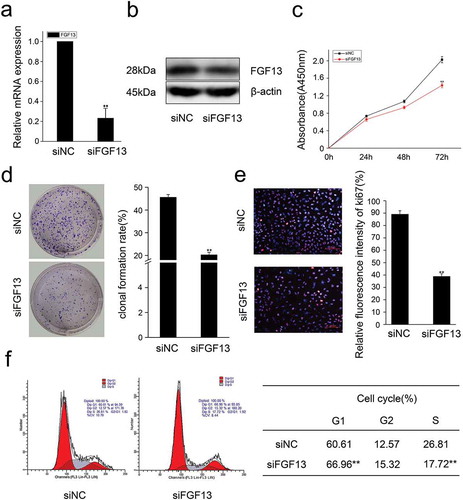
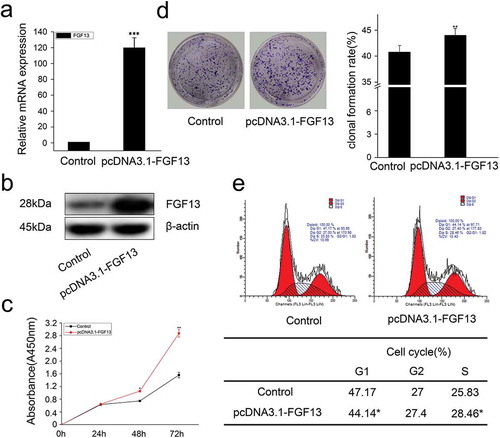
In addition, we also overexpressed FGF13 and detected cell proliferation by CCK8. As expected, the expression level of FGF13 was significantly increased in A549 cells (, b), and FGF13 enhanced the rate of cell proliferation after 48 h and 72 h compared to the control group treated with empty vector (). Moreover, FGF13 overexpression accelerated the clonogenicity of A549 cells (). Simultaneously, there displayed a decrease of cells at G1 phase and an increase of cells at S phase, detected by flow cytometry, and the differences were significantly after FGF13 was overexpressed (). Together, these results indicated that FGF13 was mainly distributed in the cytoplasm, and it was required for A549 cell proliferation and FGF13 may act as a G1/S transition regulator.
Figure 3. FGF13 accelerates cell cycle progression and promotes proliferation in A549 cells. A549 cells were transfected with pcDNA3.1-FGF13 (or Empty vector), (a) the over-expression efficiency and relative mRNA expression levels of FGF13 were measured by real-time qPCR. (b) the FGF13 relative protein levels of over-expression efficiency were detected by western blot after 48 h transfection. (c) Cell proliferation of A549 was measured by CCK8 assay at 0, 24, 48, 72 h post-transfection, respectively. (d) Cell colony formation was performed, (e) cell cycle progression was analyzed by the flow cytometer. The cell numbers of G1phase and S phase were counted in (e). Results were presented as mean± S. E. M. n = 3. *P < .05, ** P < .01
FGF13 interaction with SHCBP1 in A549 cells
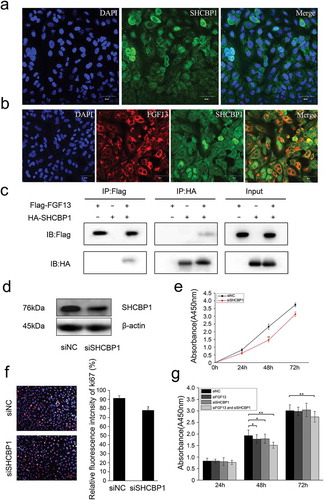
Due to the lack of a signal peptide, FGF13 cannot be secreted extracellularly, only acts as an intracellular molecule in the FGFR-independent manner.Citation6 To investigate whether FGF13 directly interacts with intracellular protein components, we performed a yeast two-hybrid screening using the pGBKT7-FGF13 as bait and a universal human-normalized library as prey. We obtained 67 clones in the yeast two-hybrid screen. According to DNA sequence analysis of the positive plasmids and remove of recurrent false positives clones, three clones remained as potential binding partners of pGBKT7-FGF13: Shcbp1, Vmp1 and Arhgap5 (). SHCBP1 played critical roles in cell proliferation, migration, adhesion, and cell cycle progression, especially in the carcinogenesis.Citation22,Citation24,Citation25 However, few studies have examined the functions of SHCBP1 in A549 cells. Therefore, we firstly investigated the localization in cells of SHCBP1. SHCBP1 was simultaneously highly expressed in the nuclear and the cytoplasmic of A549 cells by immunofluorescence staining (). Interesting, FGF13 shared a common distribution with SHCBP1 in the cytoplasm (). Next, we performed coimmunoprecipitation (CoIP) analysis using protein lysates prepared from HEK293T cells cotransfected with Flag-FGF13 and HA-SHCBP1. As expected, FGF13 readily binded to SHCBP1 in HEK293T cells. This result suggested that interaction between FGF13 and SHCBP1 in A549 cells endogenously. ().
Table 2. Proteins that interacted with FGF13 in the yeast two-hybrid system
Figure 4. FGF13 shares a common distribution with SHCBP1 in the cytoplasm, which could bind to SHCBP1. (a) Cell localization of SHCBP1 was measured in A549 cells. (b) Colocalization of FGF13 and SHCBP1 was examined in A549 cells. (c) Co-IP assay was used to determine the interaction effects of FGF13 and SHCBP1. (d)The protein expression level of SHCBP1 with the interference efficiency was detected by Western blotting. (e)The CCK8 and (f) Ki67 assay were employed to assess A549 cells proliferation after SHCBP1 silencing. (g) The CCK8 assay was performed to assess the cell proliferation rate after co-interference 24 h, 48 h, 72 h. Results were presented as mean± S. E. M. n = 3. *P < .05, ** P < .01
Colak D et alCitation31 found that SHCBP1 was highly expressed in breast cancer and significantly correlated with metastatic potential, advanced stage, and poor prognosis. Furthermore, silencing the expression of SHCBP1 through RNA interference could inhibit the proliferation of breast cancer cells.Citation24 Then, does SHCBP1 play a role alone or in combination with FGF13 in A549 cells?
We next examined the functional significance of SHCBP1 during A549 cells proliferate. Firstly, silencing of endogenous SHCBP1 was performed by siRNA and SHCBP1 protein levels were analyzed by western blotting (). Cell proliferation was measured by CCK8 assay, which showed that a significant decrease in cell growth after SHCBP1 silencing (). Ki67 staining was also performed, which showed the number of Ki67 positive cells were reduced by 14.6% (). Together, these data indicate that SHCBP1 alone can promote the proliferation of A549 cells, consistent with Romaszko’s research.Citation1 Next, we focused on investigating the effect of interaction by which co-silencing SHCBP1 and FGF13 in A549 cells. Actually, the effect of inhibition on cell proliferation was consistent with previous results with silencing FGF13 or SHCBP1. Surprisingly, the effect is more pronounced in the co-silencing group, especially at 48 h, 72 h (). Taken together, our data indicated that SHCBP1 expression is also required for proliferation in A549 cells, and SHCBP1 binding to FGF13 may play a synergistic role in promoting cell proliferation.
p21 and p27 are involved in the process of A549 cell proliferation after FGF13 and SHCBP1 interaction
During the normal cell cycle, activated CDK2/cyclin E complex promotes the progression of cells from G1 to S phase, but its diminished activity renders G1 phase arrest.Citation32,Citation33 In accordance with the previous reports, we have shown that FGF13 promoted cell proliferation by enhancing the G1/S transition during the cell cycle. Then, we focused on investigating the molecular mechanism by which FGF13 enhanced G1/S phase of the cell cycle. Depletion of FGF13 in A549 cells resulted decline of CDK2 mRNA expression level as compared to control cells ().Having demonstrated that p21 and p27 are both cell cycle kinase inhibitors, and enhanced expression levels of the two proteins can retard cell proliferation,Citation33 we next examined the functional significance of p21 and p27 in this process. Notably, the expression of p21 and p27 were more dramatically increased at both the mRNA and protein levels in A549 cells compared with the controls (), suggesting the expression levels of p21 and p27 may be regulated by FGF13 during cell proliferation. In addition, low expression of FGF13 decreased cyclin E1 protein level in A549 cells (). In contrast, there were obviously reductions in the mRNA and protein levels of p21, p27 when expressed abundant FGF13 on A549 cells. (). Meanwhile, the protein level of cyclin E1 was increased at 72 h post-transfection (). These results strongly suggested that FGF13 accelerated cell cycle progression in A549 cells by targeting CDK2, p21, p27 and cyclin E1 through alterations in the expression.
Figure 5. Interaction between FGF13 and SHCBP1 affects cell proliferation. FGF13 affects AKT signaling pathway, p21 and p27 expression levels were detected in A549 cells which overexpressed or interfered. (a) The mRNA expression levels of p21, p27 and CDK2 were analyzed on A549 cells with FGF13 silenced, the protein expression of p21, p27, cyclinE1 and FGF13 were detected with FGF13 silenced, respectively. (b) The mRNA expression levels of p21, p27 and CDK2 were evaluated in A549 cells with FGF13 overexpressed, the protein expression of p21, p27, cyclinE1 and FGF13 were detected with FGF13 overexpressed. (c) After FGF13 and SHCBP1 were silenced, the western blotting was employed to assess the expression of p21, p27 and cyclinE1, β-actin was used as a loading control. (d) FGF13 was overexpressed while SHCBP1 was interfered in A549 cells, the protein expression levels of p21, p27 and cyclinE1 were analyzed. The results of western blotting were assessed using ImageJ (mean ± SD of 3 independent experiments.*P < .05, ** P < .01)

However, the remarkable increase of p21 and p27 expression levels were found after co-silence of FGF13 and SHCBP1 (). Moreover, the expression level of cyclin E1 was also reduced compared with the control group, especially in the co-silenced cells (). Therefore, we speculated that FGF13 and SHCBP1 had a cooperative effect to accelerate the cell cycle progression depending on protein interaction. Next, western blotting was applied to verify the interdependence between FGF13 and SHCBP1 using A549 cells cotransfected either FGF13-overexpressing or FGF13-silencing and with or without the SHCBP1 expression. However, there was no significant change in the levels of p21, p27 and cyclin E1 (, d). It further demonstrated that this cooperative effect could only be verified after the interaction between FGF13 and SHCBP1.
FGF13 facilitating AKT-GSK3 activity during cell cycle progression
It has been demonstrated that the expression of p27 and p21 are regulated by AKT signaling pathways in cancer cells.Citation34 As an intersection of multiple signaling pathways in the cell, activation of serine/threonine (Ser/Thr) kinase protein kinase B (PKB/Akt) can promote cell survival through multiple pathways. To our knowledge, it has not been determined whether FGF13 promotes the proliferation of A549 cells through the mechanism discussed above. Therefore, in the present study, the expression levels of AKT, p-AKT, GSK3α, p-GSK3α, GSK3β and p-GSK3β were detected by western blotting. As shown in , depletion of FGF13 led to a significant attenuation in p-AKT (Ser473), give rise to an obviously declined in p-GSK3α (ser21) and p-GSK3β (ser9) in A549 cells. Instead, elevated levels of their phosphorylation were produced in FGF13-overexpressing cells ().
Figure 6. FGF13 activates AKT-GSK3 signaling pathway to regulate A549 cells proliferation. FGF13 and SHCBP1 co-interference or co-overexpression is performed in A549 cells, respectively. (a) After FGF13 were silenced, the protein levels of AKT, p-AKT (Ser473), GSK3α, p-GSK3α (ser21), GSK3β and p-GSK3β (ser9) were analyzed by Western blotting. (b) After FGF13 were overexpressed, AKT, p-AKT (Ser473), GSK3α,p-GSK3α(ser21), GSK3β and p-GSK3β(ser9) protein levels were detected in A549 cells.(c) FGF13 was overexpressed while SHCBP1 was interfered, the protein expression levels of AKT, p-AKT (Ser473), GSK3α, p-GSK3α (ser21), GSK3β and p-GSK3β (ser9) were analyzed., β-actin was used as a loading control. Results are presented as mean± S. E. M. n = 3. *P < .05, ** P < .01
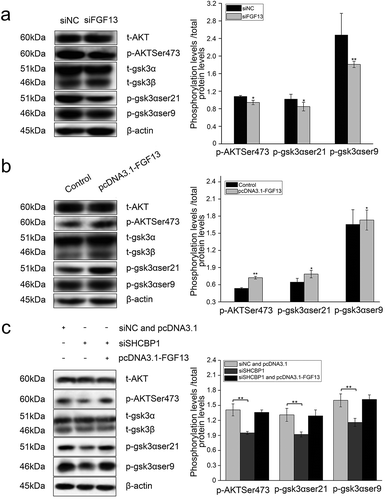
Likewise, the expression of p-AKT1, p-GSK3α (ser21) and p-GSK3β (ser9) was much lower in A549-siSHCBP1 cells than in A549-siNC cells (). However, no significant differences of the expression of p-AKT1, p-GSK3α (ser21) and p-GSK3β (ser9) were observed after cotransfected FGF13-overexpressing without the SHCBP1 expression than in FGF13-overexpressing cells (), suggesting phosphorylation of AKT, the ser21 site of GSK3α and the ser9 site of GSK3β are not activated in the absence of SHCBP1, although FGF13 is overexpressed in A549 cells.
Finally, these findings indicated that FGF13 activated the AKT/GSK3 signaling pathways after bound to SHCBP1 in A549 cells.
Discussion
FGFs and the FGF signaling pathways play crucial roles in tumor development and maintenance. Several studies show that the expression of FGFs can be involved in initiation and/or progression of lung cancer.Citation35,Citation36 FGF13 was highly expressed in some types of cancer, including pancreatic endocrine carcinoma,Citation17 melanomaCitation37 and multiple myeloma.Citation38 Hence, the high expression levels of FGF13 suggested that it plays an oncogenic role. But the mechanism of FGF13 function during cell proliferation remains largely unknown. Nevertheless, these results in our study may provide clues to its functions. In the present study, we show that FGF13 expression was markedly raised at protein levels in human NSCLC (A549) cell line compared to nonmalignant cells, and consistent with the previous research.Citation15 Furthermore, endogenous FGF13 was distributed strongly throughout the cytoplasm in A549 cell only. However, previous studies of FGF13 localization suggested that FGF13 1A was a nucleolar protein that represses ribosomal RNA transcription and attenuates protein synthesis in human NSCLC cell line H460.Citation15 Others also found that FGF13 protein was localized in both the cytoplasm and the nucleus in prostate cancer (PCa) cell lines.Citation4 Actually, alternative splicing of the FGF13 mRNA generates six transcript variants encoding five protein isoforms.Citation39 Two predominant isoforms were reported in rat cerebellum extract.Citation40 Thus, the expression patterns of FGF13 in different cell lines were reported may be due to the differential isoforms. In addition, the currently available FGF13 antibodies which require further clarification of the detailed expression of the isoform.Citation4
As an enzyme, glycogen synthase kinase 3 (GSK3) reportedly has opposing roles, which is generally considered a tumor suppressor on the one hand but maintaining cell survival and proliferation on the other.Citation41,Citation42 There are two isoforms of GSK3, GSK3α-and GSK3β.Citation43,Citation44 GSK3, kinase activity can be inactivated by phosphorylation of serine-21 in GSK3α or of serine-9 in GSK3β has been reported in cancer.Citation45,Citation46 Multiple signaling pathways feed into this site to increase the serine-phosphorylation of GSK3, which can be mediated by Akt, protein kinase A (PKA), protein kinase C, p70 S6 kinase, and other kinases.Citation42 The present study found an unknown phenomenon that increased phosphorylation levels of serine-473 in AKT, serine-21 in GSK3α, serine-9 in GSK3β were observed in FGF13 overexpressed cells, suggesting FGF13 can reduce the self-activity of GSK3α and GSK3β, which in turn contributes to A549 cells proliferation. Importantly, Akt also can be involved in the regulation of the cell cycle by negatively regulating the cyclin-dependent kinase inhibitors p27 and p21.Citation34 Indeed, p27 and p21 were markedly lower at both the mRNA and protein levels in this study. A large number of studies show that up-regulation of p21 and p27 prevents cyclin E/CDK2 activation, thereby suppressing G1/S transition in normal cells.Citation32,Citation33 Our data show that the expression level of cyclin E was controlled by p21 and p27 whether FGF13 was interfered or overexpressed in A549 cells. These results strongly suggest that the high expression of FGF13 in cytoplasmic may play important role in A549 cells proliferation.
FGF13 plays a role in microtubule regulation not only in nerve cells but also in PCa cells. Moreover, it is speculated that FGF13 may interact with other cellular components as well. Although FGF13 binding partners have been described,Citation12,Citation14,Citation15 the FGF13 interacting proteins are still not fully unexplored. The present study has verified that the interaction between FGF13 and SHCBP1 by yeast two-hybrid screening assay and co-immunoprecipitation.SHCBP1 had been previously found to specifically interact with the SH2 domain of SHC1.Citation47 SHCBP1 was the only molecule dissociated from SHC1 when EGFR is activated, which might play a thus far unknown role distinct from other SHC1 binding partners.Citation49 A previous publication showed nuclear SHCBP1 could be a causative factor for tumor progression and represent a valuable biomarker for the prognosis of NSCLC.Citation1 However, the binding of FGF13 and SHCBP1 was confirmed in A549 cells, what role does SHCBP1 play alone in A549 cells? SHCBP1 highly expressed in hepatocellular carcinoma and leukemia, breast cancer, and loss of SHCBP1 function in these cancer cell lines resulted in inhibition of cell proliferation and increase of cell apoptosis.Citation19,Citation22,Citation24,Citation25 Hereby, we disclosed that knockdown of SHCBP1 can inhibit the proliferation of A549 cells. Moreover, we also found that the expression levels of p21 and p27 were up-regulated after SHCBP1 was silenced. Likewise, Wen Feng et al. revealed that knockout of SHCBP1 can up-regulate cyclin-dependent kinase (CDK) inhibitor p21, and decrease the cyclin B1 and CDK1.Citation24 Based on this evidence, we speculate that down-regulation of SHCBP1 activates the expression levels of p21 and p27, in turn, p21 and p27 prevents cyclin E/CDK2 activation, thereby suppressing G1/S transition in A549 cells.
Intriguingly, stepwise reduction of cell proliferation is accompanied by silencing FGF13, in agreement with effects after silencing SHCBP1 by increased the expression levels of p21 and p27. And this cooperative effect could be verified after the interaction between FGF13 and SHCBP1. In addition, there were no changes in the expression levels of p21, p27, p-AKT, p-GSK3α or β upon FGF13-overexpression and SHCBP1-knockdown in A549 cells, implying the effect of FGF13 should be in the presence of SHCBP1.
Conclusion
Our results indicate that FGF13 exerted a stronger effect in promoting cell proliferation by activating AKT-GSK3 signaling pathway. Likewise, elevated FGF13 promoted A549 cells proliferation at the presence of SHCBP1. Finally, our results suggested that FGF13 may interact with SHCBP1 to affect its function, and be more favorable for A549 cells proliferation.
Disclosure of potential conflicts of interests
The authors declare that they have no conflict of interest.
Acknowledgments
This study was supported by Key Scientific Research Project of Shaanxi Education Department (Collaborative Innovation Center Project, 20JY006, 20JK0570), and all supports were gratefully acknowledged.
Additional information
Funding
- Romaszko A, Świetlik E, Doboszyńska A, Szpruch P, Luks J. Lung cancer and multiple neoplasms: a retrospective analysis. In: Pokorski M, editor. Advances in respiratory cancerogenesis. Cham: Springer International Publishing Switzerland; 2016: 53–58.
- Vinod SK, Sidhom MA, Gabriel GS, Lee MT, Delaney GP. Why do some lung cancer patients receive no anticancer treatment? J Thoracic Oncol. 2010;5(7):1025–1032. doi:10.1097/JTO.0b013e3181da85e4.
- Latimer KM. Lung cancer: clinical presentation and diagnosis. FP Essent. 2018;130:23–26.
- Yu L, Toriseva M, Tuomala M, Seikkula H, Elo T, Tuomela J, Kallajoki M, Mirtti T, Taimen P, Bostrom PJ, et al. Increased expression of fibroblast growth factor 13 in prostate cancer is associated with shortened time to biochemical recurrence after radical prostatectomy. Int J Cancer. 2016;139:140–152. doi:10.1002/ijc.30048.
- Smallwood PM, Munoz-Sanjuan I, Tong P, Macke JP, Hendry SH, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci U S A. 1996;93:9850–9857. doi:10.1073/pnas.93.18.9850.
- Kettunen P, Furmanek T, Chaulagain R, Kvinnsland IH, Luukko K. Developmentally regulated expression of intracellular Fgf11-13, hormone-like Fgf15 and canonical Fgf16, −17 and −20 mRNAs in the developing mouse molar tooth. Acta Odontol Scand. 2011;69:360–366.
- Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem. 2003;278:34226–34236. doi:10.1074/jbc.M303183200.
- Schoorlemmer J, Goldfarb M. Fibroblast growth factor homologous factors are intracellular signaling proteins. Current Biol CB. 2001;11(10):793–797. doi:10.1016/S0960-9822(01)00232-9.
- Gecz J, Baker E, Donnelly A, Ming JE, McDonald-McGinn DM, Spinner NB, Zackai EH, Sutherland GR, Mulley JC. Fibroblast growth factor homologous factor 2 (FHF2): gene structure, expression and mapping to the Borjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum Genet. 1999;104:56–63. doi:10.1007/s004390050910.
- Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16(2):215–220. doi:10.1016/j.cytogfr.2005.02.002.
- Goldfarb M, Schoorlemmer J, Williams A, Diwakar S, Wang Q, Huang X, Giza J, Tchetchik D, Kelley K, Vega A, et al. Fibroblast growth factor homologous factors control neuronal excitability through modulation of voltage-gated sodium channels. Neuron. 2007;55(3):449–463. doi:10.1016/j.neuron.2007.07.006.
- Wang C, Hennessey JA, Kirkton RD, Wang C, Bryson V, Rosenberg PB, Bursac N, Pitt GS. FGF13 is a regulator of the cardiac voltage-gated sodium channel Nav1.5. Biophys J. 2011;100(3):420a–1a. doi:10.1016/j.bpj.2010.12.2490.
- Li J, Wang Q, Wang H, Wu Y, Yin J, Chen J, Zheng Z, Jiang T, Xie L, Wu F, et al. Lentivirus mediating FGF13 enhances axon regeneration after spinal cord injury by stabilizing microtubule and improving mitochondrial function. J Neurotrauma. 2018;35(3):548–559. doi:10.1089/neu.2017.5205.
- Wu QF, Yang L, Li S, Wang Q, Yuan XB, Gao X, Bao L, Zhang X. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell. 2012;149(7):1549–1564. doi:10.1016/j.cell.2012.04.046.
- Bublik DR, Bursac S, Sheffer M, Orsolic I, Shalit T, Tarcic O, Kotler E, Mouhadeb O, Hoffman Y, Fuchs G, et al. Regulatory module involving FGF13, miR-504, and p53 regulates ribosomal biogenesis and supports cancer cell survival. Proc Natl Acad Sci U S A. 2017;114(4):E496–e505. doi:10.1073/pnas.1614876114.
- Manfredi JJ. Tumor suppression by p53 involves inhibiting an enabler, FGF13. Proc Natl Acad Sci U S A. 2017;114(4):632–633. doi:10.1073/pnas.1619815114.
- Missiaglia E, Dalai I, Barbi S, Beghelli S, Falconi M, Della Peruta M, Piemonti L, Capurso G, Di Florio A, Delle Fave G, et al. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28(2):245–255. doi:10.1200/JCO.2008.21.5988.
- Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266.
- Qu WS, Tian DS, Guo ZB, Fang J, Zhang Q, Yu ZY, Xie MJ, Zhang HQ, Lu JG, Wang W. Inhibition of EGFR/MAPK signaling reduces microglial inflammatory response and the associated secondary damage in rats after spinal cord injury. J Neuroinflammation. 2012;9(1):178. doi:10.1186/1742-2094-9-178.
- Peng C, Zhao H, Song Y, Chen W, Wang X, Liu X, Zhang C, Zhao J, Li J, Cheng G, et al. SHCBP1 promotes synovial sarcoma cell metastasis via targeting TGF-beta1/Smad signaling pathway and is associated with poor prognosis. J Exp Clin Cancer Res CR. 2017;36(1):141. doi:10.1186/s13046-017-0616-z.
- Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PloS One. 2010;5(9): e13066.
- Tao HC, Wang HX, Dai M, Gu CY, Wang Q, Han ZG, Cai B. Targeting SHCBP1 inhibits cell proliferation in human hepatocellular carcinoma cells. As Pacific J Cancer Prev APJCP. 2013;14(10):5645–5650. doi:10.7314/APJCP.2013.14.10.5645.
- Zhou Y, Tan Z, Chen K, Wu W, Zhu J, Wu G, Cao L, Zhang X, Zeng X, Li J, et al. Overexpression of SHCBP1 promotes migration and invasion in gliomas by activating the NF-kappaB signaling pathway. Molecular Carcinogenesis. 2018;57:1181–1190.
- Feng W, Li HC, Xu K, Chen YF, Pan LY, Mei Y, Cai H, Jiang YM, Chen T, Feng DX. SHCBP1 is over-expressed in breast cancer and is important in the proliferation and apoptosis of the human malignant breast cancer cell line. Gene. 2016;587(1):91–97. doi:10.1016/j.gene.2016.04.046.
- Asano E, Hasegawa H, Hyodo T, Ito S, Maeda M, Takahashi M, Hamaguchi M, Senga T. The Aurora-B-mediated phosphorylation of SHCBP1 regulates cytokinetic furrow ingression. J Cell Sci. 2013;126(15):3263–3270. doi:10.1242/jcs.124875.
- Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, Went P, Klein U, Zinzani PL, Baccarani M. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. 2007;117(3):823. doi:10.1172/JCI26833.
- Dilek C, Asmaa N, AlBandary A, Maimoona N, Hatim J, Abdelmoneim E, Taher A-T, Asma T, Dahish A, Al MO. Age-specific gene expression signatures for breast tumors and cross-species conserved potential cancer progression markers in young women. PloS One. 2013;8(5):e63204.
- Dusetti NJ, Jiang YF, Vaccaro MI, Tomasini R, Samir AA, Calvo EL, Ropolo A, Fiedler F, Mallo GV, Dagorn JC. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem Biophys Res Commun. 2002;290(2):641–649. doi:10.1006/bbrc.2001.6244.
- Folkerts H, Wierenga AT, van den Heuvel FA, Woldhuis RR, Kluit DS, Jaques J, Schuringa JJ. Elevated VMP1 expression in acute myeloid leukemia amplifies autophagy and is protective against venetoclax-induced apoptosis. Cell Death Dis 2019;10:421.
- Gen Y, Yasui K, Zen K, Nakajima T, Tsuji K, Endo M, Mitsuyoshi H, Minami M, Itoh Y, Tanaka S. A novel amplification target, ARHGAP5, promotes cell spreading and migration by negatively regulating RhoA in Huh-7 hepatocellular carcinoma cells. Cancer Lett. 2009;275(1):27–34. doi:10.1016/j.canlet.2008.09.036.
- Colak D, Nofal A, Albakheet A, Nirmal M, Jeprel H, Eldali A, Al-Tweigeri T, Tulbah A, Ajarim D, Malik OA, et al. Age-specific gene expression signatures for breast tumors and cross-species conserved potential cancer progression markers in young women. PloS One. 2013;8(5):e63204. doi:10.1371/journal.pone.0063204.
- Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18(2):753–761. doi:10.1128/MCB.18.2.753.
- Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239.
- Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3(3):245. doi:10.1038/35060032.
- Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick DT, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75(1):196–207. doi:10.1124/mol.108.049544.
- Yuan H, Li ZM, Shao J, Ji WX, Xia W, Lu S. FGF2/FGFR1 regulates autophagy in FGFR1-amplified non-small cell lung cancer cells. J Exp Clin Cancer Res CR. 2017;36(1):72. doi:10.1186/s13046-017-0534-0.
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64(15):5270–5282. doi:10.1158/0008-5472.CAN-04-0731.
- Krejci P, Mekikian PB, Wilcox WR. The fibroblast growth factors in multiple myeloma. Leukemia. 2006;20(6):1165–1168. doi:10.1038/sj.leu.2404202.
- Munoz-Sanjuan I, Smallwood PM, Nathans J. Isoform diversity among fibroblast growth factor homologous factors is generated by alternative promoter usage and differential splicing. J Biol Chem. 2000;275(4):2589–2597. doi:10.1074/jbc.275.4.2589.
- Schoorlemmer J, Goldfarb M. Fibroblast growth factor homologous factors and the islet brain-2 scaffold protein regulate activation of a stress-activated protein kinase. J Biol Chem. 2002;277(51):49111–49119. doi:10.1074/jbc.M205520200.
- Abbas S, Andrei O, Wei YZ, Bin Z, Modarressi MH, Billadeau DD, Masayoshi M, Yutaka T, Toshinari M. Deregulated GSK3beta activity in colorectal cancer: its association with tumor cell survival and proliferation. Biochem Biophys Res Commun. 2005;334:1365–1373. doi:10.1016/j.bbrc.2005.07.041.
- Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–131. doi:10.1016/j.pharmthera.2014.11.016.
- Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. Functional Redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-Catenin Signaling Shown by Using an Allelic Series of Embryonic Stem Cell Lines. Dev Cell. 2007;12(6):957–971. doi:10.1016/j.devcel.2007.04.001.
- Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. Embo J. 1990;9(8):2431–2438. doi:10.1002/j.1460-2075.1990.tb07419.x.
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–789. doi:10.1038/378785a0.
- Qingqing D, Xianghuo H, Weiya X, Jung-Mao H, Chun-Te C, Long-Yuan L, Dung-Fang L, Jer-Yen Y, Xiaoming X, Jaw-Ching L. Myeloid cell leukemia-1 inversely correlates with glycogen synthase kinase-3beta activity and associates with poor prognosis in human breast cancer. Cancer Res. 2007;67:4564. doi:10.1158/0008-5472.CAN-06-1788.
- Schmandt R, Liu S, Mcglade C. Cloning and characterization of mPAL, a novel Shc SH2 domain-binding protein expressed in proliferating cells. Oncogene. 1999;18(10):1867–1879. doi:10.1038/sj.onc.1202507.
- Yong Z, Cunjie Z, Croucher DR, Soliman MA, Nicole SD, Adrian P, Lorne T, Tate SA, Rod W, Karen C. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166. doi:10.1038/nature12308.
