ABSTRACT
LncRNA AFAP1-AS1 has been corroborated to function in diverse cancers. Our aim was to investigate the molecular mechanism of AFAP1-AS1 in PTX resistance in PCa. The levels of AFAP1-AS1, miR-195-5p, and FKBP1A were checked by qRT-PCR. 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-Diphenyltetrazolium Bromide (MTT) assay was employed to assess the resistance of PTX-resistant PCa cells to PTX. Flow cytometry was introduced to evaluate cell apoptosis. The protein levels of C-caspase 3 were determined by western blot. The starBase was used to predict the interaction between miR-195-5p and AFAP1-AS1. Xenograft tumor model was established to investigate the biological role of AFAP1-AS1 in PTX resistance in vivo. The levels of AFAP1-AS1 and FKBP1A were upregulated in PCa tissues and cells, as well as PTX-resistant PCa cells, while the expression of miR-195-5p was declined. Knockdown of AFAP1-AS1 promoted the sensitivity of PTX-resistant PCa cells to PTX, induced apoptosis of PTX-resistant PCa cells, whereas the impacts could be reversed by reducing the expression of miR-195-5p. FKBP1A overexpression could rescue the effects of miR-195-5p-mediated enhancement on the sensitivity of PTX-resistant PCa cells to PTX, promotion on apoptosis of PTX-resistant PCa cells. AFAP1-AS1 interacted with miR-195-5p and miR-195-5p could bind to the 3ʹUTR of FKBP1A. AFAP1-AS1 silencing inhibited the tumor growth in mice implanted with PC3-TXR cell. The protein level of PCNA was decreased in PC3-TXR cells transfected with sh-AFAP1-AS1, while the expression of C-caspase 3 was upregulated. AFAP1-AS1 silencing attenuated the resistance of PTX-resistant PCa cells to PTX by downregulating FKBP1A via sponging miR-195-5p.
Introduction
Prostate cancer (PCa) is a malignant tumor that occurs in men, with high incidence and mortality.Citation1 PCa cells can spread to other parts of the body, such as bones and lymph nodes, leading to development into androgen-independent and hormone-refractory PCa,Citation2–4 which is responsible for the low five-year survival rate. Chemotherapy is an effective treatment method for PCa,Citation5–7 and paclitaxel (PTX) has been verified as a good chemotherapy drug for diverse cancers,Citation8,Citation9 including PCa.Citation10 However, PTX resistance is a huge barrier for the treatment of PCa.Citation11 Hence, it is compulsory to figure out the hidden molecular mechanism of PTX resistance in PCa.
LncRNAs are a kind of RNA molecules with the length exceeding 200 nucleotides, and they cannot encode proteins.Citation12 Accumulating research has confirmed that lncRNAs play a pivotal part in chemoresistance.Citation13,Citation14 LncRNA AFAP1-AS1 was involved in various cancers, such as lung cancer,Citation15 colorectal cancer,Citation16 and nasopharyngeal carcinoma.Citation17 Yang et al. found that AFAP1-AS1 accelerated proliferation and metastasis of PCaCitation18 and another study indicated that AFAP1-AS1 modulated laryngeal carcinoma cell chemoresistance.Citation19 Nevertheless, the function of AFAP1-AS1 in modulating PTX resistance in PCa is hardly reported and the underlying mechanism is worth investigating.
MicroRNAs are small noncoding RNAs (about 22 nucleotides in length), which regulate genes expression by targeting the 3ʹ-untranslated region (3ʹUTR) at the post-transcriptional level.Citation20 Emerging reports have deeply improved our comprehension of the crucial role that miRNAs play in human cancers.Citation21–23 MiR-195-5p participated in sundry cancersCitation24,Citation25 and regulated PTX resistance.Citation26,Citation27 Moreover, miR-195-5p was found to be negatively regulated by AFAP1-AS1 in melanoma cells.Citation28 Recent research demonstrated that miR-195-5p might act as a tumor suppressor in PCa.Citation29,Citation30 However, in terms of PTX resistance, the function and mechanism of miR-195-5p in PCa remain poorly understood.
FKBP1A belongs to the immunophilin protein family and functions in protein folding and trafficking. A previous study elucidated that FKBP1A was associated with autophagy in vascular endothelial cells.Citation31 Ge et al. found that FKBP1A regulated by SNHG15/miR-338-3p axis was involved in PCa progression.Citation32 Therefore, FKBP1A may be an attractive PCa drug target and it is essential to find novel regulators that modulate the expression of FKBP1A.
In our research, we checked the levels of AFAP1-AS1, miR-195-5p, and FKBP1A in PTX-resistant PCa cells. Simultaneously, the role and underlying mechanism of AFAP1-AS1 in regulating PTX resistance were further studied by subsequent experiments. The AFAP1-AS1/miR-195-5p/FKBP1A axis might provide a promising method for solving PTX resistance in PCa.
Material and methods
Samples and cell culture
A total of 30 PCa tissues and corresponding nearby healthy tissues were acquired from Yantaishan Hospital. All patients involved in this study signed the informed consent and our research was authorized by the Ethics Committee of Yantaishan Hospital.
Human normal prostate epithelial cell line (HprEC) and PCa cell lines (PC3 and DU145) were acquired from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured using Roswell Park Memorial Institute-1640 medium (RPMI-1640, Thermo Fisher Scientific, Rockford, IL, USA) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Gibco). The corresponding PTX-resistant Pca cell lines (PC3-TXR and DU145-TXR) were established by repeatedly treating PC3 and DU145 cells with increasing concentrations of PTX according to a previous report.Citation33
Cell transfection
Small interfering RNA against AFAP1-AS1 (named as si-AFAP1-AS1) and its control (named as si-LncRNA-NC), miR-195-5p mimic (named as miR-195-5p) and its negative control (named as NC), miR-195-5p inhibitor (named as anti-miR-195-5p) and corresponding negative control (named as anti-NC) were obtained GenePharma (Shanghai, China). AFAP1-AS1 expression plasmid (named as pcDNA-AFAP1-AS1) and its negative control (named as pcDNA-NC), FKBP1A expression plasmid (named as FKBP1A) and its control (named as vector) were acquired from RiboBio (Guangzhou, China). Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was hired to transfect cells following the given protocol.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of samples was isolated by FastPure Cell/Tissue Total RNA Isolation Mini Kit (Vazyme, Nanjing, China) and then the RNA was reversely transcribed to complementary DNA (cDNA) using PrimeScript™ RT Master Mix kit (Takara, Dalian, China). The qRT-PCR was performed by AceQ® Universal SYBR qPCR Master Mix (Vazyme) and the data were analyzed by using 2−ΔΔCt method. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as the endogenous controls. Corresponding primers:
AFAP1-AS1 (forward 5ʹ-AGCCTGTTGAATCAGCCAACT-3ʹ, reverse 5ʹ-GGTTCATACCAGCCCTGTCC-3ʹ); MiR-195-5p (forward 5ʹ-GATAGCAGCACAGAAATATTGGC-3ʹ, reverse 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ); FKBP1A (forward 5ʹ-AGCCTGTTGAATCAGCCAACT-3ʹ, reverse 5ʹ-GGTTCATACCAGCCCTGTCC-3ʹ); GAPDH (forward 5ʹ-ACACCCACTCCTCCACCTTT-3ʹ, reverse 5ʹ-TTACTCCTTGGAGGCCATGT-3ʹ); U6 (forward 5ʹ-TCCGGGTGATGCTTTTCCTAG-3ʹ, reverse 5ʹ- GTGCAGGGTCCGAGGT-3ʹ).
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
PTX resistance was evaluated by MTT assay. 1 × 104 PC3-TXR and DU145-TXR cells were seeded in 96-well plates and incubated for 24 h. After transfection for 48 h, cells were treated with various doses of PTX for 24 h. Then, MTT solution (Sigma, St Louis, MO, USA) was added to the cells and incubated for another 4 h. Subsequently, 200 μL dimethyl sulfoxide (DMSO, Thermo Fisher Scientific) was added after discarding the medium. Optical density values were examined at 490 nm wavelength using the microplate reader (Bio-Rad, Richmond, CA, USA). The concentration of PTX causing 50% inhibition of growth (IC50) was determined by the relative survival curve.
Transwell assay
Transwell chamber precoated with Matrigel (Corning Life Sciences, Corning, NY, USA) or not was employed to check the capacity of cell invasion or migration. PC3-TXR and DU145-TXR cells were seeded in 6-well plates. After 48 h transfection, 5 × 104 cells were added to the upper chamber, and the medium containing 2% was added to the lower chamber. An inverted microscope (MultiskanEX, Lab Systems, Helsinki, Finland) was used to analyze the migrated cells treated with crystal violet (Solarbio, Beijing, China).
Flow cytometry
Cell apoptosis was checked using Annexin Apoptosis Detection Kit (Sigma) according to the given procedure. 48 h after transfection, PC3-TXR and DU145-TXR cells were exposed with 100 nM PTX for 24 h. Then, cells were washed with PBS and then resuspended in binding buffer. 5 μL Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and 5 μL propidium iodide (PI) were added to the buffer for incubating for 5 min in the dark. The stained cells were analyzed by Attune NxT (Thermo Fisher Scientific).
Luciferase reporter assay
The interaction between miR-195-5p and AFAP1-AS1 or FKBP1A was predicted by starBase.Citation34 Sequences of wild type AFAP1-AS1 (AFAP1-wt), mutant AFAP1-AS1 (AFAP1-mut), wild type FKBP1A 3ʹUTR (FKBP1A-wt) and its mutant (FKBP1A-mut) harboring the predicted binding sites of miR-195-5p were synthesized and cloned into the pGL3 luciferase reporter vector (Promega, Madison, WI, USA). And then, the vectors with miR-195-5p or its negative control were co-transfected into PC3-TXR and DU145-TXR cells by Lipofectamine 2000 (Invitrogen). The luciferase activity was examined by the Dual-Glo® Luciferase Assay System (Promega).
RNA immunoprecipitation (RIP) assay
RIP was carried out using Magna RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) following the protocols. Briefly, harvested cells were lysed and incubated with magnetic beads conjugated with anti-Argonaute 2 (Anti-Ago2) antibody (Millipore) and immunoglobulin G (IgG) (Millipore) was used as a negative control. The protein was removed by Proteinase K. The immune precipitated RNA was purified and analyzed by qRT-PCR.
RNA pull-down assay
Biotin-labeled probe against miR-195-5p (Bio-miR-195-5p) and its negative control (Bio-NC) were obtained from Sangon (Shanghai, China). Transfected cells were lysed and incubated with streptavidin-coupled beads (Sangon). After being treated by proteinase K, AFAP1-AS1 was isolated and checked by qRT-PCR.
Western blot analysis
RIPA buffer (Thermo Fisher Scientific) was employed to extract proteins from tissues and cells. The protein concentration was checked by BCA Protein Quantification Kit (Vazyme). Proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto the polyvinylidene difluoride (PVDF) membranes (Millipore), which were blocked with 5% skimmed milk. Afterward, the membranes were washed by Phosphate-buffered saline (PBS) and then incubated with the primary antibodies: C-caspase 3 (1:1500, ab2302, Abcam, Cambridge, United Kingdom), FKBP1A (1:1500, ab2918, Abcam), or GAPDH (1:2500, ab181602, Abcam) overnight. After being washed again, the membranes were incubated with the secondary antibody (1:3000, ab205718, Abcam) for 3 h. The membranes were analyzed by the ChemiDoc™ MP Imaging System (Bio-Rad) after being treated with ECL kit (Beyotime, Shanghai, China).
Xenograft mouse model
The animal experiment was approved by the Animal Care and Use Committee of Yantaishan Hospital and carried out following the guidelines of the national animal protection and ethics institute. The short hairpin RNA targeting AFAP1-AS1 (named as sh-AFAP1-AS1), as well as the negative control (named as sh-lncRNA-NC) was constructed by BioSETTIA (San Diego, CA, USA). Six-week-old nude mice were purchased from the Shanghai Experimental Animal Center (Shanghai, China). PC3-TXR cells transfected with sh-lncRNA-NC or sh-AFAP1-AS1 were injected subcutaneously into the flank of the nude mice. The tumor volume was measured with a caliper every 5 d according to the formula: volume = 0.5 × length × width.Citation2 The tumor weight was checked after the mice were euthanized.
Statistical analysis
Experimental data were calculated by GraphPad Prism 7 (La Jolla, CA, USA) and the data were presented by mean ± standard deviation (SD). Two independent groups were compared by Student’s t-test. For more than two groups, the one-way analysis of variance (ANOVA) was used to evaluate the difference. Every experiment was repeated at least three times independently. P < .05 represented statistical significance.
Results
AFAP1-AS1 was sharply upregulated in PTX-resistant PCa cells and AFAP1-AS1 silencing sensitized PTX-resistant PCa cells to PTX
To explore the role of AFAP1-AS1 in PCa, we first detected its expression level in PCa tissues. The data showed that AFAP1-AS1 was significantly elevated in PCa tissues compared with nearby healthy tissues (). Next, PTX-resistant PCa cells were established to study the function of AFAP1-AS1 in chemoresistance. The results indicated that the expression of AFAP1-AS1 was obviously increased in PCa cells (compared with normal cells) and PTX-resistant PCa cells (compared with PCa cells) (). Our data showed a successful knockdown efficiency of si-AFAP1-AS1 in PC3-TXR and DU145-TXR cells (). MTT assay illustrated that IC50 value of PTX in both PC3-TXR and DU145-TXR cells transfected with si-AFAP1-AS1 was remarkably declined compared with corresponding controls (,). Moreover, cell apoptosis, migration, and invasion were measured in PC3-TXR and DU145-TXR cells after the treatment of 100 nM PTX for 24 h. Knockdown of AFAP1-AS1 induced apoptosis of PTX-resistant PCa cells () and the protein level of C-caspase 3 was upregulated (). Transwell assay disclosed that the downregulation of AFAP1-AS1 suppressed the migration and invasion of PTX-resistant PCa cells (,). These results suggested that AFAP1-AS1 was associated with PTX resistance and AFAP1-AS1 silencing could promote the sensitivity of PTX-resistant PCa cells to PTX.
Figure 1. AFAP1-AS1 silencing promoted PTX sensitivity and apoptosis and repressed migration and invasion of PTX-resistant PCa cells. (a) The expression level of AFAP1-AS1 was detected by qRT-PCR in prostate cancer tissues (n = 30) and normal tissues (n = 30). (b) The level of AFAP1-AS1 in the human normal prostate epithelial cell line (HPrEC), PCa cell lines (PC3 and DU145) and paclitaxel resistance PCa cells (PC3-TXR and DU145-TXR) was measured by qRT-PCR. (c) The expression of AFAP1-AS1 in transfected cells and corresponding control was checked by qRT-PCR. (d, e) MTT assay was used to evaluate the resistance of PC3-TXR and DU145-TXR cells to PTX. (f) Flow cytometry was employed to check apoptosis. (g) The protein level of C-caspase 3 in transfected PTX-resistant PCa cells was determined by western blot. (h) Transwell assay was hired to assess the ability of migration and invasion, and the cell numbers of migration or invasion were calculated. *P < .05

AFAP1-AS1 interacted with miR-195-5p and negatively modulated the expression of miR-195-5p in PTX-resistant PCa cells
To investigate the function of miR-195-5p in PCa, we measured the expression level of miR-195-5p in PCa tissues. The data showed that miR-195-5p expression was conspicuously downregulated in PCa tissues compared with adjacent healthy tissues () and negatively associated with AFAP1-AS1 (). The expression of miR-195-5p in PCa cells and PTX-resistant PCa cells was also checked and the result revealed that the level of miR-195-5p was markedly decreased in PCa cells (compared with normal cells) and PTX-resistant PCa cells (compared with PCa cells) (). Past research has declared the fact that the interaction between lncRNA and miRNA functions in regulating PTX resistance in cancers. By using starBase, we found that AFAP1-AS1 could interact with miR-195-5p () and the interaction was validated by the dual-luciferase reporter assay (). RIP assay indicated that the relative enrichment of AFAP1-AS1 was higher in Anti-Ago2 group than control group (). Simultaneously, RNA pull-down assay showed that AFAP1-AS1 was notably enriched in PTX-resistant PCa cells transfected with Bio-miR-195-5p (). Moreover, AFAP1-AS1 overexpression significantly reduced the expression of miR-195-5p, whereas knockdown of AFAP1-AS1 strikingly upregulated the level of miR-195-5p in PC3-TXR and DU145-TXR cells (). Taken together, these results manifested that AFAP1-AS1 bound to miR-195-5p and negatively regulated the expression of miR-195-5p in PTX-resistant PCa cells.
Figure 2. AFAP1-AS1 interacted with miR-195-5p and negatively regulated the expression of miR-195-5p. (a) The expression level of miR-195-5p was checked by qRT-PCR in tumor tissues (n = 30) and nearby healthy tissues (n = 30). (b) The correlation between AFAP1-AS1 and miR-195-5p in cancer tissues was calculated by Pearson’s correlation coefficient. (c) The level of miR-195-5p in HPrEC, PCa cells and PTX-resistant PCa cells was measured by qRT-PCR. (d) The interaction between AFAP1-AS1 and miR-195-5p was predicted by starBase. (e) The dual-luciferase reporter assay was used to check the luciferase activity of cells cotransfected with the miR-195-5p and AFAP1-AS1-wt or AFAP1-AS1-mut. (f) The RIP assay was conducted using Anti-Ago2 to investigate the relationship between AFAP1 and miR-195-5p and Anti-IgG was used as the control. (g, h) RNA pull-down was performed and the relative enrichment of AFAP1-AS1 in samples was detected by qRT-PCR. *P < .05
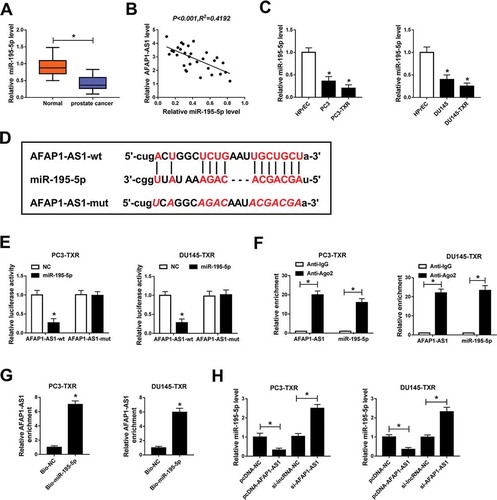
Downregulation of miR-195-5p rescued the effects of AFAP1-AS1 silencing mediated on PTX sensitivity, apoptosis, migration and invasion of PTX-resistant PCa cells
To deeply understand the potential mechanism of AFAP1-AS1 and miR-195-5p in regulating PTX resistance in PCa, we examined the expression of miR-195-5p in PTX-resistant PCa cells transfected with si-AFAP1-AS1 and si-AFAP1-AS1 + anti-miR-195-5p, as well as the matched controls. The data showed that abrogation of AFAP1-AS1 elevated the expression of miR-195-5p in PTX-resistant PCa cells, but the effect could be reversed by lessening the expression of miR-195-5p (). MTT assay indicated that downregulation of miR-195-5p impaired the effect of AFAP1-AS1-mediated inhibition on resistance of PTX-resistant PCa cells to PTX (,). Similarly, reducing the expression of miR-195-5p suppressed apoptosis of PTX-resistant PCa cells () and weakened the protein level of C-caspase 3 in PTX-resistant PCa cells (). In addition, downregulating the expression of miR-195-5p reversed the effects of AFAP1-AS1 silencing mediated suppression on migration and invasion of PTX-resistant PCa cells (,). These results suggested that miR-195-5p and AFAP1-AS1 played opposite roles in regulating the resistance of PTX-resistant PCa cells to PTX, and miR-195-5p inverted the impacts of AFAP1-AS1-mediated promotion on RTX sensitivity, apoptosis, and inhibition on migration and invasion.
Figure 3. Downregulation of miR-195-5p reversed the effect of AFAP1-AS1 silencing mediated promotion on the sensitivity of PTX-resistant cells to PTX. (a) The level of miR-195-5p in PC3-TXR and DU145-TXR cells transfected with si-AFAP1-AS1 and si-AFAP1-AS1 + anti-miR-195-5p, as well as their match controls, was detected by qRT-PCR. (b, c) The resistance of transfected PTX-resistant PCa cells to PTX was assessed by MTT assay. (d) Flow cytometry was introduced to evaluate apoptosis of transfected PTX-resistant PCa cells. (e) The protein level of C-caspase 3 in PTX-resistant PCa cells was measured by western blot. (f) Migration and invasion of transfected cells were checked by transwell assay, and the corresponding cell numbers were calculated. *P < .05
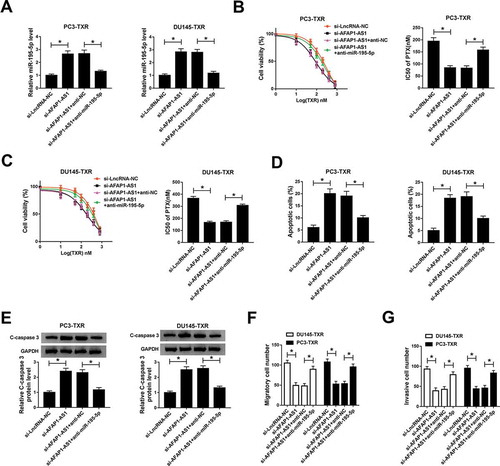
Mir-195-5p targeted the 3ʹUTR of FKBP1A and negatively modulated the expression of FKBP1A via interacting with AFAPA1-AS in PTX-resistant PCa cells
In order to explore the function of FKBP1A in PCa, we evaluated the expression level of FKBP1A using qRT-PCR. The data showed that the mRNA level of FKBP1A was significantly increased in PCa tissues compared with normal tissues () and was negatively correlated with the expression of miR-195-5p (). At the same time, the protein level of FKBP1A was measured by western blot and the result indicated that FKBP1A was obviously upregulated in PCa cells (compared with normal cells) and PTX-resistant PCa cells (compared with PCa cells) (). Interestingly, starBase also showed that miR-195-5p could target the 3ʹUTR of FKBP1A () and the dual-luciferase reporter assay was employed to confirm the prediction. The results demonstrated that miR-195-5p diminished the luciferase activity of FKBP1A-wt in PC3-TXR and DU145-TXR cells, whereas the activity of FKBP1A-mut was not changed (). Besides, RIP assay also corroborated the interaction between miR-195-5p and FKBP1A (). Further studies manifested that elevating the expression of miR-195-5p downregulated the protein level of FKBP1A in the two RTX-resistant PCa cells, whereas the effect could be rescued by the overexpression of AFAP1-AS1 (). All in all, these results illustrated that FKBP1A was upregulated in RTX-resistant PCa cells and was a target of miR-195-5p, which negatively regulated the expression of FKBP1A via interacting with AFAP1-AS1.
Figure 4. MiR-195-5p targeted the 3ʹUTR of FKBP1A and AFAP1-AS1 could regulate the expression of FKNP1A by sponging miR-195-5p. (a) The mRNA expression of FKBP1A was measured by qRT-PCR in cancer tissues (n = 30) and adjacent healthy tissues (n = 30). (b) The correlation between FKBP1A and miR-195-5p in cancer tissues was calculated by Pearson’s correlation coefficient. (c) The protein level of FKBP1A in HPrEC, PCa cells and PTX-resistant PCa cells was measured by western blot. (d) The interaction between FKBP1A and miR-195-5p was predicted by starBase. (e) The dual-luciferase reporter assay was performed to assess the luciferase activity of cells cotransfected with the miR-195-5p and FKBP1A-wt or FKBP1A-mut. (f) The RIP assay was carried out and the relative enrichment of samples was measured by qRT-PCR. (g) The protein level of FKBP1A in transfected cells was detected by qRT-PCR. *P < .05
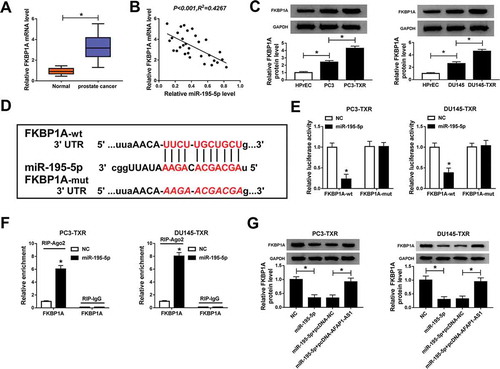
Overexpression of FKBP1A reversed the effects of miR-195-5p-mediated on PTX sensitivity, apoptosis, migration and invasion of PTX-resistant PCa cells
To further investigate the relationship between miR-195-5p and FKBP1A in modulating PTX resistance, we first detected the protein level of FKBP1A in PTX-resistant PCa cells transfected with miR-195-5p and miR-195-5p + FKBP1A, as well as their corresponding controls. The results showed that upregulating the expression of miR-195-5p reduced the protein level of FKBP1A, but this effect could be alleviated by FKBP1A overexpression (). MTT assay indicated that FKBP1A overexpression boosted the IC50 value of PTX in PTX-resistant PCa cells in the case of high expression of miR-195-5p (,). Moreover, overexpression of FKBP1A inhibited apoptosis of PTX-resistant PCa cells () and reduced the protein level of C-caspase 3 in PTX-resistant PCa cells (). Further study showed that overexpression of FKBP1A could reverse the impacts of miR-195-5p-mediated suppression on migration and invasion of PTX-resistant PCa cells (,). To sum up, these results illuminated that FKBP1A overexpression abolished the effects of miR-195-5p-mediated promotion on PTX sensitivity and apoptosis of PTX-resistant PCa cell, as well as the inhibition on migration and invasion of PTX-resistant PCa cells.
Figure 5. FKBP1A overexpression alleviated the effects of miR-195-5p-mediated on PTX sensitivity, apoptosis, migration and invasion of PTX-resistant PCa cells. (a) The protein level of FKBP1A in PC3-TXR and DU145-TXR cells transfected with miR-195-5p and miR-195-5p + FKBP1A, as well as corresponding controls, was detected by western blot. (b, c) The resistance of transfected PTX-resistant PCa cells to PTX was assessed by MTT assay. (d) Flow cytometry was used to check cell apoptosis. (e) The protein level of C-caspase 3 in PTX-resistant PCa cells was measured by western blot. (f) Transwell assay was performed to check cell migration and invasion, and the corresponding cell numbers were counted. *P < .05
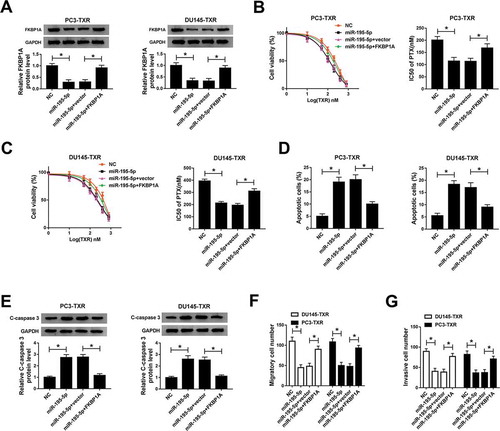
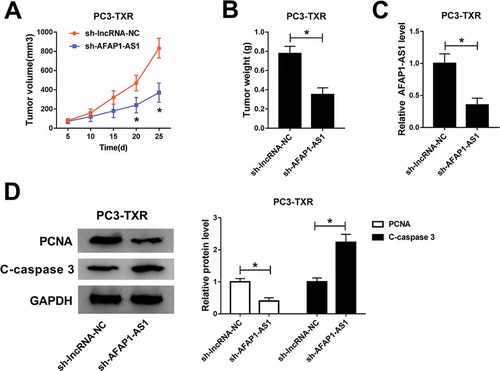
Knockdown of AFAP1-AS1 inhibited tumor growth in vivo
To verify the function of AFAP1-AS1 in PTX-resistant cells in vivo, we established the xenograft mouse model using the PC3-TXR cells transfected with sh-lncRNA-NC and sh-AFAP1-AS1. The data showed that knockdown of AFAP1-AS1 led to the obvious shrink in tumor volume () and decline in tumor weight (). The knockdown efficiency of AFAP1-AS1 was assessed by qRT-PCR (). Besides, the protein levels of PCNA and C-caspase 3 were measured and the results illustrated that PCNA was conspicuously decreased while C-caspase 3 was notably increased in PC3-TXR cell transfected with sh-AFAP1-AS1 (). Collectively, these results suggested that knockdown of AFAP1-AS1 could suppress the tumor growth in vivo.
Figure 6. AFAP1-AS1 silencing repressed the tumor growth in vivo. (a) Tumor volume was measured every 5 d after subcutaneous injection. (b) Weight of the resected tumor was examined after the mice were killed. (c) The level of AFAP1-AS1 in transfected PC3-TXR cell was measured by qRT-PCR. (d) The protein levels of PCNA and C-caspase 3 were determined by western blot
Discussion
PCa threatens men’s health and PTX may be an effective chemotherapy drug in clinical practice.Citation10 However, chemoresistance attenuates the efficacy of chemotherapies and PTX resistance is a big obstacle for the treatment of PCa.Citation11 Growing evidence has confirmed that lncRNAs are involved in the regulation of chemoresistance. Gu et al. found that lncRNA CRNDE regulated colorectal cancer cell chemoresistanceCitation13 and Cai et al. disclosed that lncRNA GBCDRlnc1 induced chemoresistance of gallbladder cancer cells by activating autophagy.Citation35 A previous report indicated that lncRNA AFAP1-AS1 regulated HEp-2 cell chemoresistance.Citation19 However, the possible role of AFAP1-AS1 in modulating PTX resistance in PCa has not been uncovered.
In the present study, we measured the level of AFAP1-AS1 in PCa cells and PTX-resistant PCa cells and investigated the mechanism of AFAP1-AS1 in modulating PTX resistance. The results showed that the level of AFAP1-AS1 was obviously upregulated in PCa cells (compared with normal cells) and PTX-resistant PCa cells (compared with PCa cells). Knockdown of AFAP1-AS1 decreased IC50 value of PTX, indicating that AFAP1-AS1 could promote the resistance of PTX-resistant PCa cells to PTX. Moreover, AFAP1-AS1 silencing promoted apoptosis and elevated the protein level of C-caspase 3. Transwell assay illustrated that abrogation of AFAP1-AS1 repressed metastasis of PTX-resistant PCa cells. Further study demonstrated that knockdown of AFAP1-AS1 inhibited the tumor growth in xenograft mice in vivo and reduced the protein level of PCNA. These results illustrated that AFAP1-AS1 might function as an oncogene in the progression of PCa and could reduce the sensitivity of PTX-resistant PCa cells to PTX.
MiR-195-5p functioned in modulating PTX resistance.Citation26,Citation27 In this study, we confirmed that miR-195-5p was downregulated in PCa cells compared with normal cells, which was in line with previous reports.Citation29,Citation30 And we also observed that miR-195-5p was obviously declined in PTX-resistant PCa cells compared with PCa cells. The interactions between lncRNAs and miRNAs were reported in many studies associated with chemoresistance.Citation13,Citation19 In this research, we found that AFAP1-AS1 could interact with miR-195-5p and negatively modulated the expression of miR-195-5p. Further studies disclosed that downregulating miR-195-5p rescued the impact of AFAP1-AS1 silencing mediated inhibition on the resistance of PTX-resistant PCa cells to PTX. Simultaneously, inhibiting the expression of miR-195-5p repressed apoptosis and promoted migration and invasion of PTX-resistant cells in the case of AFAP1-AS1 silencing. Our results elucidated that miR-195-5p might act as a tumor suppressor and could promote the sensitivity of PTX-resistant PCa cells to PTX.
FKBP1A was reported to participate in the progression of PCa.Citation32 Interestingly, we found that miR-195-5p could target the 3ʹUTR of FKBP1A and negatively modulated the expression of FKBP1A. Moreover, AFAP1-AS1 could modulate the expression of FKBP1A in PTX-resistant PCa cells by sponging miR-195-5p. Further studies disclosed that miR-195-5p decreased the IC50 value and induced apoptosis, whereas overexpression of FKBP1A elevated the IC50 value of PTX and inhibited apoptosis of PTX-resistant PCa cells. Besides, upregulating FKBP1A rescued the effects of miR-195-5p-mediated inhibition on migration and invasion of PTX-resistant PCa cells. These results demonstrated that AFAP1-AS1 silencing enhanced the sensitivity of PTX-resistant PCa cells to PTX by downregulating FKBP1A via sponging miR-195-5p.
In conclusion, our research showed that AFAP1-AS1 was significantly upregulated in PTX-resistant PCa cells and modulated PTX resistance by miR-195-5p/FKBP1A axis. The novel mechanism might provide potential targets for improving chemotherapy of PCa.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387.
- Loblaw DA, Virgo KS, Robert N, Somerfield MR, Edgar BJ, Mendelson DS, Richard M, Sharp SA, Smith TJ, James T. Initial hormonal management of androgen-sensitive metastatic, recurrent, or progressive prostate cancer: 2006 update of an American society of clinical oncology practice guideline. J Clin Oncol. 2016;25(12):1596–1605. doi:10.1200/JCO.2006.10.1949.
- Fang Y, Wu J, Li T, Luo Y, Qiu Q, Quan X, Gao L, Liu W. Biological effects of novel “combi-targeting” molecule and its effect on DNA repair pathway in hormone-refractory prostate cancer. Am J Cancer Res. 2014;5(8):2387–2395. PMID:26396914.
- Lin E, Chen MC, Huang CY, Hsu SL, Huang WJ, Lin MS, Wu JC, Lin H. All-trans retinoic acid induces DU145 cell cycle arrest through Cdk5 activation. Cell Physiol Biochem. 2014;33(6):1620–1630. doi:10.1159/000358724.
- Eisenberger MA. Chemotherapy for endocrine resistant cancer of the prostate. Prog Clin Biol Res. 1990;359:155–164. PMID:2284289.
- Lu G, Shuai C, Yuwei W, Akira W, Laura B, Zhen S, Yuzhuo W, Haojie H. Inhibition of the androgen receptor as a novel mechanism of taxol chemotherapy in prostate cancer. Cancer Res. 2009;69(21):8386–8394. doi:10.1158/0008-5472.CAN-09-1504.
- Yagoda A, Petrylak D. Cytotoxic chemotherapy for advanced hormone-resistant prostate cancer. Cancer. 2015;71(S3):1098–1109. doi:10.1002/1097-0142(19930201)71:3+<1098::aid-cncr2820711432>3.0.co;2-g.
- Armstrong DK, Bundy B, Winzel L, Huang HQ, Baergen R, Lele S, Copeland LJ, Walker JL, Burger RA. Gynecologic oncology group. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354(1):34–43. doi:10.1056/NEJMoa052985.
- Ferretti G, Felici A, Cognetti F. HER2 and response to paclitaxel in node-positive breast cancer. N Engl J Med. 2008;358(2):197;author reply 198. doi:10.1056/NEJMc073152.
- Pazdur R, Kudelka AP, Kavanagh JJ, Cohen PR, Raber MN. The taxoids: paclitaxel (Taxol) and docetaxel (Taxotere). Cancer Treat Rev. 1993;19(4):351–386. doi:10.1016/0305-7372(93)90010-o.
- Mahon KL, Henshall SM, Sutherland RL, Horvath LG. Pathways of chemotherapy resistance in castration-resistant prostate cancer. Endocr Relat Cancer. 2011;18(4):R103–123. doi:10.1530/ERC-10-0343.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521.
- Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16(1):9. doi:10.1186/s12943-017-0583-1.
- Hu YR, Yu YC, You S, Li K, Tong X, Chen S, Chen E, Lin XZ, Chen Y. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16(1):174. doi:10.1186/s12943-017-0743-3.
- Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X, Zhang W, Deng H, Zhou M, Peng S, et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37(1):729–737. doi:10.1007/s13277-015-3860-x
- Feng W, Ni H, Feng S, Min L, Lin C. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother. 2016;81:152–159. doi:10.1016/j.biopha.2016.04.009.
- Hao B, Zhaojian G, Wenling Z, Xiayu L, Yong Z, Qianjin L, Pan C, Lei S, Yu L, Yizhou J. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6(24):20404–20418. doi:10.18632/oncotarget.4057.
- Yang ZT, An F, Hu JD, Zhao WH. Long noncoding RNA AFAP1-AS1 accelerates the proliferation and metastasis of prostate cancer via inhibiting RBM5 expression. Eur Rev Med Pharmacol Sci. 2019;23(8):3284–3290. doi:10.26355/eurrev_201904_17690.
- Yuan Z, Xiu C, Song K, Pei R, Miao S, Mao X, Sun J, Jia S. Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell stemness and chemoresistance. J Cell Mol Med. 2018;22(9):4253–4262. doi:10.1111/jcmm.13707.
- Jovanovic M, Hengartner MO. miRNAs and apoptosis: rNAs to die for. Oncogene. 2006;25(46):6176–6187. doi:10.1038/sj.onc.1209912.
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H. MicroRNA signatures in human ovarian cancer. Cancer Res. 2009;67(18):8699–8707. doi:10.1158/0008-5472.CAN-07-1936.
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2010;120(5):1046–1054. doi:10.1002/ijc.22394.
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TLJ, Tapio V. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi:10.1158/0008-5472.CAN-07-0533.
- Wang T, Ren Y, Liu R, Ma J, Shi Y, Zhang L, Bu R. miR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed Res Int. 2017;7378148. doi:10.1155/2017/7378148.
- Chang M, Lin H, Fu H, Wang B, Han G, Fan M. MicroRNA‐195‐5p regulates osteogenic differentiation of periodontal ligament cells under mechanical loading. J Cell Physiol. 2017;232(12):3762–3774. doi:10.1002/jcp.25856.
- Liu R, Liu X, Zheng Y, Jie GU, Xiong S, Jiang P, Jiang X, Huang E, Yang Y, Di GE. MicroRNA-7 sensitizes non-small cell lung cancer cells to paclitaxel. Oncol Lett. 2014;8(5):2193–2200. doi:10.3892/ol.2014.2500.
- Weiner-Gorzel K, Dempsey E, Milewska M, McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A, Sharpe D. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med. 2015;4(5):745–758. doi:10.1002/cam4.409.
- Liu F, Hu L, Pei Y, Zheng K, Wang W, Li S, Qiu E, Shang G. Zhang J and Zhang X. Long non-coding RNA AFAP1-AS1 accelerates the progression of melanoma by targeting miR-653-5p/RAI14 axis. BMC Cancer. 2020;20(1):258. doi:10.1186/s12885-020-6665-2.
- Cai C, He H, Duan X, Wu W, Mai Z, Zhang T, Fan J, Deng T, Zhong W, Liu Y, et al. miR-195 inhibits cell proliferation and angiogenesis in human prostate cancer by downregulating PRR11 expression. Oncol Rep. 2018;39(4):1658–1670. doi:10.3892/or.2018.6240
- Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y, Liu Y, Li S, Liang Z, Xu X, et al. MicroRNA-195-5p, a new regulator of Fra-1, suppresses the migration and invasion of prostate cancer cells. J Transl Med. 2015;13(1):289. doi:10.1186/s12967-015-0650-6
- Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z, Zhao J, Su L, Zhang S, Zhang Y, et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy. 2014;10(6):957–971. doi:10.4161/auto.28363
- Zhang Y, Zhang D, Lv J, Wang S, LncRNA ZQ. SNHG15 acts as an oncogene in prostate cancer by regulating miR-338-3p/FKBP1A axis. Gene. 2019;705:44–50. doi:10.1016/j.gene.2019.04.033.
- McDermott M, Eustace AJ, Busschots S, Breen L, Crown J, Clynes M, O’Donovan N, Stordal B. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: a practical guide with case studies. Front Oncol. 2014;4:40. doi:10.3389/fonc.2014.00040.
- Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Databaseissue):D92–97. doi:10.1093/nar/gkt1248.
- Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang J, Tang Z, Quan Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer. 2019;18(1):82. doi:10.1186/s12943-019-1016-0.
