ABSTRACT
Background
Multiple functions of miR-199b-5p in diseases have been demonstrated by existing studies. However, never has the correlation between miR-199b-5p and multiple myeloma (MM) been established.
Methods
qRT-PCR analyzed RNA expression and western blot measured protein expression. Cell proliferation ability was tested via colony formation and EdU assays, and apoptosis was determined via TUNEL, flow cytometry and detection of apoptosis-related proteins. Position of LRRC75A antisense RNA 1 (LRRC75A-AS1) was recognized by FISH assay. RIP, RNA pull-down and luciferase reporter experiments explored the molecular interplay.
Results
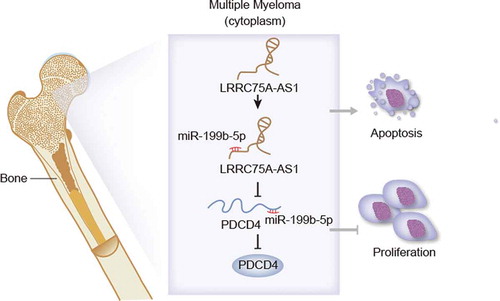
GEO (Gene Expression Omnibus) data revealed miR-199b-5p upregulation in MM specimens, and qRT-PCR data verified miR-199b-5p upregulation in MM cells. Inhibiting miR-199b-5p markedly impeded MM cell proliferation and stimulated apoptosis. Moreover, we demonstrated the mechanism that miR-199b-5p was decoyed by LRRC75A-AS1 and miR-199b-5p targeted programmed cell death 4 (PDCD4) to repress its expression. Further, LRRC75A-AS1 was verified to repress proliferation and prompt apoptosis in a PDCD4-dependent way in MM cells.
Conclusion
Our data displayed that miR-199b-5p was sequestered by LRRC75A-AS1 so that PDCD4 was released to repress MM, implying the targeting miR-199b-5p as a novel thought for improving MM therapy.
KEY WORDS:
Introduction
Multiple myeloma (MM), a carcinoma in hematology, is featured by clonally proliferative malignant plasma cells in the bone marrow.Citation1,Citation2 MM takes up about 1% of cancers and 13% of hematologic tumors.Citation3 Statistics show approximately 80,000 diagnoses of MM each year in the globe. Patients with MM suffer from end-organ damages, such as hypercalcemia, anemia, osteolytic bone disease, and renal insufficiency.Citation4 Regardless of the improved diagnosis and treatment of MM, most patients turn out to relapse with a poor prognosis.Citation5 The etiology of MM remains complicated and still needs clearer explanation, so exploring the possible mechanisms in MM is vitally important.
Non-coding RNAs (ncRNAs) are transcripts without protein-coding ability. MicroRNAs (miRNAs) are a class of ncRNAs with approximately 23 nucleotides (nt) in length and are illustrated to play various roles in mediating cellular processes relating to cancer.Citation6 MiRNAs can act as oncogenes or repressors in numerous carcinomas. For example, miR-202 induces cell apoptosis in esophageal squamous cell carcinoma via targeting HSF2.Citation7 MiR-101-3p inhibits HOTAIR-activated proliferation and invasion through direct target of SRF in gastric cancer.Citation8 MiR-92a facilitates the proliferation in cervical cancer via inhibition of p21 and promotion of cell cycle.Citation9 MiR-302 c-3p inhibits the migration and invasion in hepatocellular carcinoma by TRAF4.Citation10
Past studies support that miR-199b-5p plays different roles in multiple cancers. Chen et al. demonstrate that miR-199b-5p accelerates osteosarcoma development through regulation of HER2.Citation11 Ren et al. illustrate that miR-199b-5p targets Stonin 2 to regulate epithelial-to-mesenchymal transition and metastases in papillary thyroid cancer.Citation12 Wu et al. report that miR-199b-5p represses the proliferation, migration and invasion in triple negative breast cancer via targeting DDR1.Citation13 However, no research has investigated and demonstrated the role of miR-199b-5p in MM.
Long non-coding RNA (lncRNA) is recognized as a functional molecule than can serve as an upstream decoyer for miRNA. Several miRNAs are reported to be blocked by lncRNAs in MM, such as oncogenic miR-410 sequestered by OIP5-AS1,Citation14 and tumor-suppressor miR-1271-5p was targeted by UCA1.Citation15 Small nucleolar RNA host gene 29 (SNHG29), also known as LRRC75A-AS1, is supported to repress the proliferation and migration in colorectal cancer.Citation16 However, relation of LRRC75A-AS1 with miR-199b-5p and MM is unclear.
Programmed cell death 4 (PDCD4) is a well-established tumor-repressor gene. For instance, PDCD4 restrains cell invasion in breast cancer via enhancing tissue inhibitor of metalloproteinases-2.Citation17 Reduced PDCD4 accelerates cell growth in non-small cell lung cancer by PI3K/Akt signaling.Citation18 Several miRNAs are found to target PDCD4 to elicit carcinogenic influences, such as miR-21 targeting PDCD4-STAT3 to modulate the progression of salivary adenoid cystic carcinoma.Citation19 In MM, it is proved that miR-182 and miR-21 strengthen drug resistance of MM cells by repressing PDCD4.Citation20,Citation21 Nevertheless, association of PDCD4 with miR-199b-5p and LRRC75A-AS1 has never been established.
This research aims to probe the expression pattern, role and mechanism related to miR-199b-5p in MM.
Materials and methods
Cell lines and transfection plasmids
Four human MM cell samples (RPMI-8266, NCI-H929, OPM-2, U266) and normal plasma cells (nPCs) were all procured from ATCC (Manassas, VA) and kept in the RPMI-1640 medium (Invitrogen, Carlsbad, CA) at 37°C with 5% CO2. 1% penicillin-streptomycin solution (Sigma, St. Louis, MO) and 10% FBS (Invitrogen) served as the supplements for culture medium. For transfection of U266 and OPM-2 cells, the miR-199b-5p inhibitor (oligonucleotides targeting miR-199b-5p), miR-199b-5p mimics and their relative negative controls (NC) miR-NC were produced by GeneChem (Shanghai, China). The pc-LRRC75A-AS1, pc-PDCD4, NC pcDNA3.1 vectors, the PDCD4-specific shRNAs, and NC (shNC) were designed by Shanghai Genepharm Co. Ltd (Shanghai, China). Plasmid transfection was achieved by Lipofectamine 2000 Reagent (Invitrogen).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) assay
Total RNAs were isolated from U266 and OPM-2 cell samples by use of TRIzol reagent (Invitrogen) as per the guidebook, and then reverse-transcribed into cDNA. SYBR Premix Ex Taq II kit (Perfect Real-Time; Takara, Tokyo, Japan) was applied for amplifying cDNA. Gene expression was calculated by the delta-delta Ct method, standardizing to U6 or GAPDH.
Colony formation
MM cell samples in the 6-well plates (600 cells/well) were cultivated for 14 d, with medium changed every 3 d. Clones were fixed and stained with methanol and 0.2% crystal violet. After washing in PBS, colonies consisting of >50 cells were counted.
5-ethynyl-2ʹ-deoxyuridine (EdU) staining
MM cell samples on the sterile coverslips were plated into the 24-well plates for treatment with EdU incorporation assay kit (Ribobio, Guangzhou, China). EdU-positive cells were visualized by laser confocal microscopy (Olympus, Tokyo, Japan). Nuclei were double-stained with DAPI solution and EdU as positively proliferative cells.
Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) staining
MM cell samples after transfection and fixation were permeabilized in 0.2% Triton X-100 and prepared for incubation with dUTP-end labeling (Clontech, Mountain View, CA). Following nuclear staining with DAPI, TUNEL-positive cells were analyzed by laser confocal microscopy.
Flow cytometer for apoptosis
Annexin V apoptosis detection kit (Life technologies, Grand Island, NY) was applied for analyzing cell apoptosis. 2 × 105 cell samples were rinse in cold PBS and suspended in the Binding buffer adding 10 ml of Annexin V-FITC and 10 μL of PI for 15 min. Samples were finally subjected to flow cytometry (BD Biosciences, San Jose, CA).
MS2-RNA immunoprecipitation (RIP) assay
Cell samples of U266 and OPM-2 were cultured for co-transfection with pMS2-GFP, pSL-MS2 and pSL-MS2-miR-199b-5p for 48 h. RIP assay was implemented on the basis of the protocol of RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) and qRT-PCR was successively conducted.
Fluorescence in situ hybridization (FISH) assay
MM cell samples were fixed with 4% formaldehyde for 15 min in PBS and hybridized with FISH probe synthesized for LRRC75A-AS1 (Ribobio). Cell nuclei were treated with DAPI solution for counterstaining, and then analyzed by confocal microscopy.
RNA pull-down assay
The wild type (WT) and mutant (Mut) sequences of miR-199b-5p covering the predictive LRRC75A-AS1 or PDCD4 binding sites were synthesized and biotinylated into Bio-miR-199b-5p-WT/Mut, and then cultured with the protein extracts from U266 and OPM-2 cell samples. After the beads were added for 2 h, the pull-downs were analyzed through qRT-PCR.
Dual-luciferase reporter assays
Recombinant luciferase reporter vectors were acquired by cloning LRRC75A-AS1, PDCD4 or other indicated genes (ZBTB18, PHACTR4, ARHGAP21, MYRF, STK4) containing the wild type or mutant miR-199b-5p-binding sites into pmirGLO reporter vectors (Promega, Madison, WI). The miR-199b-5p mimics and miR-NC were co-transfected with the above vectors into MM cell samples for 48 h, then analyzed by Dual-Luciferase Reporter Assay System (Promega).
Western blot
Protein lysates of MM cell samples were collected for electrophoresis on 12% SDS-PAGE, and then shifted onto PVDF membranes and cultured with 5% nonfat milk. The primary antibodies against control GAPDH, Bcl-2, Bax, Total-caspase-3/9, Cleaved-caspase-3/9, PDCD4, and the appropriate secondary antibodies labeled with HRP were purchased from Abcam (Cambridge, MA) and employed after dilution. Following washing in the TBST, the band density was detected using the enhanced chemiluminescence (ECL) detection system (Bio-Rad, Hercules, CA).
In vivo assay
The 6-week-old BALB/c nude mice were supplied by Vital River Laboratory Animal Technology (Beijing, China). U266 cells (5 × 106 cells/mouse) were injected subcutaneously in mice to produce xenograft model. The tumor volume (calculated as length×width2/2) was monitored every 4 d following injection. Twenty-eight days after, xenografts were excised from the sacrificed mice and weight of tumors were recorded. MiR-199b-5p level tumors was tested by qRT-PCR. All animal experiments were approved by the Animal Ethics Committee of Rongcheng People’s Hospital of Shandong Province.
Statistical analysis
Experimental results were given as the mean ± standard deviation (SD) of more than three independent bio-repeats. PRISM 6 (GraphPad, San Diego, CA) was applied for developing data analysis through t test and one-way analysis of variance (ANOVA), with p-value <0.05 indicating statistical significance.
Results
MiR-199b-5p exerted oncogenic function in MM
To determine the functional microRNA in multiple myeloma (MM), we browsed GEO datasets and discovered that miR-199b-5p is an up-regulated miRNA in specimens of MM patients in comparison to samples of healthy controls (). Based on this prediction, we sought to study the role of miR-199b-5p in MM. MiR-199b-5p was observed to be distinctly overexpressed in four MM cells compared to that in normal nPCs (). Then, miR-199b-5p inhibitor was transfected into U266 and OPM-2 cells that exhibited the highest miR-199b-5p expression. MiR-199b-5p level was markedly decreased after the transfection (). Besides, miR-199b-5p mimic was transfected into RMPI-8266 and NCI-H929 cells which exhibited relatively low miR-199b-5p level. As confirmed, miR-199b-5p level was obviously increased after miR-199b-5p mimic transfection (Figure S1A). Colony formation assay displayed that miR-199b-5p repression significantly weakened the proliferation of U266 and OPM-2 cells (), and miR-199b-5p mimic elicited opposite impacts on RMPI-8266 and NCI-H929 cells (Figure S1B). Cells that were EdU-positive were decreased by miR-199b-5p down-regulation (), but the EdU-labeled MM cells were increased under miR-199b-5p overexpression (Figure S1C). As for cell apoptosis, TUNEL assay demonstrated that cell apoptosis was stimulated when miR-199b-5p was silenced (), and was hindered when miR-199b-5p was overexpressed (Figure S1D). Flow cytometry analysis indicated that miR-199b-5p silencing induced cell apoptosis (), and miR-199b-5p overexpression impeded apoptosis (Figure S1E).
Figure 1. MiR-199b-5p was oncogenic in MM. (a) GEO data analysis showed miR-199b-5p up-regulation in MM. (b) MiR-199b-5p expression in nPCs, RPMI-8266, NCI-H929, OPM-2 and U266 cells was estimated via qRT-PCR. (c) The transfection efficacy of miR-199b-5p inhibitor in OPM-2 and U266 cells was determined through qRT-PCR. (d-e) Cell proliferation of OPM-2 and U266 cells was tested by colony formation and EdU assays. (f-g) Cell apoptosis of OPM-2 and U266 cells was examined via TUNEL and flow cytometry analysis. (h) Western blotting of apoptosis-related proteins including Bcl-2, Bax, total and cleaved caspase-3 and caspase-9. *P < .05 and **P < .01 vs control group
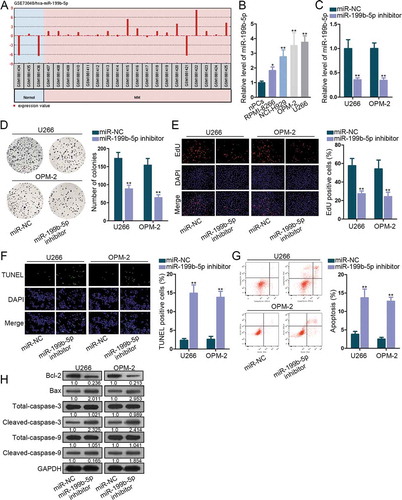
Additionally, when miR-199b-5p was inhibited, Bcl2 protein expression was reduced, and the protein expression of Bax, cleaved-caspase-3 and cleaved-caspase-9 was increased. However, levels of total caspase-3 or total caspase-9 did not vary (). Under miR-199b-5p overexpression, Bcl-2 level increased, and the levels of Bax, cleaved-caspase-3, and cleaved-caspase-9 declined, with total caspase-3 and caspase-9 unchanged (Figure S1F).
Moreover, we injected mice with U266 cells transfected with NC inhibitor or miR-199b-5p inhibitor to test miR-199b-5p function in vivo. As a result, U266 cells with miR-199b-5p knockdown presented slower tumor growth in mice compared with control (Figure S2A). Also, at 28d, we observed smaller volume and lighter weight of xenografts in mice with miR-199b-5p knockdown (Figure S2B-C). Accordingly, xenografts with miR-199b-5p knockdown presented lower miR-199b-5p level as presented by qRT-PCR data (Figure S2D). Western blot data depicted that xenografts in mice showed lower level of Bcl-2 and higher level of Bax, cleaved-caspase-3 and cleaved-caspase-9 under miR-199b-5p knockdown versus the control in vivo, with total caspase-3 and caspase-9 unchanged (Figure S2E). These data illustrated that cell proliferation and apoptosis resistance were repressed by inhibition of miR-199b-5p in vitro and in vivo.
MiR-199b-5p bound with LRRC75A-AS1 in MM
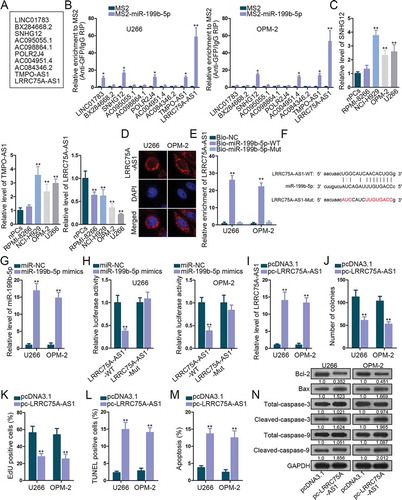
Since abundant microRNAs (miRNAs) are involved in ceRNA model in carcinomas, we decided to figure out the lncRNAs binding with miR-199b-5p in MM. Under the standard of strict stringency based on CLIP data, StarBase v3.0 predicted ten lncRNAs potentially targeting miR-199b-5p (). According to MS2-RIP assay, three lncRNAs (SNHG12, TMPO-AS1, and LRRC75A-AS1) were enriched in the precipitates of MS2-miR-199b-5p group in both U266 and OPM-2 cells (), indicating that the three lncRNAs interacted with miR-199b-5p in both two cell lines. Levels of these lncRNAs in MM cells versus nPCs were analyzed by qRT-PCR. Only LRRC75A-AS1 exhibited dramatically low expression in MM cells, whereas SNHG12 and TMPO-AS1 were found upregulated in MM cells (). Thus, we picked LRRC75A-AS1 to further investigate. FISH assay localized LRRC75A-AS1 in the cytoplasm of U266 and OPM-2 cells (), further suggesting the possibility that LRRC75A-AS1 participated in the ceRNA network. Later, we tested the interplay between miR-199b-5p and LRRC75A-AS1. Shown by RNA pull-down result, biotinylated miR-199b-5p-WT, rather than miR-199b-5p-Mut, pulled down LRRC75A-AS1 in both cell lines (). The sequences of LRRC75A-AS1-WT, LRRC75A-AS1-Mut and miR-199b-5p for luciferase reporter assay are depicted in . MiR-199b-5p expression was elevated after transfection of miR-199b-5p mimics (). The luciferase activity of LRRC75A-AS1-WT, but not LRRC75A-AS1-Mut, was impaired by miR-199b-5p overexpression (). Thus, LRRC75A-AS1 was confirmed to interact with miR-199b-5p. Furthermore, effect of LRRC75A-AS1 in MM was explored. LRRC75A-AS1 was up-regulated by pc-LRRC75A-AS1 transfection in U266 and OPM-2 cells (). As detected by function assays, overexpressing LRRC75A-AS1 impaired cell proliferation and strengthened apoptosis of U266 and OPM-2 cells (-N). Taken together, miR-199b-5p bound to LRRC75A-AS1 and LRRC75A-AS1 developed suppressive effects on MM cellular activities.
Figure 2. The interplay between LRRC75A-AS1 and miR-199b-5p. (a) Ten lncRNAs were screened out by starBase v3.0. (b) MS2-RIP screening of lncRNAs interacting with miR-199b-5p in OPM-2 and U266 cells. (c) qRT-PCR analysis of SNHG12, TMPO-AS1 and LRRC75A-AS1 levels in nPCs and MM cells. (d) FISH assay for the location of LRRC75A-AS1 in OPM-2 and U266 cells. (e) RNA pull-down analysis of miR-199b-5p interaction with LRRC75A-AS1. (f) The sequences of LRRC75A-AS1-WT/Mut and miR-199b-5p. (g) qRT-PCR analysis of miR-199b-5p overexpression in U266 and OPM-2 cells. (h) Luciferase reporter experiments were employed to validate the binding of miR-199b-5p to LRRC75A-AS1. (i) LRRC75A-AS1 was increased by pc-LRRC75A-AS1 as confirmed by qRT-PCR. (j-k) Colony formation and EdU assays examined the proliferation of OPM-2 and U266 cells. (l-n) TUNEL, flow cytometry and western blotting measured the apoptosis of OPM-2 and U266 cells. *P < .05 and **P < .01 vs control group
PDCD4 was the target of miR-199b-5p
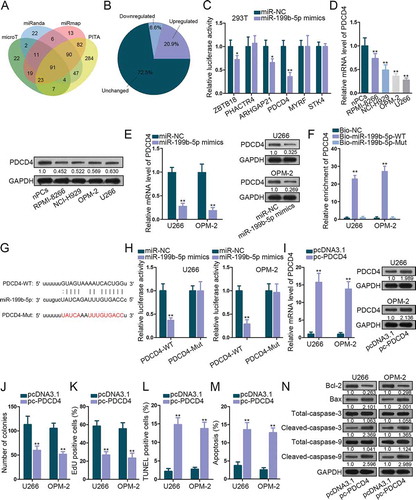
To find the downstream targets for miR-199b-5p, we scanned starBase v3.0. 91 miR-199b-5p target genes were identified under specific selecting condition in Starbase3.0 (4 prediction programs: microT, miRanda, miRmap and PITA, strict stringency, 4 cancer types) (). Subsequently, levels of these were dissected in MM cells versus normal cells. Shown by the pie chart, 6.6% genes (6 of 91) were significantly down-regulated (). The luciferase reporter assay was utilized to test the impact of miR-199b-5p on these targets. We found that the luciferase activity of PDCD4 was decreased the most significantly by miR-199b-5p mimic (). Thus, we speculated PDCD4 as a promising target for miR-199b-5p. qRT-PCR and western blot verified the down-regulation of PDCD4 mRNA and protein in MM cells (). When overexpressing miR-199b-5p, PDCD4 mRNA and protein levels were obviously lessened (). Sequentially, the interplay between miR-199b-5p and PDCD4 mRNA was determined. We observed the enrichment of PDCD4 in the pulldown products of bio-miR-199b-5p-WT group and the weakened PDCD4-WT luciferase activity under miR-199b-5p mimic transfection (-H). Next, we investigated the role of PDCD4 in MM. As depicted by qRT-PCR and western blot, PDCD4 level was promoted by pc-PDCD4 (). Colony formation and EdU experiments confirmed that ectopic PDCD4 expression inhibited cell proliferation (-k). TUNEL assay, flow cytometry analysis and western blot testified that apoptosis in MM cells was stimulated by the up-regulation of PDCD4 (-n). These results proved that miR-199b-5p targeted PDCD4 and PDCD4 was a repressor in MM.
Figure 3. MiR-199b-5p targeted PDCD4 in MM cells. (a) The Venn diagram revealed the targets of miR-199b-5p identified by 4 prediction programs in Starbase. (b) Pie chart of the expression change rates of these genes in nPCs and MM cells. (c) Luciferase reporter assay in 293 T cells to test the binding affinity of six down-regulated genes in MM cells with miR-199b-5p. (d-e) qRT-PCR and western blotting detections of PDCD4 expression in MM cells in comparison with control cells or in MM cells transfected with miR-199b-5p mimics or miR-NC. (f) The binding between miR-199b-5p and LRRC75A-AS1 was testified by RNA pull-down. (g-h) Sequences of PDCD4-WT/Mut and miR-199b-3p. Luciferase reporter experiments was used for confirming the binding of miR-199b-3p at predicted sites in PDCD4. (i) The overexpression efficiency of pc-PDCD4 in U266 and OPM-2 cells was evaluated via qRT-PCR and western blotting. (j) Quantification of colonies formed by MM cells. (k) Quantification of EdU-labeled MM cells under PCDC4 overexpression. (l) Quantification of TUNEL-labeled MM cells upon PCDC4 overexpression. (m) Quantification of apoptotic MM cells in flow cytometry analysis. (n) Western blot of apoptosis-related genes in MM cells with PDCD4 overexpression. *P < .05 and **P < .01 vs control group
LRRC75A-AS1/miR-199b-5p/PDCD4 axis functioned in MM
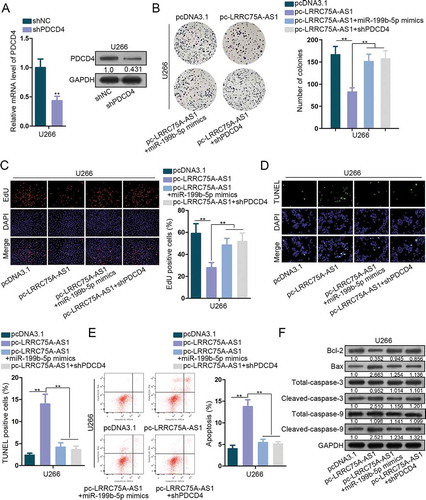
In the end, rescue experiments were implemented to interrogate the function of LRRC75A-AS1/miR-199b-5p/PDCD4 axis. shPDCD4 was transfected into U266 cells and qRT-PCR and western blot validated the knockdown of PDCD4 (). Colony generation of U266 cells was restrained by pc-LRRC75A-AS1 and was rescued by miR-199b-5p mimic or shPDCD4 (). Number of EdU-labeled cells was decreased by overexpressing LRRC75A-AS1, and such decrease was reversed by overexpressing miR-199b-5p or silencing PDCD4 (). TUNEL assay and flow cytometry analysis elucidated that miR-199b-5p mimic or shPDCD4 reversed the increase of cell apoptosis caused by LRRC75A-AS1 overexpression (-e). Besides, protein levels of Bcl-2 were lowered and that of Bax, cleaved-caspase-3 and cleaved-caspase-9 were improved by pc-LRRC75A-AS1, and these phenomena were abolished under co-transfection of miR-199b-5p mimics or shPDCD4 (). Thus, LRRC75A-AS1 repressed MM cell proliferation and enhanced apoptosis via miR-199b-5p/PCDC4.
Figure 4. LRRC75A-AS1/miR-199b-5p/PDCD4 axis functioned in MM. (a) The inference efficiency of shPDCD4 in U266 cells for rescue assays was analyzed through both qRT-PCR and western blot. (b-c) MM cells were under transfection of pcDNA3.1, pc-LRRC75A-AS1, pc-LRRC75A-AS1 + miR-199b-5p mimics, or pc-LRRC75A-AS1 + shPDCD4. The proliferation of U266 cells in four groups was dissected via colony formation and EdU assays. (d-f) The apoptosis of U266 cells under four treatments was estimated via TUNEL, flow cytometry and western blot. **P < .01 vs control group
Discussion
The modulation of miRNAs on MM has been demonstrated by previous researches. For instance, ibrutinib attenuates miR-21 in multiple myeloma through decreasing NF-κB and STAT3.Citation22 MiR-631 re-sensitizes multiple myeloma cells to bortezomib through inhibition of UbcH10.Citation23 MiR-199b-5p is previously studied to be a tumor repressor or oncogenic marker in human carcinomas, such as triple negative breast cancer, papillary thyroid carcinoma and osteosarcoma.Citation11–13 These data indicated that function of miR-199b-5p might vary in different cancer type. According to GEO data, miR-199b-5p showed upregulation in MM samples, indicating its involvement in MM. Consistently, in vitro data of our research proved that miR-199b-5p level was markedly elevated in MM cells. Function assays suggested the oncogenic role of miR-199b-5p in MM cells through validating that miR-199b-5p strengthened proliferation and inhibited apoptosis and that inhibiting miR-199b-5p reduced tumorigenesis in vivo.
Recent reports have explained that in the ceRNA network, lncRNAs exhausts miRNAs to de-repress miRNAs target.Citation24–26 Several lncRNAs are found as miRNA decoyers in MM as well. For example, MALAT1 facilitates MM tumorigenesis by sponging miR-1271-5p,Citation27 lncRNA IRAIN sequesters miR-125b to impair MM progression.Citation28 Previously, LRRC75A-AS1 has been once exposed to inhibit colorectal carcinoma.Citation16 In this study, we first identified LRRC75A-AS1, also known as small nucleolar RNA host gene 29 (SNHG29), as the lncRNA binding to miR-199b-5p. In subsequence, mechanism assays confirmed the interaction between LRRC75A-AS1 and miR-199b-5p. LRRC75A-AS1 level was substantially decreased in MM cells, and LRRC75A-AS1 up-regulation hindered cell proliferation and apoptosis resistance in MM cells.
Thereafter, programmed cell death 4 (PDCD4) was screened out to be the target of miR-199b-5p in MM. The suppressive function of PDCD4 in tumors has been widely demonstrated, such as in non-small cell lung cancer, breast cancer and salivary adenoid cystic carcinoma.Citation17–19 Concordantly, our data elucidated the targeting and suppression of PDCD4 by miR-199b-5p. Besides, the inhibitory role of PDCD4 in MM cells was also co firmed. In the end, data from rescue assays supported the role of LRRC75A-AS1/miR-199b-5p/PDCD4 pathway in MM cells.
In conclusion, miR-199b-5p was up-regulated in MM cells and developed carcinogenic property in MM. In addition, we first revealed the LRRC75A-AS1/miR-199b-5p/PDCD4 axis in MM (). These data indicated that targeting miR-199b-5p might be an innovative thought for MM therapy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (3.5 MB)Acknowledgments
All supports from participants in this research were undeniable.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Spitzer TR, Sachs DH, Cosimi B. Multiple myeloma. N Engl J Med. 2011;364:2364. author reply.
- Becker N. Epidemiology of multiple myeloma. Recent Results Cancer Res. 2011;183:25–35.
- Alexander DD, Mink PJ, Adami HO, Cole P, Mandel JS, Oken MM, Trichopoulos D. Multiple myeloma: a review of the epidemiologic literature. Int J Cancer. 2007;120(Suppl 12):40–61.
- Anderson KC. Progress and paradigms in multiple myeloma. Clin Cancer Res. 2016;22:5419–5427.
- Naymagon L, Abdul-Hay M. Novel agents in the treatment of multiple myeloma: a review about the future. J Hematol Oncol. 2016;9:52.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233.
- Meng X, Chen X, Lu P, Ma W, Yue D, Song L, Fan Q. miR-202 promotes cell apoptosis in esophageal squamous cell carcinoma by targeting HSF2. Oncol Res. 2017;25:215–223.
- Wu X, Zhou J, Wu Z, Chen C, Liu J, Wu G, Zhai J, Liu F, Li G. MiR-101-3p suppresses HOX transcript antisense RNA (HOTAIR)-induced proliferation and invasion through directly targeting SRF in gastric carcinoma cells. Oncol Res. 2017.(8):1383–1390
- Su Z, Yang H, Zhao M, Wang Y, Deng G, Chen R. MicroRNA-92a promotes cell proliferation in cervical cancer via inhibiting p21 expression and promoting cell cycle progression. Oncol Res. 2017;25:137–145.
- Yang L, Guo Y, Liu X, Wang T, Tong X, Lei K, Wang J, Huang D, Xu Q. The tumor suppressive miR-302c-3p inhibits migration and invasion of hepatocellular carcinoma cells by targeting TRAF4. J Cancer. 2018;9:2693–2701.
- Chen Z, Zhao G, Zhang Y, Ma Y, Ding Y, Xu N. MiR-199b-5p promotes malignant progression of osteosarcoma by regulating HER2. J BUON. 2018;23:1816–1824.
- Ren L, Xu Y, Qin G, Liu C, Yan Y, Zhang H. miR-199b-5p-Stonin 2 axis regulates metastases and epithelial-to-mesenchymal transition of papillary thyroid carcinoma. IUBMB Life. 2018.(1):28–40.
- Wu A, Chen Y, Liu Y, Lai Y, Liu D. miR-199b-5p inhibits triple negative breast cancer cell proliferation, migration and invasion by targeting DDR1. Oncol Lett. 2018;16:4889–4896.
- Yang N, Chen J, Zhang H, Wang X, Yao H, Peng Y, Zhang W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017;8:e2975.
- Yang Y, Chen L. Downregulation of lncRNA UCA1 facilitates apoptosis and reduces proliferation in multiple myeloma via regulation of the miR-1271-5p/HGF axis. J Chin Med Assoc. 2019.(9):699–709
- Chen J, Lan J, Ye Z, Duan S, Hu Y, Zou Y, Zhou J. Long noncoding RNA LRRC75A-AS1 inhibits cell proliferation and migration in colorectal carcinoma. Exp Biol Med (Maywood). 2019:1535370219874339.(14):1137–1143
- Nieves-Alicea R, Colburn NH, Simeone AM, Tari AM. Programmed cell death 4 inhibits breast cancer cell invasion by increasing tissue inhibitor of metalloproteinases-2 expression. Breast Cancer Res Treat. 2009;114:203–209.
- Zhen Y, Li D, Li W, Yao W, Wu A, Huang J, Gu H, Huang Y, Wang Y, Wu J, et al. Reduced PDCD4 Expression Promotes Cell Growth Through PI3K/Akt Signaling in Non-Small Cell Lung Cancer. Oncol Res. 2016;23:61–68.
- Jiang LH, Ge MH, Hou XX, Cao J, Hu SS, Lu XX, Han J, Wu YC, Liu X, Zhu X, et al. miR-21 regulates tumor progression through the miR-21-PDCD4-Stat3 pathway in human salivary adenoid cystic carcinoma. Lab Invest. 2015;95:1398–1408.
- Wu Y, Zhu X, Shen R, Huang J, Xu X, He S. miR-182 contributes to cell adhesion-mediated drug resistance in multiple myeloma via targeting PDCD4. Pathol Res Pract. 2019:152603. doi: 10.1016/j.prp.2019.152603.
- Luo X, Gu J, Zhu R, Feng M, Zhu X, Li Y, Fei J. Integrative analysis of differential miRNA and functional study of miR-21 by seed-targeting inhibition in multiple myeloma cells in response to berberine. BMC Syst Biol. 2014;8:82.
- Ma J, Gong W, Liu S, Li Q, Guo M, Wang J, Wang S, Chen N, Wang Y, Liu Q, et al. Ibrutinib targets microRNA-21 in multiple myeloma cells by inhibiting NF-kappaB and STAT3. Tumour Biol. 2018;40:1010428317731369.
- Xi H, Li L, Du J, An R, Fan R, Lu J, Wu YX, Wu SX, Hou J, Zhao LM. hsa-miR-631 resensitizes bortezomib-resistant multiple myeloma cell lines by inhibiting UbcH10. Oncol Rep. 2017;37:961–968.
- Zhang L, Fang F, He X. Long noncoding RNA TP73-AS1 promotes non-small cell lung cancer progression by competitively sponging miR-449a/EZH2. Biomed Pharmacother. 2018;104:705–711.
- Li Y, Lv M, Song Z, Lou Z, Wang R, Zhuang M. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed Pharmacother. 2018;103:939–946.
- Lan X, Liu X. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med. 2019;23:2351–2361.
- Liu N, Feng S, Li H, Chen X, Bai S, Liu Y. Long non-coding RNA MALAT1 facilitates the tumorigenesis, invasion and glycolysis of multiple myeloma via miR-1271-5p/SOX13 axis. J Cancer Res Clin Oncol. 2020.(2):367–379
- Jiang Y, Chen J, Chen G. Long noncoding RNA IRAIN acts as tumor suppressor via miR-125b in multiple myeloma. Oncol Lett. 2019;18:6787–6794.