ABSTRACT
Cancer diagnosis and therapy is quickly moving from the traditional histology-based approaches to genomic stratification, providing a huge opportunity for radiogenomics, associating imaging features with genomic data. Genome sequencing is time consuming, expensive and invasive whereas 18F-FDG PET/CT is readily available, fast and noninvasive. The aim of this study was to determine the relationship between the maximum standardized uptake value (SUVmax) and the frequency of 11 common oncogenic anomalies determined by specific common genomic alterations in tissue biopsies from patients with cancer. We retrospectively studied 102 consecutive untreated patients with gastrointestinal, lung, and breast cancer who underwent 18F-FDG PET/CT imaging, shortly prior to molecular testing by a biopsy for genomic profiling that consisted of 11 common DNA alterations: (1) TP53, (2) DNA repair, (3) EGFR, (4) PI3K/AKT/MTOR (PAM) pathway including PTEN, PIK3CA, AKT, TSC, CCNB1, MTOR, FBXW2, and NF2, (5) MEK, (6) CYCLIN including CCND,CDK, CDKN, and RB, (7) WNT, (8) ALK, (9) MYC, (10) MET, and (11) FGF/FGFR. Higher SUVmax was associated with the presence of TP53 and PAM genomic anomalies (p < .05), but not the other 9 gene groups (p > .05). More importantly, SUVmax was positively correlated with total number of oncogenic anomalies (r = 0.27, p = .005). We propose higher SUVmax as an indicator for total number of common oncogenic anomalies. This finding is a step forward in noninvasive stratification of cancer patients, in terms of the overall load of oncogenic anomalies, based on their SUVmax.
Introduction
The relatively new field of cancer genome sequencing is changing the landscape of oncology.Citation1 Radiogenomics plays an important role by finding relationship and creating link between imaging profiles and genomics.Citation2 Radiogenomics has been used mainly in computer tomography (CT) and magnetic resonance imaging (MRI) to draw a link between imaging features and tumor genotypes.Citation3 However, radiogenomics in the field of nuclear medicine is not explored to the same extent, although 18F-fluorodeoxyglucose positron emission tomography (18F-FDG PET) is regularly used for cancer diagnosis and treatment follow up.Citation4–6 Standardized uptake value (SUVmax) is routinely used as a quantitative indicator of 18F-FDG uptake, thus glucose metabolism rate (GMR). SUVmax has been shown to correlate with P53 alterations and its relationship with a variety of genomic alteration has been studied previously; however, it remains mostly nonspecific at the molecular and genetic level.Citation7–9
There are several common genomic anomalies that play role in tumorigenesis.Citation1 Determining the presence of each genomic alteration is necessary for targeted therapy. The total number of common oncogenic anomalies is also crucial information, as an indicator of the overall load of oncogenic anomalies.Citation10,Citation11
Increased SUVmax is a hallmark of cancer. Although commonly explained as a result of increased proliferation rate, the genetic underpinning of increased SUVmax in cancer is still not clearly understood.Citation12,Citation13 Oncogenic anomalies are proposed to be associated with metabolic reprogramming in cancer cells, causing increased GMR; however, such an association has not yet been established, clinically.Citation14 In our prior study, we showed a potential relationship between SUVmax and the number of genomic anomalies of the tumor; however, we analyzed only a small subset of patients (N = 3) at the two extremes of SUVmax.Citation13 Herein, we sought to study if higher total number of these genomic anomalies is associated with higher SUVmax, in the biopsied lesions. Genome sequencing is time consuming, expensive and invasive whereas 18F-FDG PET/CT is readily available, fast and noninvasive. Our hypothesis is that the total number of oncogenic anomalies is related to SUVmax, in the biopsied lesion.
Results
Patients
We studied 102 untreated patients including 42 (41.2%) breast, 26 (25.5%) gastrointestinal, and 34 (33.3%) lung cancer patients. Due to the retrospective nature of this study, the patients were in different stages of their cancers and the lesion biopsied included primary or metastatic lesions.
Associations between individual genes/gene families and SUVmax
Forty seven of the 102 patients (46%) had anomalies in TP53 and a significantly higher mean SUVmax of 9.0 compare to patients with unaltered TP53 which had a mean SUVmax of 6.3 (p = .002). Forty of the 102 patients (39%) had abnormalities in PAM pathway genes (PTEN, PIK3CA, AKT, TSC, CCNB1, MTOR, FBXW2, and NF2). The mean SUVmax for these 40 patients was 8.7, significantly higher than 6.8 in patients without abnormal PAM pathway genes (p = .042).
Four of 102 patients (4%) had DNA repair gene (BRCA, BRIP, ATM, MMR, MSH, MLH) anomalies. The mean SUVmax of these 4 patients was 9.7, not significantly different from 7.4 in those without DNA repair gene anomalies (p = .28). Twenty eight of 102 patients (27%) had EGFR gene anomalies. The mean SUVmax of these 28 patients was 7.6, not significantly different from 7.5 in those without EFGR gene anomalies (p = .98). Eighteen of the 102 patients (18%) had abnormalities in MEK genes (RAS, RAF, MAPK, and CNAS). The mean SUVmax for these 18 patients was 8.9, not significantly higher than 7.2 in patients without abnormal MEK pathway genes (p = .16). Thirty nine of the 102 patients (38%) had abnormalities in CYCLIN pathway genes (CCND, CDK, CDKN, RB) and a mean SUVmax of 7.8 compared to 7.4 in the patient without CYCLIN abnormality. These means were not significantly different (p = .68). Seventeen of the 102 patients (17%) had abnormalities in WNT genes (APC, CTNNB, NOTCH) and a mean SUVmax of 8.2 compared to 7.4 in the patient without WNT abnormality. These means were not significantly different (p = .52). Three of the 102 patients (3%) had abnormalities in ALK gene and a mean SUVmax of 11.9 compared to 7.4 in the patient without ALK abnormality. These means were not significantly different (p = .09). Fourteen of the 102 patients (14%) had MYC gene anomalies and a corresponding mean SUVmax of 8.7, not significantly different compared to the 88 patients without MYC gene anomalies with a mean SUVmax of 7.4 (p = .31). Two of the 102 patients (2%) had MET gene anomalies and a corresponding mean SUVmax of 6, not significantly different compared to the 100 patients without MET gene anomalies with a mean SUVmax of 7.6 (p = .63). Twenty one of the 102 patients (21%) had FGF/FGFR gene anomalies and a mean SUVmax of 6.8 as compared to the 81 patient without anomalies and a mean SUVmax of 7.7. This was not significant (p = .40).
Total number of main oncogenic anomalies
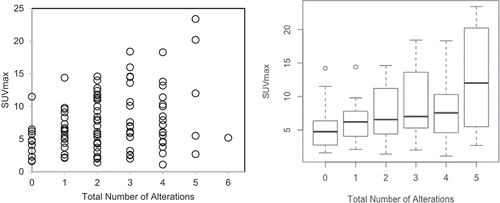
Total number of oncogenic anomalies from the aforementioned 11 genetic groups was calculated for each patient from summation of the individual genomic anomalies. This number actually ranged from 0 to 6, as no patient had more than 6 oncogenic anomalies. In fact, 12 patients (12%) had no genetic anomalies, 19 patients (19%) had 1 genetic alteration, 30 patients (29%) had 2 total anomalies, 17 patients (17%) had 3 total anomalies, 18 patients(18%) had total of 4 anomalies, 5 patients (5%) had total of 5 anomalies, and 1 patient (1%) had 6 oncogenic anomalies (). No patient had more than 6 oncogenic anomalies. The SUVmax was positively correlated to the total of oncogenic anomalies (r = 0.27, p = .005).
Figure 1. Relationship between SUVmax and the total number of oncogenic alterations with Pearson correlation coefficient r = 0.27 (p = .005). Left panel shows the scatter plot of all patients (n = 102) with circles represent individual datapoints. Right panel shows the box plot for all patients excluding the group with 6 oncogenic alterations because there was only one patient in that group. The central thick black line indicates the median, and the bottom and top of the rectangle are the 25th (Q1) and 75th (Q3) percentiles. The circles represent outlier SUVmax, defined as either larger than Q3 + 1.5 × IQR or smaller than Q1 – 1.5 × IQR, where IQR = Q3 – Q1 is the interquartile range. The horizontal “whiskers” represent the largest and smallest non-outlier observations in the data set
Discussion
Radiogenomics, defined as associating imaging features with genomic data, is gaining attention as the precision medicine is rapidly evolving.Citation15,Citation16 18F-FDG PET/CT is standard of care for cancer staging/restaging, to guide therapeutic decisions and monitor therapeutic response. In our prior study, we used a simple method where we ranked the patients based on SUVmax. Then we selected three patients with the highest SUVmax and three patients with the lowest SUVmax, and speculated that the patients with the highest SUVmax group had more genomic anomalies.Citation13 In the current study, we performed a rigorous statistical analysis of a more homogenous subset of the previous study population, containing only patients with breast, lung and gastrointestinal tumors. In addition, the analysis of the relationship between SUVmax and total number of genetic anomalies was performed on all 102 patients. The aim of this study was to determine the relationship of the SUVmax of the biopsied lesion, with the sum of 11 common oncogenic anomalies including TP53, EGFR, ALK, MYC, MET, FGF/FGFR, DNA repair, PI3K/Akt/mTOR (PAM), MEK, CYCLIN, and WNT determined by specific common genomic anomalies in tissue biopsies from breast, gastrointestinal and lung cancer patients. Our speculation was that higher number of total oncogenic anomalies cause metabolic reconfiguration,Citation14 thus increased GMR and SUVmax.
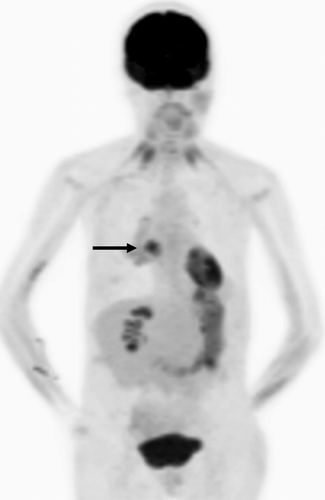
SUVmax was positively correlated to the total number of oncogenic anomalies (r = 0.27, p = .005), in the biopsied lesion (). The SUVmax was also positively correlated with TP53, as previously shown,Citation13 and also with PAM anomalies (). Relationship between SUVmax and other 9 individual oncogenic abnormalities, individually, was not statistically significant (). Representative images from two patients with total number of oncogenic anomalies of 0 and 5 and respective SUVmax of 4.9 and 23.6 are shown ().
Table 1. Mean SUVmax and number of patients with or without genomic alteration (n = 102)
Figure 2. FDG PET projection image in a patient with adenocarcinoma of the lung. The biopsied right lung hypermetabolic lesion had SUVmax of 20.2 (arrow). This lesion had a total number of main oncogenic alterations of 5
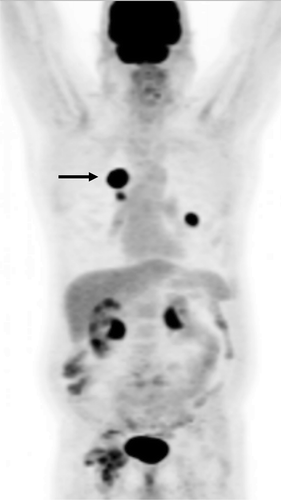
Figure 3. FDG PET projection image in a patient with adenocarcinoma of the lung. The biopsied right lung hypermetabolic lesion had SUVmax of 4.9 (arrow). This lesion had a total number of main oncogenic alterations of 0
Our finding suggests that higher SUVmax is an indicator of total number of oncogenic anomalies. We speculate that higher number of oncogenic anomalies cause metabolic reprogramming by stimulating glucose uptake, and channeling glucose to aerobic glycolysis,Citation14,Citation17,Citation18 therefore increase SUVmax. We suggest that SUVmax merits further study as an accessible and noninvasive surrogate for the total number of oncogenic anomalies derived from sequencing a tissue biopsy.
This study had several limitations including its retrospective nature, relatively small number of patients, and lack of a definite underlying mechanism. Also, the imaging and molecular testing were not exactly concurrent, because of the retrospective nature of the study.
Conclusions
18F-FDG PET/CT SUVmax is positively correlated with total number of oncogenic anomalies (r = 0.27, p = .005). This finding suggests that SUVmax can estimate the total number of oncogenic anomalies, noninvasively. Since mutational load has been implicated as a predictive factor for immunotherapy response,Citation1,Citation9
Patients and methods
Patient selection
We studied 102 consecutive untreated patients including 42 breast, 26 gastrointestinal and 34 lung cancer patients who underwent 18F-FDG PET/CT, within six months before a biopsy for genomic profiling, a subpopulation of our prior workCitation13. A 6-month cutoff was chosen to avoid a false positive or a false negative SUVmax secondary to post-biopsy inflammatory changes or long time-lapse, respectively. Although concurrent imaging and molecular testing is ideal, the retrospective nature of this study precluded achieving concurrent imaging and molecular testing. Genomic profiling included 11 common DNA anomalies: (1) TP53, (2) DNA repair, (3) EGFR, (4) PI3K/AKT/MTOR (PAM) pathway including PTEN, PIK3CA, AKT, TSC, CCNB1, MTOR, FBXW2, and NF2, (5) MEK, (6) CYCLIN including CCND,CDK, CDKN, and RB, (7) WNT, (8) ALK, (9) MYC, (10) MET, and (11) FGF/FGFR. This study was performed in accordance with the guidelines of the UCSD Internal Review Board (PREDICT [Profile Related Evidence Determining Individualized Cancer Therapy], protocol; NCT02478931).
18F-FDG PET-CT imaging
All patients were asked to fast for at least six hours prior to their scan. Blood glucose levels were measured immediately before the FDG injection and no patient had a blood glucose level >160 mg/dl. Patients were intravenously injected with 370–740 MBq FDG, within a 5–10 second interval. Following an uptake period of approximately 1 hour in a quiet room at rest, a multi-station 3-dimensional (3D) whole body PET acquisition with CT, for attenuation correction, was performed for approximately 60 min, using a GE Discovery VCT scanner (GE, Waukesha, WI). Whole-body CT covers a region ranging from the head to the mid-thigh. PET images were acquired, after the CT scan, at a rate of 2 minutes/bed position, in the 3D acquisition mode. CT images were then reconstructed onto a 512 × 512 matrix. PET images were reconstructed using a standard whole-body 3D iterative reconstruction: 2 iterations; 28 subsets onto a 128 × 128 matrix with attenuation correction, decay correction, and scatter correction. The photon energy window was 425–650 keV. Slice thickness was 3.27 mm and reconstruction diameter was 70 cm. Pixel size was 5.47 mm × 5.47 mm with spatial resolution of 5 mm. 18F-FDG PET/CT images were generated for review on a workstation.
Image analysis
All PET images were reviewed and further analyzed on the institution’s pictures archiving and communication system (PACS), (AGFA Impax 6.3, Mortsel Belgium) by a board certified academic nuclear medicine physician. The lesions that were later biopsied were selected for imaging analysis. Focal activities of the biopsied lesions were manually identified on PET images. SUVs of the biopsied lesions were obtained by manually placing a circular region of interest (ROI) at the site of the maximum FDG uptake in the PET images and the maximal activity (SUVmax) was recorded. SUV was calculated as decay-corrected activity of tissue volume (kBq/mL)/injected FDG activity per body mass (kBq/g).
Genomic analysis
Genomic analysis was performed on the biopsy samples, using a clinical next generation sequencing (NGS) based assay (182 to 315 genes) (FoundationOne™, Foundation Medicine Inc., Cambridge, MA), to interrogate for DNA alterations including detection of base substitutions, insertions, deletions, copy number alterations, and selected gene fusions in 11 classes of genomic pathways: TP53, EGFR, ALK, MYC, MET, FGF/FGFR, DNA repair, PI3K/Akt/mTOR (PAM), MEK, CYCLIN, and WNT pathway genes were analyzed.
Statistical analysis
Statistical analysis was done in R, version 3.5.2 and Microsoft Excel. Differences between SUVmax in oncogene positive versus negative group patients were analyzed by the paired t-test (two-tailed) and considered to be significant at a p value less than 0.05. The Pearson correlation coefficient (r) was used in the linear relationship between the SUVmax and total number of oncogenic anomalies.
Ethical approval and consent to participate
This study was performed in accordance with the guidelines of the UCSD Internal Review Board (PREDICT [Profile Related Evidence Determining Individualized Cancer Therapy], protocol; NCT02478931).
Disclosure of potential conflicts of interest
Razelle Kurzrock receives research funding from Genentech, Incyte, Merck Serono, Pfizer, Sequenom, Foundation Medicine, Grifols, Konica Minolta, Omniseq and Guardant, as well as consultant fees from Loxo, X Biotech, NeoMed, and Actuate Therapeutics, speaker fees from Roche, and an equity interest in IDby DNA and Curematch Inc.
Additional information
Funding
References
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558.
- Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol. 2015;12:862–866. doi:10.1016/j.jacr.2015.04.019.
- Jansen RW, van Amstel P, Martens RM, Kooi IE, Wesseling P, de Langen AJ, Menke-van der Houven van Oordt CW, Jansen BHE, Moll AC, Dorsman JC, et al. Non-invasive tumor genotyping using radiogenomic biomarkers, a systematic review and oncology-wide pathway analysis. Oncotarget. 2018;9(28):20134–20155. doi:10.18632/oncotarget.24893.
- Dhingra VK, Mahajan A, Basu S. Emerging clinical applications of PET based molecular imaging in oncology: the promising future potential for evolving personalized cancer care. Indian J Radiol Imaging. 2015;25:332–341. doi:10.4103/0971-3026.169467.
- Lee JW, Lee SM. Radiomics in oncological PET/CT: clinical applications. Nucl Med Mol Imaging. 2018;52:170–189. doi:10.1007/s13139-017-0500-y.
- Suárez-Piñera M, Belda-Sanchis J, Taus A, Sánchez-Font A, Mestre-Fusco A, Jiménez M, Pijuan L. FDG PET-CT SUVmax and IASLC/ATS/ERS histologic classification: a new profile of lung adenocarcinoma with prognostic value. Am J Nucl Med Mol Imaging. 2018;8:100–109
- Heiden BT, Chen G, Hermann M, Brown RKJ, Orringer MB, Lin J, Chang AC, Carrott PW, Lynch WR, Zhao L, et al. 18F-FDG PET intensity correlates with a hypoxic gene signature and other oncogenic abnormalities in operable non-small cell lung cancer. PLoS One. 2018;13(7):e0199970. doi:10.1371/journal.pone.0199970.
- Nair VS, Gevaert O, Davidzon G, Napel S, Graves EE, Hoang CD, Shrager JB, Quon A, Rubin DL, Plevritis SK, et al. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res. 2012;72(15):3725–3734. doi:10.1158/0008-5472.CAN-11-3943.
- Caicedo C, Garcia-Velloso MJ, Lozano MD, Labiano T, Vigil Diaz C, Lopez-Picazo JM, Gurpide A, Zulueta J, Richter Echevarria JA, Perez Gracia JL, et al. Role of [1⁸F]FDG PET in prediction of KRAS and EGFR mutation status in patients with advanced non-small-cell lung cancer. Eur J Nucl Med Mol Imaging. 2014;41:2058–2065. doi:10.1007/s00259-014-2833-4.
- Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi:10.1186/s13073-017-0424-2.
- Ock CY, Hwang JE, Keam B, Kim SB, Shim JJ, Jang HJ, et al. Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nat Commun. 2017;8:1050.
- Ahn KS, Kang KJ, Kim YH, Kim TS, Song BI, Kim HW, O’Brien D, Roberts LR, Lee JW, Won KS, et al. Genetic features associated with 18F-FDG uptake in intrahepatic cholangiocarcinoma. Ann Surg Treat Res. 2019;96:153–161. doi:10.4174/astr.2019.96.4.153.
- Chang GH, Kurzrock R, Tran L, Schwaederle M, Hoh CK. mutations and number of alterations correlate with maximum standardized uptake value (SUVmax) determined by positron emission tomography/computed tomography (PET/CT). Oncotarget. 2018;9:14306–14310.
- Choi H, Na KJ. Pan-cancer analysis of tumor metabolic landscape associated with genomic alterations. Mol Cancer. 2018;17:150. doi:10.1186/s12943-018-0895-9.
- Ghasemi M, Nabipour I, Omrani A, Alipour Z, Assadi M. Precision medicine and molecular imaging: new targeted approaches toward cancer therapeutic and diagnosis. Am J Nucl Med Mol Imaging. 2016;6:310–327.
- Vaidya T, Agrawal A, Mahajan S, Thakur MH, Mahajan A. The continuing evolution of molecular functional imaging in clinical oncology: the road to precision medicine and radiogenomics (Part I). Mol Diagn Ther. 2019;23:1–26. doi:10.1007/s40291-018-0366-4.
- Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16:635–649. doi:10.1038/nrc.2016.77.
- Granja S, Pinheiro C, Reis RM, Martinho O, Baltazar F. Glucose addiction in cancer therapy: advances and drawbacks. Curr Drug Metab. 2015;16:221–242.
