ABSTRACT
Problem and aim
The overexpression of MAP17 has been reported in various human carcinomas. However, its molecular mechanism in non-small cell lung cancer (NSCLC) has not been fully understood. Our study aimed to reveal the molecular mechanism of NSCLC that involved MAP17 and identify its target miRNA.
Methods
RT-qPCR and immunoblot assays were conducted to measure the expression of mRNA and protein in NSCLC tissues and cell lines. Meanwhile, the A549 cells (an NSCLC cell line) were randomly assigned to the MAP17 overexpression group, the MAP17 knockdown group and negative control group to study the roles of MAP17 in cell viability, cell proliferation, migration, invasion, and apoptosis by performing Trypan blue exclusion, MTT, colony formation, transwell, wound healing and flow-cytometric apoptosis assays. The luciferase reporter assay was conducted to confirm the target relationship between MAP17 and miR-27a-3p.
Results
The upregulation of MAP17 mRNA and protein was observed in NSCLC tissues and cell lines. In vitro, the positive roles of MAP17 on cell viability, migration, and invasion were confirmed in A549 cells. It was also found that MAP17 could inhibit cell apoptosis by suppressing the activation of the p38 pathway. This research eventually proved the target relationship between MAP17 and miR-27a-3p, and that miR-27a-3p reversed the effects of MAP17 in A549 cells by directly targeting MAP17.
Conclusions
MAP17 plays an oncogenic role in NSCLC by suppressing the activation of the p38 pathway. Apart from that, the miR-27a-3p can inhibit the expression of MAP17 to suppress the NSCLC progression.
Introduction
The high incidence and mortality of lung carcinoma are the primary causes of cancer death.Citation1 Small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) are two main histologically distinct groups of lung carcinoma. NSCLC, as the primary type of lung carcinoma, can be divided into different subtypes; lung adenocarcinoma was the most prevalent subtype to be studied.Citation2–4 Because of the limitation of effective systemic treatments, most of the patients with NSCLC exhibited poor prognosis, and the 5-year survival rate of the patients remains approximately 18%.Citation5 Therefore, exploring the oncogenic molecular processes and metastatic pathways in NSCLC might help improve the therapy outcome of NSCLC.
MAP17 (PDZK1IP1) is a small non-glycosylated membrane-associated protein of 17 kDa and is primarily located on the plasma membrane and Golgi apparatus.Citation6 MAP17 contains a hydrophobic N-terminus encoding one PDZ-binding domain and two transmembrane regions.Citation7 In 1995, Kocher et al. for the first time found the overexpression of MAP17 in various human carcinomas such as breast cancer, colon cancer, and lung cancer.Citation8 The MAP17 research drew the attention of other scholars to explore this field, and many studies have proved that overexpression of MAP17 can increase proliferation, reduce apoptosis and promote migration in tumor cells.Citation9–14 Despite the frequent overexpression of MAP17 in human carcinomas, no report suggests a possible role of MAP17 in NSCLC except the study carried out by Irene et al. In their study, Irene et al. collected samples from different NSCLC patient cohorts and proved that MAP17 expression was upregulated in NSCLC, especially in lung adenocarcinoma.Citation3 Nonetheless, Irene et al. did not reveal the role of MAP17 in cancer cell phenotypes.
MicroRNAs (miRNAs) containing 18–25 nucleotides are endogenous and non-coding RNAs, that can bind to 3’-UTR of mRNAs to inhibit the target gene expression.Citation15 Many studies have proved that miRNAs are closely associated with the differentiation, morphogenesis, and carcinogenesis of tissues or cells.Citation16–19 Recently, miRNAs have been reported to act as oncogenes or tumor inhibitors in various human carcinomas to regulate cell proliferation, migration, and invasion.Citation20–24 For instance, the role of miR-27a in the progression of carcinomas has been reported. Researches have proved that miR-27a is a tumor promoter in breast cancer, gastric adenocarcinoma, hepatocellular carcinoma, pancreatic cancer and renal cell carcinoma.Citation25–28 As for miR-27a-3p, Chang et al. found that miR-27a-3p knockdown could inhibit the proliferation of HCT-116 cells, thereby suggesting that miR-27a-3p could contribute to the cell proliferation in colon cancer.Citation29 The similar results also occurred in gastric cancer.Citation30 However, miR-27a-3p may play a different role in lung carcinoma compared with other human carcinomas. The reason was that Yang et al. treated A549 cells (an NSCLC cell line) with genistein for two days; then, miR-27a was upregulated to show the anti-cancer effect.Citation31 Therefore, it is suspected that miR-27a-3p played a suppressing role in NSCLC progression.
In the current study, we first explored the potential effect of MAP17 on A549 cell line (an NSCLC cell line) phenotypes including cell proliferation, migration, invasion and apoptosis using in vitro experiments. We then confirmed the binding relationship between MAP17 and miR-27a-3p predicted by TargetScan Human 7.2 using the dual-luciferase reporter gene assay. Our findings in this study may help with the efficient development of therapy for NSCLC.
Materials and methods
NSCLC tissue specimens
Samples were collected in sterile containers and immediately frozen in liquid nitrogen, such as 20 pairs of cancerous and adjacent healthy tissues from NSCLC patients (12 male patients and 8 female patients; age range: 38–53 yrs) as well as 10 non-NSCLC lung tissues from patients with pulmonary tuberculosis (n = 4, age range: 45–55 yrs), idiopathic pulmonary fibrosis (n = 4, age range: 44–56 yrs) and empyema (n = 2, age range: 50 and 55 yrs). All patients were admitted to Jianghan University Affiliated Wuhan NO. 6 Hospital (China) and provided written informed content. All procedures were, most importantly, performed strictly according to the requirements of the Ethics Committee. Besides, before tumor resection, none of the patients had received any anticancer treatments such as chemotherapy or radiotherapy. The clinical characteristics of all participants are listed in .
Table 1. Clinical parameter of 20 patients with NSCLC and 10 non-NSCLC patients in this study
Cell culture
All the NSCLC cell lines (A549, Calu-3, H1975, H2228), and MRC-5 cell line (immortalized normal lung cell line) were purchased from ATCC (USA). MRC-5 cell line and Calu-3 cell line were cultured in DMEM media (Gibco, USA), A549 cell line was cultured in F-12 K medium (Gibco, USA), and H1975 and H2228 cells were cultured in RPMI 1640 media (Gibco, USA). All the media for cell growth was added with 10% fetal bovine serum (FBS, Gibco, USA), and 1% antibiotic-antimycotic solution (Gibco, USA). Cells were incubated at 37°C in a humidified incubator with 5% CO2.
Cell transfection
50 nM MAP17 small interfering RNA (siRNA), 25 nM miR-27a-3p mimic, and negative control RNA purchased from GenePharma Co., Ltd (Shanghai, China) were diluted using the serum-free medium (Gibco, USA), and transfected into cells using lipofectamine 2000 (Invitrogen, USA). The cells were transfected with the product mentioned above based on the manufacturer’s protocol after culturing the cells to 70% confluence. After six hours’ incubation in 5% CO2 at 37°C, the serum-free medium in each 60-mm dish was replaced by medium containing 10% FBS for another 24 h incubation.
qRT-PCR
Total RNA was isolated from tissue specimens and cells using TRNzol reagent (Cat#: DP405-02, TIANGEN, China). A NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, USA) was used to assess the quality of RNA isolation. RNA samples were reverse-transcribed into cDNAs using the SuperScript III Reverse Transcriptase (Thermo Fisher Scientific, USA) followed by qRT-PCR using the SuperScript IV One-Step RT-PCR System (Cat#:12594025, Applied Biosystems, USA). The MAP17 mRNA (forward primer: CGTCGGAAACAAGGCAGATG, reverse primer: TCACTGGACCTGAAACTGGC) expression was normalized to GAPDH. The comparative CT method (2−ΔΔCT) was employed for the calculation of relative mRNA expression.Citation32
Western blot
Cells or tissues were suspended in lysis buffer containing dithiothreitol, ethylenediamine tetraacetic acid, pH8.0 Tris-HCl and 2% sodium dodecyl sulfate (SDS) for 30 min in ice. By centrifuging the resulting lysates, the supernatant containing the total protein was collected. A 12% SDS-polyacrylamide gel was then used to electrophorese total proteins. The protein was blotted onto PVDF membranes (Membrane Solutions, China). Subsequently, the PVDF membranes were dipped in 5% defatted milk in TBST solution containing 10 mmol/L Tris–HCl (pH8.0) and 0.1% Tween 20 for 60 min. After washing the blocked PVDF membranes for four times, primary antibodies against MAP17 (Cat#: 199540, Abcam, UK), p38 (Cat#: ab170099, Abcam, UK), phosphor-p38 (Cat#: ab170099, Abcam, UK) and GAPDH (Cat#: ab9485, Abcam, UK) were respectively used to incubate with PVDF membranes at 4°C overnight. After washing with TBST for four times (8 min each time), the Goat Anti-Rabbit IgG (Cat#ab6721, Abcam, UK) was added to incubate with the PVDF membranes at room temperature for 90 min. Lastly, the membranes were washed for four times with TBST, and exposed to the Pierce ECL Western Blotting Substrate (Thermo Fischer, USA). The intensity was measured using ImageJ software.
Trypan blue exclusion assay
Trypan blue exclusion assay was conducted to assess cell viability by discriminating viable and non-viable cells. Viable cells cannot take up trypan blue dye whilst dead cells can. Briefly, A549 cells were seeded in a six-well plate at 2 × 105 cells per well. Then, 0.4% Trypan Blue Solution (Cat#15250061, Thermo Fischer Scientific) was used to treat cells for 3–5 min. Cell viability and total cell numbers were assessed at 0 h, 24 h, 48 h, and 72 h after transfection using a hemacytometer under a microscope at 400× magnification. The total cell number and blue staining cell number were counted. The number of non-viable cells was defined as the number of total cells-the number of blue staining cells.
Methylthiazole tetrazolium assay (MTT assay)
The viability capacity of A549 cells was also evaluated by MTT assay. First, the cells (5000 cells/well) were seeded into 96‑well plates, and transfected with MAP17 siRNA according to the protocol mentioned above, and continued to culture for 1-day, 2-day, 3-day, 4-day, and 5-day. Subsequently, 20 µl of 5 mg/ml MTT reagent (Sigma-Aldrich, USA) was added to each well. After another 4 h incubation, the optical density (OD) was assessed at 490 nm using a Varioskan Flash microplate reader (Thermo Fisher Scientific, USA).
Colony formation assay
A549 cells were transfected and harvested at the logarithmic phase. Then cells were digested by mild trypsinization (Sigma-Aldrich, USA) and plated in a 6-well plate for colony formation at approximately 1000 cells/well. The medium was replaced three times a week during the colony formation period. After incubation for two weeks, colonies were fixed for 30 min using 4% paraformaldehyde (Sigma-Aldrich). Subsequently, 0.1% crystal violet solution was used to stain the colonies for one hour, and the colonies were counted using an IX73 inverted microscope system (Olympus, Japan).
Transwell invasion assay
The A549 cells were divided into three groups to respectively transfect with MAP17 overexpression, miR-27a-3p mimic, MAP17 overexpression+miR-27a-3p, MAP17 knockdown, and NC vectors. The matrigel (BD Bioscience, USA) was diluted in RPMI-1640 medium in a volume ratio of 1:3 and added in the upper surface of a filter membrane in the upper chamber of transwell inserts (BD Bioscience, USA) for incubation at 37°C overnight. Then 2 × 105 cells in the logarithmic growth phase were added into the upper chamber of a transwell insert (Corning, USA) for incubation in medium without serum. At the same time, the bottom chamber was filled with the medium containing 10% FBS as the chemoattractant. After incubation for one day, the cells at the downside of the membrane were fixed in 4% formaldehyde for 20 min. The membranes were then immerged in PBS and 0.1% crystal violet to wash and stain the invading cells. The number of cells fixed to the lower surface of the membrane was photographed using the IX73 inverted microscope (Olympus, Japan). The average cell number in five random fields was calculated and represented the invading capability.
Wound healing assay
A549 cells were cultured in 6-well plates until the confluence was approximatively 100%. A wound of around 1-mm in width was created by scraping every cell layer using the tip of a 20 μL pipette. Subsequently, the cells were washed and incubated in a medium without serum for one day. The remaining cells were cultured in a serum-free medium for 24 h, and the migration of cells into the wound area was observed at 0 h and 24 h under the inverted microscope at 400× magnification. The migration rate was defined as the scratch width at 24 h divided by scratch width at 0 h.
Flow cytometric apoptosis assay
The transfected cells were fixed with 70% ice-cold ethanol (Sigma, USA), and stained with a total of 1.25 μL Annexin V-FITC and 3 μL propidium iodide (eBiosciences, USA) for 20 min in the dark. The apoptotic cells were examined within 60 min using the cytometer FC500 flow cytometer (BD Biosciences). The percentage of the right panels was defined as the apoptosis rate in this study.
Luciferase reporter assay
Wild-type 3′UTR of MAP17 mRNA that contains the binding site of miR-27a-3p was amplified by PCR from A549 cell genomic DNA and inserted into a pGL3-control vector (Promega, USA). The following PCR primers were used: Forward: TGCCTATGAGAATGTGCCCG; Reverse: GACATCCATCCCATGTGCCT. A site-directed mutagenesis kit (Agilent Technologies, USA) was employed to generate the mutant-type 3′UTR of MAP17 mRNA by overlap-extension PCR method. These constructed vectors, namely pGL3-MAP17-Wt or pGL3-MGMT-Mut, was used to transfect the A549 cells using the lipofectamine 2000 reagent (Invitrogen, USA). A549 cells were seeded into 24-well plates for one-day incubation. Subsequently, the cells were co-transfected with the pGL3-MAP17-Wt or pGL-MAP17-Mut, the Renilla vector (pGL4.74), and miR-27a-3p mimic (0.4 μg) or NC (0.4 μg). After 48 h culture, the luciferase activity was measured using a luminometer (Beckman, USA).
Statistical analysis
All the data from at least three individual experiments in cell lines were represented as mean ± standard deviation (SD) and analyzed using One-way ANOVA or two-way ANOVA with Dunnett’s multiple comparisons test. Data from tissue specimen were analyzed using the non-parametric Kruskal-Wallis ANOVA test method. P< .05 represents a significant difference. GEPIA 2 was used to determine the expression level of MAP17 in lung squamous carcinoma (LUSC) and lung adenocarcinoma (LUAD) and its association with the stage of LUSC and LUAD.
Result
MAP17 was overexpressed in NSCLC tissues and cells
We used RT-qPCR and western blotting assays to respectively detect the expression of MAP17 mRNA and protein in NSCLC tissues, adjacent healthy tissues and non-NSCLC lung tissues. The result indicated that MAP17 was significantly upregulated in carcinomas compared with adjacent healthy lung tissues (P< .05, ). The western blot assay results showed that MAP17 protein expression was significantly upregulated in tumor tissues compared with adjacent healthy tissues (P= .0074, ). Also, the GEPIA 2 algorithm (http://gepia2.cancer-pku.cn/#index) showed that MAP17 was significantly associated with the cancer stage in LUSC: MAP17 expression was the highest at stage IV () and MAP17 was significantly upregulated in LUAD (). It was even observed that the isoform MAP17-001 was the most abundant isoform in both LUAD and LUSC (). The expression of MAP17 mRNA was higher in NSCLC cell lines (A549, Calu-3, H1975, and H2228), especially in A549 cell line than in normal human lung cell line MRC-5 (P< .05, ). MAP17 protein was significantly up-regulated in NSCLC cell lines (A549, Calu-3, H1975, and H2228) (). Accordingly, our data demonstrated that the upregulation of MAP17 was observed in NSCLC tissues and cells, especially in the A549 cell line. For this reason, the A549 cell line was chosen in the following experiments in vitro.
Figure 1. The expression of MAP17 was upregulated in NSCLC tissues. (a) The relative expression of MAP17 mRNA was examined in tumor specimens (n = 20), adjacent specimens (n = 20) and normal lung specimens (n = 10) using the RT-qPCR assay method. Every bar represents mean±SD in triplicate. (b) The expression of MAP17 protein was analyzed in tumor specimens (n = 20), adjacent specimens (n = 20) and normal lung specimens (n = 10) after performing a western blot assay. Every bar represents mean±SD in triplicate. (c) The stage plot indicates the association between MAP17 expression and LUSC (lung squamous carcinoma) and LUAD (lung adenocarcinoma) produced by the GEPIA 2 algorithm. (d) The box plot shows the expression of MAP17 in LUAD and LUSC produced by the GEPIA 2 algorithm. (e) The abundance of three isoforms of MAP17 in LUAD and LUSC was MAP17-001, according to the GEPIA 2 algorithm
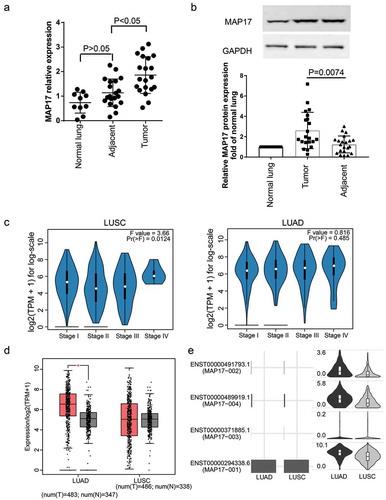
Figure 2. The expression of MAP17 was upregulated in NSCLC cell lines. (a) The relative expression of MAP17 was assessed by RT-qPCR in four NSCLC cell lines (A549, Calu-3, H1975, and H2228), including one normal lung cell line (MRC-5). (b) MAP17 protein expression was verified using the western blot assay kit in four NSCLC cell lines (A549, Calu-3, H1975, and H2228), including one normal lung cell line (MRC-5). Every bar represents the mean±SD of three repetitive experiments. The P-value was calculated using one-way ANOVA (*P< .05, **P< .001, compared with the normal lung cell line MRC-5)
MAP17 promoted viability and inhibited apoptosis of the A549 cell line in vitro
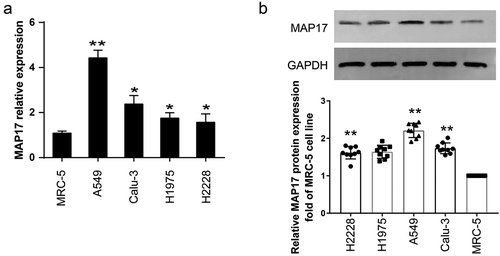
To evaluate the influence of MAP17 on NSCLC cell viability and apoptosis phenotypes, we performed Trypan blue exclusion, MTT, colony formation, and flow-cytometric apoptosis assays on the A549 cell line with enforced MAP17 overexpression (OE) and MAP17 knockdown (KD). The Trypan blue exclusion assay results demonstrated that the percentage of cell death in the MAP17 KD group was significantly higher than that in the NC group at 72 h. However, it was found that the percentage of cell death in the MAP17 OE group was significantly lower than that in the NC group at 48 h and 72 h (P< .05, ). Similarly, on the 4th and 5th day of the MTT assay, it was observed that cell viability in the MAP17 OE group was obviously enhanced (P< .05), whereas the cell viability in the MAP17 KD group was significantly reduced (P< .05) in A549 cells compared with the NC group (). Also, the colony formation assay results showed that A549 cells transfected with MAP17 OE vectors formed more cell colonies, and those transfected with MAP17 KD vectors formed fewer cell colonies compared with the NC group: statistical analysis indicated that the MAP17 OE promoted the colony formation of A549 cells by 70%. On the contrary, the MAP17 KD suppressed the colony formation of A549 cells by 46% (P< .05, ). Besides, we measured the apoptosis of A549 cells by performing a flow cytometry assay. As presented in , MAP17 OE significantly decreased the apoptosis rate (by 16%, P< .05), whereas the apoptosis rate of the MAP17 KD group was almost 3-fold of that of the NC group. Our data strongly proved that MAP17 played an oncogenic role in A549 cells.
Figure 3. MAP17 enhanced A549 cell viability and suppressed cell apoptosis. (a) A trypan blue exclusion assay was carried out to detect the cell viability induced by MAP17 overexpression (OE) or MAP17 knockdown (KD). (b) An MTT assay was employed to evaluate the viability ability in A549 cells transfected with NC, MAP17 OE, or KD plasmids. The OD value was detected every day at 490 nm for five days. (c) A colony formation assay kit was used to measure the colony-formation ability in the A549 cells transfected. (d) The apoptosis rate of A549 cells transfected with NC, MAP17 OE or KD plasmids was assessed using flow cytometry. The error bars represent mean±SD from three independent experiments. The P-value was calculated with one-way ANOVA (*P< .05, **P< .001, compared with the NC group)
MAP17 promoted invasion and migration of A549 cell line in vitro
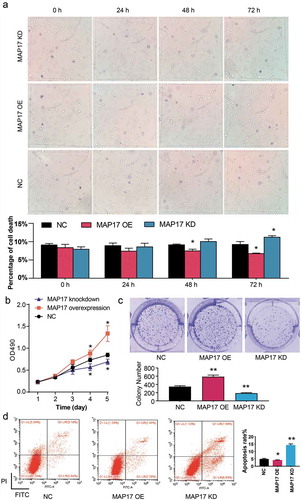
To detect the effect of MAP17 on A549 cell mobility phenotypes, transwell invasion and wound healing assays were conducted. Transwell invasion assay results showed that the invading cell number of the MAP17 OE group was 1.33-fold of that of the NC group. Nonetheless, MAP17 knockdown resulted in a 41% decrease in the invading cell number (P< .05, ). The migratory rate increased by 45% in the MAP17 OE group and decreased by 40% in the MAP17 KD group compared with the NC group (P< .05, ). Our data further strengthened that MAP17 played an oncogenic role in A549 cells.
Figure 4. MAP17 enhanced A549 cell invasion and migration. (a) The invasion ability of A549 cells with MAP17 overexpression (OE) or knockdown (KD) was measured using the transwell assay. (b) A wound healing assay was conducted to assess the migration ability of A549 cells with MAP17 OE or KD. The migration rate was defined as the ratio of migration distance at 24 h to cell spacing at 0 h. MAP17 OE represented MAP17 overexpression, while MAP17 KD represented MAP17 knockdown. The error bars represent mean±SD from three independent experiments. The P-value was calculated with one-way ANOVA (*P< .05, compared with NC group)
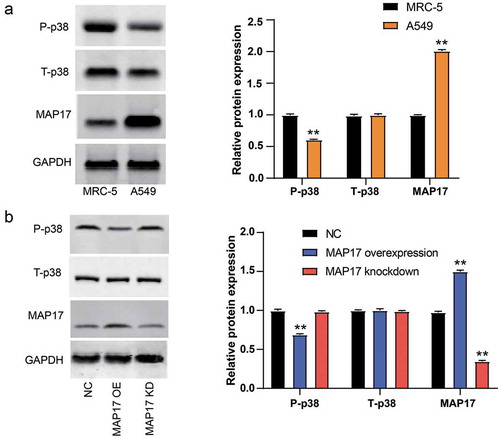
MAP17 suppressed phospho-p38 protein expression in A549 cells
To detect the p38 signaling pathway activation in NSCLC, the western blotting experiment was conducted to measure the expression of relevant proteins. As shown in , we found that the protein level of phospho-p38 was lower (P< .001), whereas the protein level of MAP17 was higher (P< .001) in A549 cell line compared with the MRC-5 cell line. Subsequently, the effect of forced MAP17 overexpression and knockdown in A549 cells on the protein level of phospho-p38 was detected. The results displayed that the protein level of phospho-p38 was reduced in the MAP17 overexpression group and increased in the MAP17 knockdown group (P< .05, ). Interestingly, the total p38 did not significantly differ among the groups. These results indicated that MAP17 might promote NSCLC by inhibiting p38 phosphorylation.
Figure 5. MAP17 upregulation reduced the expression of phospho-p38 protein in A549 cell line. (a) MAP17, total-p38, and phospho-p38 protein expression in MRC-5 cell line (a normal lung cell line) and A549 cell line. (b) A549 cells with MAP17 OE and KD, respectively, showed a lower level and a higher level of phospho-p38 protein. MAP17 OE and MAP17 KD represented MAP17 and MAP17 knockdown, respectively. All the bars represent mean±SD from three independent experiments. The P value was calculated with one-way ANOVA (**P< .001, compared with the control group: MARC5 in (a) and NC group in (b))
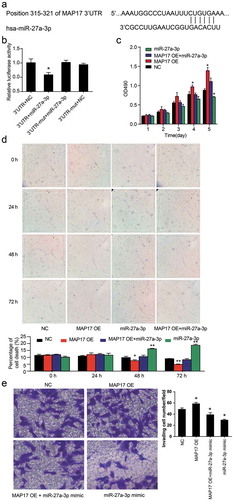
miR-27a-3p directly targeted MAP17 in A549 cells
The TargetScan Human 7.2 algorithm (http://targetscan.org/vert_72/) was used to predict the binding site of miR-27a-3p on MAP17 mRNA 3’UTR. As shown in , the MAP17 mRNA 3’UTR has a conserved sequence in the position 315–321 that could bind to miR-27a-3p. The dual-luciferase reporter assay results displayed that miR-27a-3p mimic transfection caused a 40% decrease in the relative luciferase activity in the wild-type MAP17 mRNA 3’UTR group (P< .05, ). Next, MTT, trypan blue exclusion, and transwell assays were conducted to further research how MAP17 affected A549 cells by binding with miR-27a-3p. MTT assay results showed that cell viability was significantly inhibited by the gain of function of miR-27a-3p. However, the inhibited cell viability was recovered by co-transfection with MAP17 OE vectors (P< .05, ). In the trypan blue exclusion assay, miR-27a-3p upregulation significantly enhanced the cell death at 48 h and 72 h, and it compromised the decreased apoptosis caused by MAP17 overexpression (P< .05, ). The transwell invasion assay also confirmed that decreased cell invasion caused by overexpressed miR-27a-3p was reversed by MAP17 OE co-transfection (P< .05, ). These results displayed that miR-27a-3p could suppress the influence of MAP17 on A549 cell phenotypes by targeting the 3’ UTR of MAP17 mRNA.
Figure 6. miR-27a-3p suppressed the viability and invasion of the A549 cell line by targeting MAP17. (a) This figure illustrates the binding scheme between MAP17 mRNA 3’UTR and miR-27a-3p predicted by the TargetScan Human 7.2 database. (b) A luciferase reporter gene assay was performed using the A549 cell line to validate the target relationship between miR-27a-3p and MAP17. 3’UTR. Wild-type MAP17 mRNA 3’UTR was constructed to fit the pGL3 vector. NC; miR-27a-3p mimicked the negative control. 3’UTR-mut: mutated MAP17 mRNA 3’UTR was constructed to fit the pGL3 vector. The Renilla luciferase plasmids (pGL4.74) were also transfected as the internal control,*P< .05, compared with the 3’UTR+NC group. (c) MTT assay results showed that miR-27a-3p fully decreased the promotive effect of MAP17 on the viability of A549 cells at days 4 and 5. The cells were transfected with NC, miR-27a-3p mimic, MAP17 OE plasmids, and MAP17 OE plasmids+miR-27a-3p mimic. The OD value was detected at 490 nm and represented as mean±SD from three independent experiments. (d) Trypan blue exclusion assay results also showed that miR-27a-3p alleviated the effect of MAP17 overexpression on the cell viability of A549 cells. (e) Transwell invasion assay results showed that miR-27a-3p inhibited the promotive effect of MAP17 on the invasion of A549 cells. Every bar represents the mean±SD from three independent experiments. The P-value was calculated with one-way ANOVA (*P< .05, **P< .01, compared with NC group)
Discussion
The effects of genes’ aberrant expressions on NSCLC have dual characteristics,Citation33,Citation34 and the signaling gene-induced lung cancer is usually complicated.Citation35–37 Therefore, identifying the impact and molecular mechanism of the MAP17 gene in NSCLC is essential to improve the therapeutic strategies for NSCLC. Our study proved that the MAP17 overexpression exhibited a promotive effect on cell viability, proliferation, invasion, and migration in A549 cells by suppressing the p38 signaling pathway. Besides, we proved that the promotive effects of MAP17 on A549 cells could be compromised by miR-27a-3p.
The overexpression of MAP17, a small 17 kDa non-glycosylated membrane protein, has been reported to be related to human carcinomas.Citation9,Citation10,Citation38,Citation39 In our study, we found that both the mRNA level and the protein level of MAP17 in NSCLC tissues and cells was upregulated compared to adjacent healthy lung tissues and cells. Hence, we believe that our findings may extend the relationship between MAP17 and cancers to NSCLC. MAP17 can enhance the abilities of proliferation, migration, and invasion in various human carcinomas.Citation14,Citation38,Citation40 Only a few studies have investigated the effect of MAP17 on cell viability in NSCLC. However, in our study, we found that forced MAP17 expression in A549 cells not only significantly enhanced the abilities of cell viability, but also promoted proliferation, invasion and migration. The results proved that MAP17 played an oncogenic role in NSCLC.
We also found that the protein expression of phosphor-p38 was significantly reduced in A549 cells with the MAP17 upregulation. The tumor-suppressing role of the p38 pathway has been identified in human cancer cells.Citation41–43 The activation of p38 could limit the tumorigenic capability of MAP17 in the breast cancer cells with the high intracellular ROS level.Citation14 In A549 cells, Xia et al.Citation44 found that increased phospho-p38 levels inactivated the p38 MAPK pathway to promote invasion and migration. The p38 signaling pathway in different cancer cells exhibited different functions. In our study, we found that the protein expression of phospho-p38 but not total p38 was significantly downregulated in A549 cells with MAP17 overexpression. Our data suggested that the MAP17 overexpression could inactivate the p38 signaling pathway to regulate the cell phenotypes of A549 cells.
Widely exist in human, miRNAs have strong organization specificities. In recent years, a growing body of researches have reported that miRNAs as underlying gene expression regulators are associated with the pathogenesis of human carcinomas.Citation45–48 For instance, after using various concentrations of genistein to treat A549 cells to confirm the anti-cancer effect of genistein, Yang et al.Citation31 found that the genistein suppressed the proliferation of A549 cells through the upregulation of miR-27a. The downregulation of miR-27a was also detected in the tissue specimens from the NSCLC patients.Citation49 In this study, miR-27a-3p was validated to be a regulator of MAP17 mRNA, and it was shown to suppress A549 cell phenotypes including viability, proliferation and mobility. In addition, miR-27a-3p compromised the cancer-promotive effects of MAP17 in A549 cells. These findings proved that miR-27a-3p could inhibit the MAP17 expression thus suppressing the NSCLC malignancy.
Conclusion
In conclusion, the binding relationship between miR-27a-3p and the 3’ UTR of MAP17 mRNA was proved. The upregulation of MAP17 in A549 cells could promote cell viability by inhibiting the activation of the p38 pathway. miR-27a-3p could suppress NSCLC malignancy by suppressing MAP17. Our findings reveals the molecular mechanism of MAP17 and miR-27a-3p in NSCLC, although animal model experiments are needed to generalize our findings.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. doi:10.3322/caac.21492.
- Yu N, Yong S, Kim HK, Choi YL, Jung Y, Kim D, Seo J, Lee YE, Baek D, Lee J, et al. 2019. Identification of tumor suppressor miRNAs by integrative miRNA and mRNA sequencing of matched tumor-normal samples in lung adenocarcinoma. Mol Oncol. 13:1356–1368. doi:10.1002/1878-0261.12478.
- Ferrer I, Quintanal-Villalonga A, Molina-Pinelo S, Garcia-Heredia JM, Perez M, Suarez R, Ponce-Aix S, Paz-Ares L, Carnero A. 2018. MAP17 predicts sensitivity to platinum-based therapy, EGFR inhibitors and the proteasome inhibitor bortezomib in lung adenocarcinoma. J Exp Clin Cancer Res. 37:195. doi:10.1186/s13046-018-0871-7.
- Chen S, Wang Q, Zhou XM, Zhu JP, Li T, Huang M. 2016. MicroRNA-27b reverses docetaxel resistance of non-small cell lung carcinoma cells via targeting epithelial growth factor receptor. Mol Med Rep. 14:949–954. doi:10.3892/mmr.2016.5332.
- Siegel RL, Miller KD, Jemal A. 2016. Cancer statistics, 2016. CA Cancer J Clin. 66:7–30. doi:10.3322/caac.21332.
- Guijarro MV, Link W, Rosado A, Leal JF, Carnero A. 2007. MAP17 inhibits Myc-induced apoptosis through PI3K/AKT pathway activation. Carcinogenesis. 28:2443–2450. doi:10.1093/carcin/bgm154.
- Jaeger C, Schaefer BM, Wallich R, Kramer MD. 2000. The membrane-associated protein pKe#192/MAP17 in human keratinocytes. J Invest Dermatol. 115:375–380. doi:10.1046/j.1523-1747.2000.00071.x.
- Kocher O, Cheresh P, Brown LF, Lee SW. Identification of a novel gene, selectively up-regulated in human carcinomas, using the differential display technique. Clin Cancer Res. 1995;1:1209–1215.
- Garcia-Heredia JM, Jekyll CA, Hyde M. 2018. MAP17’s up-regulation, a crosspoint in cancer and inflammatory diseases. Mol Cancer. 17:80. doi:10.1186/s12943-018-0828-7.
- de Miguel-luken MJ, Chaves-Conde M, de Miguel-luken V, Munoz-Galvan S, Lopez-Guerra JL, Mateos JC, Pachon J, Chinchon D, Suarez V, Carnero A. 2015. MAP17 (PDZKIP1) as a novel prognostic biomarker for laryngeal cancer. Oncotarget. 6:12625–12636. doi:10.18632/oncotarget.3470.
- Guijarro MV, Leal JF, Blanco-Aparicio C, Alonso S, Fominaya J, Lleonart M, Castellvi J, Ramon Y Cajal S, Carnero A. 2007. MAP17 enhances the malignant behavior of tumor cells through ROS increase. Carcinogenesis. 28:2096–2104. doi:10.1093/carcin/bgm124.
- Shao Y, Lv H, Zhong DS, Zhou QH. 2018. EGFR-TKI resistance and MAP17 are associated with cancer stem cell like properties. Oncol Lett. 15:6655–6665. doi:10.3892/ol.2018.8129.
- Perez M, Peinado-Serrano J, Garcia-Heredia JM, Felipe-Abrio I, Tous C, Ferrer I, Martin-Broto J, Saez C, Carnero A. 2016. Efficacy of bortezomib in sarcomas with high levels of MAP17 (PDZK1IP1). Oncotarget. 7:67033–67046. doi:10.18632/oncotarget.11475.
- Guijarro MV, Vergel M, Marin JJ, Munoz-Galvan S, Ferrer I, Ramon Y Cajal S, Roncador G, Blanco-Aparicio C, Carnero A. 2012. p38alpha limits the contribution of MAP17 to cancer progression in breast tumors. Oncogene. 31:4447–4459. doi:10.1038/onc.2011.619.
- Ambros V. 2004. The functions of animal microRNAs. Nature. 431:350–355. doi:10.1038/nature02871.
- Moghaddam T, Neshati Z. 2019. Role of microRNAs in osteogenesis of stem cells. J Cell Biochem. 120:14136–14155. doi:10.1002/jcb.28689.
- Li L, Li Y, Tang C. 2019. The role of microRNAs in the involvement of vascular smooth muscle cells in the development of atherosclerosis. Cell Biol Int. 43:1102–1112. doi:10.1002/cbin.11164.
- Wu F, Yang Z, Li G. 2009. Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys Res Commun. 386:549–553. doi:10.1016/j.bbrc.2009.06.075.
- Skaftnesmo KO, Prestegarden L, Micklem DR, Lorens JB. MicroRNAs in tumorigenesis. Curr Pharm Biotechnol. 2007;8:320–325. doi:10.2174/138920107783018390.
- Liu ZM, Wu ZY, Li WH, Wang LQ, Wan JN, Zhong Y. 2019. MiR-96-5p promotes the proliferation, invasion and metastasis of papillary thyroid carcinoma through down-regulating CCDC67. Eur Rev Med Pharmacol Sci. 23:3421–3430. doi:10.26355/eurrev_201904_17706.
- Bai TL, Liu YB, Li BH. 2019. MiR-411 inhibits gastric cancer proliferation and migration through targeting SETD6. Eur Rev Med Pharmacol Sci. 23:3344–3350. doi:10.26355/eurrev_201904_17697.
- Xu Q, Cheng L, Chen J, Lu W, Wang P. 2019. miR-376a inhibits the proliferation and invasion of osteosarcoma by targeting FBXO11. Hum Cell. 32:390–396. doi:10.1007/s13577-019-00256-2.
- Zhang J, Zhu Y, Hu L, Yan F, Chen J. 2019. miR-494 induces EndMT and promotes the development of HCC (Hepatocellular carcinoma) by targeting SIRT3/TGF-beta/SMAD signaling pathway. Sci Rep. 9:7213. doi:10.1038/s41598-019-43731-4.
- He S, Wang Z, Tang H, Dong J, Qu Y, Lv J. 2019. MiR-217 inhibits proliferation, migration, and invasion by targeting SIRT1 in osteosarcoma. Cancer Biother Radiopharm. 34:264–270. doi:10.1089/cbr.2017.2394.
- Peng H, Wang X, Zhang P, Sun T, Ren X, Xia Z. miR-27a promotes cell proliferation and metastasis in renal cell carcinoma. Int J Clin Exp Pathol. 2015;8:2259–2266.
- Guttilla IK, White BA. 2009. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 284:23204–23216. doi:10.1074/jbc.M109.031427.
- Huang S, He X, Ding J, Liang L, Zhao Y, Zhang Z, Yao X, Pan Z, Zhang P, Li J, et al. 2008. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 123:972–978. doi:10.1002/ijc.23580.
- Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. 2007. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 67:11001–11011. doi:10.1158/0008-5472.CAN-07-2416.
- Su C, Huang DP, Liu JW, Liu WY, Cao YO. 2019. miR-27a-3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting BTG1. Oncol Lett. 18:2825–2834. doi:10.3892/ol.2019.10629.
- Zhou L, Liang X, Zhang L, Yang L, Nagao N, Wu H, Liu C, Lin S, Cai G, Liu J. 2016. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget. 7:51943–51954. doi:10.18632/oncotarget.10460.
- Yang Y, Zang A, Jia Y, Shang Y, Zhang Z, Ge K, Zhang J, Fan W, Wang B. 2016. Genistein inhibits A549 human lung cancer cell proliferation via miR-27a and MET signaling. Oncol Lett. 12:2189–2193. doi:10.3892/ol.2016.4817.
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. doi:10.1006/meth.2001.1262.
- Yoon D, Bae K, Kim JH, Choi YK, Yoon KA. Oncogenic effect of the novel fusion gene VAPA-Rab31 in lung adenocarcinoma. Int J Mol Sci. 2019;20. doi:10.3390/ijms20092309.
- Tian H, Zhang Y, Zhang Q, Li S, Liu Y, Han X. 2019. Effects of BENC-511, a novel PI3K inhibitor, on the proliferation and apoptosis of A549 human lung adenocarcinoma cells. Biosci Trends. 13:40–48. doi:10.5582/bst.2019.01006.
- Luo J, Yao Y, Ji S, Sun Q, Xu Y, Liu K, Diao Q, Qiang Y, Shen Y. 2019. PITX2 enhances progression of lung adenocarcinoma by transcriptionally regulating WNT3A and activating Wnt/beta-catenin signaling pathway. Cancer Cell Int. 19:96. doi:10.1186/s12935-019-0800-7.
- Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, Chen PH, Boroughs LK, Rodriguez MLM, Zhang W, et al. 2019. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 79:3251–3267. doi:10.1158/0008-5472.CAN-18-3527.
- Luo J, Liu Z. 2019. Long non-coding RNA TTN-AS1 promotes the progression of lung adenocarcinoma by regulating PTEN/PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 514:140–147. doi:10.1016/j.bbrc.2019.04.050.
- Chen X, Liao Y, Yu Y, Zhu P, Li J, Qin L, Liao W, Huang Z. 2017. Elevation of MAP17 enhances the malignant behavior of cells via the Akt/mTOR pathway in hepatocellular carcinoma. Oncotarget. 8:92589–92603. doi:10.18632/oncotarget.21506.
- Perez M, Praena-Fernandez JM, Felipe-Abrio B, Lopez-Garcia MA, Lucena-Cacace A, Garcia A, Lleonart M, Roncador G, Marin JJ, Carnero A. 2013. MAP17 and SGLT1 protein expression levels as prognostic markers for cervical tumor patient survival. PLoS One. 8:e56169. doi:10.1371/journal.pone.0056169.
- Guijarro MV, Leal JF, Fominaya J, Blanco-Aparicio C, Alonso S, Lleonart M, Castellvi J, Ruiz L, Ramon YCS, Carnero A. 2007. MAP17 overexpression is a common characteristic of carcinomas. Carcinogenesis. 28:1646–1652. doi:10.1093/carcin/bgm083.
- Marzi L, Combes E, Vie N, Ayrolles-Torro A, Tosi D, Desigaud D, Perez-Gracia E, Larbouret C, Montagut C, Iglesias M, et al. 2016. FOXO3a and the MAPK p38 are activated by cetuximab to induce cell death and inhibit cell proliferation and their expression predicts cetuximab efficacy in colorectal cancer. Br J Cancer. 115:1223–1233. doi:10.1038/bjc.2016.313.
- Hong B, Li H, Zhang M, Xu J, Lu Y, Zheng Y, Qian J, Chang JT, Yang J, Yi Q. 2015. p38 MAPK inhibits breast cancer metastasis through regulation of stromal expansion. Int J Cancer. 136:34–43. doi:10.1002/ijc.28958.
- Bulavin DV, Fornace AJ Jr. 2004. p38 MAP kinase’s emerging role as a tumor suppressor. Adv Cancer Res. 92:95–118. doi:10.1016/S0065-230X(04)92005-2.
- Xia ZX, Li ZX, Zhang M, Sun LM, Zhang QF, Qiu XS. 2016. CARMA3 regulates the invasion, migration, and apoptosis of non-small cell lung cancer cells by activating NF-small ka, CyrillicB and suppressing the P38 MAPK signaling pathway. Exp Mol Pathol. 100:353–360. doi:10.1016/j.yexmp.2015.10.004.
- Xu Q, Tong JL, Zhang CP, Xiao Q, Lin XL, Xiao XY. 2017. miR-27a induced by colon cancer cells in HLECs promotes lymphangiogenesis by targeting SMAD4. PLoS One. 12:e0186718. doi:10.1371/journal.pone.0186718.
- Ow SH, Chua PJ, Bay BH. 2018. miR-149 as a potential molecular target for cancer. Curr Med Chem. 25:1046–1054. doi:10.2174/0929867324666170718102738.
- Zhang HD, Jiang LH, Sun DW, Li J, Ji ZL. 2017. The role of miR-130a in cancer. Breast Cancer. 24:521–527. doi:10.1007/s12282-017-0776-x.
- Danza K, Silvestris N, Simone G, Signorile M, Saragoni L, Brunetti O, Monti M, Mazzotta A, De Summa S, Mangia A, et al. 2016. Role of miR-27a, miR-181a and miR-20b in gastric cancer hypoxia-induced chemoresistance. Cancer Biol Ther. 17:400–406. doi:10.1080/15384047.2016.1139244.
- Li J, Wang Y, Song Y, Fu Z, Yu W. 2014. miR-27a regulates cisplatin resistance and metastasis by targeting RKIP in human lung adenocarcinoma cells. Mol Cancer. 13:193. doi:10.1186/1476-4598-13-193.
