ABSTRACT
The aim of the present study was to identify the potential risk of circulating-HPV-DNA in non-small cell lung cancer (NSCLC) and to analyze abnormally expressed miRNAs in circulating HPV-DNA-positive NSCLC. HPV universal primers were used to detect the presence of HPV-DNA in the peripheral blood of 100 patients with NSCLC. The relationship between circulating-HPV-DNA and NSCLC patients characteristics was analyzed. Then, eight differentially expressed miRNAs in NSCLC were screened based on the TCGA database. The levels of miRNAs in circulating HPV-DNA-positive NSCLC patients were detected by real-time quantitative PCR. ROC curves were generated to evaluate the diagnostic performance. Circulating-HPV-DNA was found in 16 patients. The proportion of HPV-DNA-positive patients with poorly differentiated NSCLC, advanced lung cancer and lymph node metastasis was higher than that of HPV-DNA-negative patients. The levels of miR-183, miR-210 and miR-182 were significantly higher and miR-144 was significantly lower in HPV-DNA-positive NSCLC than those in HPV-DNA-negative NSCLC patients. When using a single miRNA to identify circulating HPV-DNA-positive NSCLC patients, miR-210 had a higher area under the ROC curve (AUC) than other miRNAs, and its sensitivity and specificity were also higher. In addition, the combination of two miRNAs was more effective than a single miRNA. Among them, miR-210+ miR-144 had the highest AUC value and showed the best prediction performance. Circulating-HPV-DNA may serve as a risk factor in NSCLC. Plasma miR-183, miR-210, miR-182 and miR-144 can be used as reliable biomarkers to identify circulating HPV-DNA-positive NSCLC.
Introduction
Non-small cell lung cancer (NSCLC) has become one of the most common deadly cancers in the world, with a 5-year survival rate of less than 15%.Citation1 Among all carcinogenic factors, tobacco exposure has been widely recognized as the most relevant causative factor.Citation2 However, there are still many nonsmoking lung cancer patients, especially women.Citation3 Numerous studies have reported that human papillomavirus (HPV) infection is also an important pathogenic factor in lung cancer,Citation4 especially in South Korea, Japan and Taiwan, but the Mainland China is still lacking such studies.
HPVs are a group of double-stranded closed-loop small DNA viruses. More than 200 genotypes have been isolated and identified. According to their pathogenic ability, they are divided into two groups: high-risk HPVs, including HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73 and 82, which mainly cause malignant lesions, and low-risk HPVs, including HPV6, 11, 40, 42, 43, 44, 45, 54, 61, 72, and 81, which are often associated with benign lesions.Citation5 Previous studies have suggested that tissue HPV-DNA is closely related to the occurrence and development of both reproductive and non-reproductive system tumors, such as cervical cancer, lung cancer,Citation6 breast cancer,Citation7 head and neck squamous cell carcinoma,Citation8 etc. Recent studies showed that HPV-DNA also exists in the blood circulation. And circulating HPV-DNA is a new predictor to the severity and recurrence of various cancers.Citation9,Citation10 However, the relationship between circulating HPV-DNA and NSCLC is still unclear.
MicroRNAs (miRNAs) are small non-coding RNAs of approximately 22 nucleotides in length that play a key role in a variety of biological events.Citation11 MiRNAs can interact with the 3ʹ untranslated region of the target mRNA to perform complete or incomplete base complementation, thereby negatively regulating the expression of the target mRNA at the post-transcriptional level.Citation12 Numerous studies have shown that miRNAs are involved in tumor development, progression and metastases. The expression of different miRNAs can be upregulated or downregulated, which can play a role in promoting tumors or suppressing tumors.Citation13 Numerous studies have reported that plasma-circulating miRNAs can reflect the biological characteristics of tumors and can be used as potential predictors in tumor diagnosis, prognosis and response to treatment.Citation14 Previous research has shown that there are abnormally expressed miRNAs in HPV-associated tumors.Citation15,Citation16 Therefore, we speculated that plasma miRNAs may have detection value in circulating HPV-DNA-positive NSCLC.
In this study, we evaluated the detected rate of circulating HPV-DNA in patients with NSCLC and verified it in an independent cohort. Then, the TCGA database was used to screen differentially expressed miRNAs in lung adenocarcinoma and squamous cell carcinoma. Plasma miRNAs of circulating HPV-DNA-positive/negative NSCLC patients were further detected to explore their potential diagnostic value. Overall, we hope to provide new insight for the early detection of circulating HPV-DNA-positive NSCLC and novel ideas for the pathogenesis studies of NSCLC.
Results
Detection of the presence of circulating HPV-DNA in patients with NSCLC
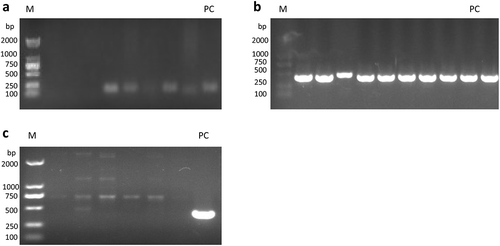
Among 100 NSCLC patients, HPV-DNA was detected in 16 blood samples with the HPV universal primers. The circulating HPV-DNA detection rate was 16% ()). Further detection with HPV16- and HPV18-specific nested-PCR primers revealed that all positive samples were HPV16 ()), and HPV18 was not detected ()). The gene sequencing results were consistent with the PCR results. 32 patients were enrolled in the independent validation cohort (Supplementary Table S1). Circulating HPV-DNA was detected in 5 patients, while 6 tissue samples were HPV-DNA positive which were all HPV type 16 (Supplementary Table S2) . The sensitivity was 66.7% (95% CI: 22.3%-95.7%) and the specificity was 96.2% (95% CI: 80.4% −99.9%) (P< .05, Fisher’s exact test).
Figure 1. Detection of HPV-DNA in the peripheral blood of patients with non-small cell lung cancer. (a)HPV universal primer amplification electrophoresis map; (b)HPV16-specific primer amplification electropherogram; (c)HPV18-specific primer amplification electropherogram. Note: M: marker; PC: positive control
Relationships between the presence of HPV-DNA in blood circulation and clinical-pathological parameters of patients with NSCLC
Circulating HPV-DNA-positive patients were compared with HPV-DNA-negative patients to analyze the relationships between the presence of HPV-DNA in blood circulation and clinical-pathological parameters of patients with NSCLC (). Circulating HPV-DNA-positive NSCLC patients were significantly different from HPV-DNA-negative ones in terms of tumor pathological type, degree of differentiation, tumor stage, and lymph node metastasis (P< .05). In age, gender, and primary tumor diameter, there were no significant differences between the two groups (P> .05). The pathological types of HPV-DNA-positive patients were more common with adenocarcinoma, and the proportion of poorly differentiated cancer was higher than that of HPV-DNA-negative patients. In terms of tumor stage and lymph node metastasis, the proportion of HPV-DNA-positive patients with advanced lung cancer and lymph node metastasis was higher than that of HPV-DNA-negative patients, but there was no significant difference in tumor diameter between the two groups (P> .05).
Table 1. Comparison of clinical-pathological parameters betweenHPV16-DNA-positive and HPV16-DNA-negative non-small cell lung cancer patients
Screening of differentially expressed miRNAs in lung adenocarcinoma and squamous cell carcinoma based on TCGA
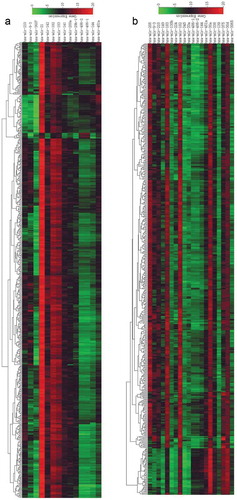
According to significance and expression intensity, 14 miRNAs with significant differences in expression in lung adenocarcinoma were screened ( and )), of which 10 were upregulated and 4 were downregulated. There were 20 miRNAs that were differentially expressed in lung squamous cell carcinoma ( and )), of which 10 were upregulated and 10 were downregulated. Four miRNAs were upregulated in both that included miR-183, miR-9-3, miR-210 and miR-182. Four miRNAs were downregulated, namely, miR-144, miR-451a, miR-486-1 and miR-486-2.
Table 2. Differentially expressed miRNAs in lung adenocarcinoma
Table 3. Differentially expressed miRNAs in lung squamous cell carcinoma
Figure 2. Heat map clustering of differentially expressed miRNA microarray data. (a) Heat map of differentially expressed miRNAs in lung adenocarcinoma; (b) heat map of differentially expressed miRNAs in lung squamous cell carcinoma. Red represents upregulated miRNAs; the darker the color, the more pronounced the upregulation. Green represents downregulated miRNAs; the darker the color, the more pronounced the downregulation
Levels of differentially expressed miRNAs in non-small cell lung cancer in plasma
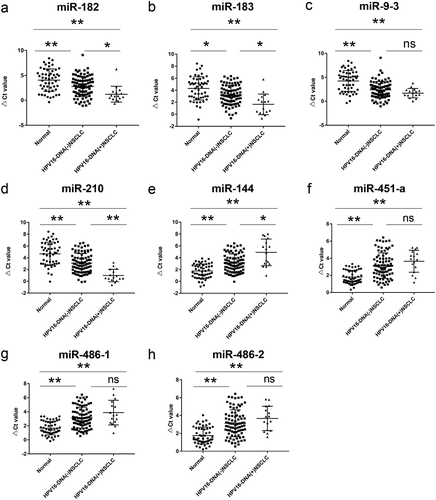
Eight circulating miRNAs were detected by real-time quantitative PCR(RT-qPCR) to analyze their differential expression in patients with NSCLC (). The expression levels of miR-183, miR-9-3, miR-210 and miR-182 in patients with NSCLC were significantly higher than those in healthy control (P< .01), while the expression levels of miR-144, miR-451a, miR-486-1 and miR-486-2 were significantly downregulated (P < .01). The expression levels of miR-9-3, miR-451a, miR-486-1 and miR-486-2 in HPV-DNA-positive NSCLC patients were not significantly different from those in HPV-DNA-negative NSCLC patients (P> .05), while the expression levels of miR-183, miR-210 and miR-182 in HPV-DNA-positive patients were significantly higher than those in HPV-DNA-negative patients (P< .05). The expression level of miR-144 was significantly lower in HPV-DNA-positive patients than HPV-DNA-negative NSCLC patients (P< .01).
Figure 3. The expression levels of eight miRNAs in the plasma of normal human subjects, HPV16-DNA (-) NSCLC patients, and HPV16-DNA (+) NSCLC patients were measured. RT-qPCR was used to analyze the plasma levels of miR-182 (a), miR-183 (b), miR-9-3 (c), miR-210 (d), miR-144 (e), miR-451-a (f), miR-486-1 (g), and miR-486-2 (h) in 52 healthy subjects, 84 HPV16-DNA (-) NSCLC patients, and 16 HPV16-DNA (+) NSCLC patients. Note: *: P< .05, **: P < .01, ns: P > .05; the smaller the ΔCt value, the higher the expression
The assays with single and combined miRNAs to differentiate circulating HPV-DNA-positive NSCLC patients from healthy individuals
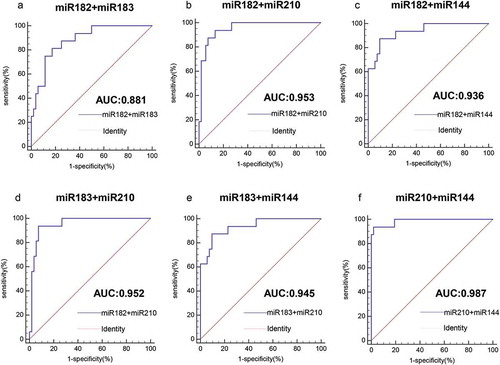
As shown by the ROC curve, we evaluated the ability of four miRNAs to distinguish circulating HPV-DNA-positive NSCLC patients from healthy individuals by AUC value. As shown in , the AUC value of miR-182 was 0.858 (95% confidence interval (CI): 0.752–0.931, P< .0001), the AUC value of miR-183 was 0.843 (95% CI: 0.735–0.920, P< .0001), the AUC value of miR-210 was 0.950 (95% CI: 0.868–0.988, P < .0001), and the AUC value of miR-144 was 0.881 (95% CI: 0.780–0.947, P < .0001). The results showed that all four miRNAs could effectively distinguish between healthy controls and HPV-DNA-positive NSCLC patients. Among them, miR-210 had the highest AUC value and had good diagnostic value, and miR-144 was the second most common miRNA. The diagnostic values of miR-182 and miR-183 were comparable. Moreover, miR-210 had the highest specificity of 86.54%, and miR-210 and miR-182 had higher sensitivity, which was 93.75% ( and ). When two miRNAs were combined to detect HPV-DNA-positive NSCLC, their AUC values were all greater than 0.9, which were significantly higher than those of single miRNAs. Among them, miR-210+ miR-144 had the highest AUC value of 0.987 (95% CI: 0.923–0.997, P< .0001) ( and ). Moreover, the sensitivity and specificity of the combined detection of the two miRNAs were also significantly increased.
Table 4. The efficacy of plasma miR-182, miR-183, miR-210 and miR-144 to distinguish HPV16-DNA-positive NSCLC cases from healthy controls
Figure 4. Analysis of the value of four miRNAs for the diagnosis of HPV16-DNA-positive NSCLC and healthy individuals. (a-d) The ROC curve was used to assess the value of miR-182, miR-183, miR-210 and miR-144 in the diagnosis of HPV16-DNA-positive NSCLC. (miR-182, 0.858, P < .0001; miR-183, 0.843, P < .0001; miR-210, 0.950, P < .0001; and miR-144, 0.881, P < .0001.)
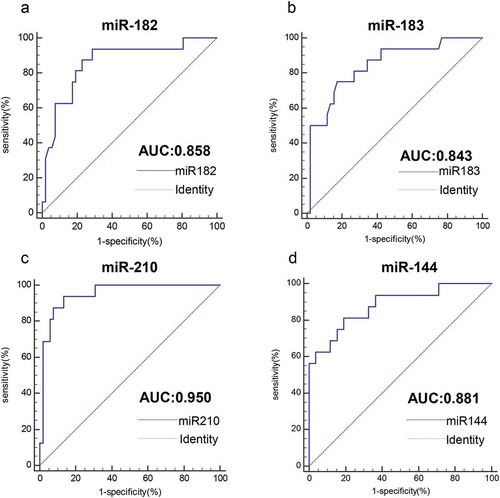
Figure 5. Analysis of the value of combined miRNAs for the diagnosis of HPV16-DNA-positive NSCLC and healthy individuals. (a-f) The ROC curve shows that for the distinction between HPV16-DNA-positive NSCLC and healthy individuals, the combined detection of the two miRNAs is more accurate than the single miRNA
Assays with single and combined miRNAs to differentiate circulating HPV-DNA-positive NSCLC patients from circulating HPV-DNA-negative NSCLC patients
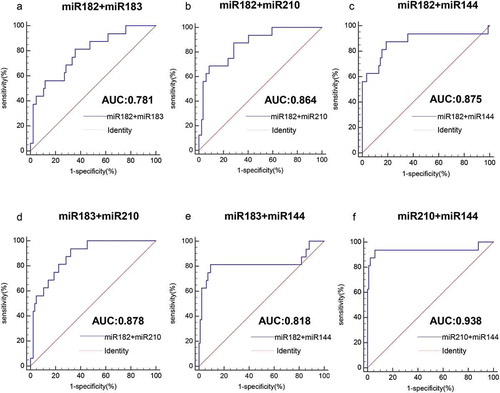
We evaluated the ability of four miRNAs to distinguish HPV-DNA-positive NSCLC and HPV-DNA-negative NSCLC patients by area under the ROC curve (AUC). As shown in , the AUC value of miR-182 was 0.746 (95% CI: 0.649–0.828, P< .0001), the AUC value of miR-183 was 0.753 (95% CI: 0.657–0.834, P< .0001), the AUC value of miR-210 was 0.865 (95% CI: 0.782–0.925, P< .0001), and the AUC value of miR-144 was 0.729 (95% CI.: 0.631–0.813, P< .0001). The results showed that all four miRNAs could differentiate HPV-DNA-positive NSCLC and HPV-DNA-negative NSCLC patients to some extent. Among them, miR-210 had the highest AUC value and better diagnostic value; miR-182, miR-183 and miR-144 also had certain diagnostic value. Moreover, miR-210 had the highest specificity of 92.86%, and miR-182 had the highest sensitivity, reaching 87.50% ( and ). When two miRNAs were combined to detect HPV-DNA-positive NSCLC and HPV-DNA-negative NSCLC patients, their AUC values were slightly higher than those of single miRNAs. Among them, miR-210+ miR-144 had the highest AUC value of 0.938 (95% CI: 0.871–0.976, P< .0001) ( and ). The combined AUC values of the remaining miRNAs were all below 0.9. Moreover, the detection sensitivity of the combinations miR-210+ miR-144 and miR-183+ miR-210 was 93.75%, and the detection specificity of the combination miR-210+ miR-144 was the highest, reaching 94.05%.
Table 5. Efficacy of plasma miR-182, miR-183, miR-210 and miR-144 to distinguish HPV16-DNA-positive NSCLC patients from HPV16-DNA-negative NSCLC patients
Figure 6. Value analysis of four miRNAs for the diagnosis of HPV16-DNA-positive NSCLC patients and HPV16-DNA-negative NSCLC patients. (a-d) The ROC curve was used to assess the value of miR-182, miR-183, miR-210 and miR-144 in the diagnosis of HPV16-DNA-positive NSCLC. (miR-182, 0.746, P< .0001; miR-183, 0.753, P < .0001; miR-210, 0.865, P < .0001; miR-144, 0.729, P < .0001.)
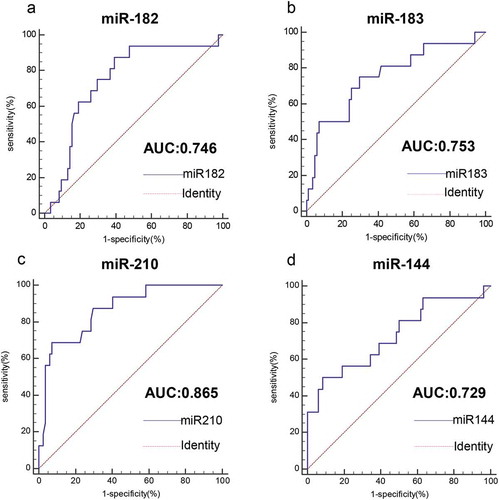
Figure 7. Analysis of the value of combined miRNAs for the diagnosis of HPV16-DNA-positive NSCLC patients and HPV16-DNA-negative NSCLC patients. (a-f) The ROC curve shows that for the distinction between HPV16-DNA-positive NSCLC and HPV16-DNA-negative NSCLC patients, the combined detection of the two miRNAs is more accurate than the single miRNA
Discussions
In this study, 100 patients with newly diagnosed NSCLC were included. Circulating HPV-DNA was detected in 16 patients by PCR. The results also showed that the detected HPV type of the NSCLC patients in this region was HPV16. After the analysis of tumor pathological type, differentiation, tumor stage, and lymph node metastasis, significant differences were found between the circulating HPV-DNA-positive and negative NSCLC patients. And disease severity of positive patients was worse than that of negative patients. Then, miR-183, miR-9-3, miR-210, miR-182, miR-144, miR-451a, miR-486-1 and miR-486-2 were screened by the TCGA database and further verification was conducted by RT-qPCR. We found that miRNAs are closely related to circulating HPV-DNA-positive NSCLC. MiR-210 as a single detection index or miR-210+ miR-144 as a combined detection indicator has a better predictive performance to diagnose circulating HPV-DNA-positive NSCLC.
HPV-DNA-positive lung cancer was first discovered by detecting the respiratory epithelium in patients with lung squamous cell carcinoma,Citation17 so most believed that squamous epithelial cells in the bronchi, which are similar to the cervical squamous-column transition zone, are a preferred cell type for HPV infection. HPV16 and HPV18 are the main types detected in patients with lung cancer, but the infection rate and infection type in different regions of the world are different.Citation18 South America has the highest HPV infection rate, followed by Europe, North America and Asia.Citation3 In this study, only HPV16-DNA was detected. Some regions have also been reported to have other HPV types, such as HPV18, 30, 31, 33, and 39.Citation4 However, some studies have also reported the absence of HPV-DNA in lung cancer.Citation19 The reasons may be (1) the low HPV-DNA positive rate of lung cancer patients in the area, (2) the degradation of DNA in paraffin-embedded samples.
In addition to tumor tissues, HPV-DNA can also be detected in blood cells, plasma, serum and umbilical arterial blood.Citation4 The association between HPV-DNA in tissues and in blood is still unknown. In the validation cohort, HPV-DNA was detected in the tissues instead of blood circulation in 2 patients. Several studies suggested that the presence of viral DNA in both blood and tissues indicates the exists of active virus, which further confirms that circulating HPV-DNA detection is of some significance in the evaluation of cancer patients.Citation20 With the deepening of virology and oncology-related research, the value of circulating-HPV- DNA has also received increasing attention. And an increasing number of reports showed closely relation between circulating HPV-DNA and cancers. Chiou et al successfully detected HPV-DNA in the blood of patients with non-small cell lung cancer in Taiwan, China, and found that the concentration of HPV-DNA was synergistic with the development of lung cancer.Citation21 Since blood sample collection is more acceptable to patients than histological samples, HPV-DNA is expected to become a potential circulating marker for detecting and monitoring tumor progress. This study found a significant increase in the proportion of tumor metastasis in circulating HPV-DNA-positive NSCLC patients, but the tumor diameter did not change obviously, suggesting that circulating-HPV-DNA may be closely related to tumor migration and invasion. However, more experimental evidence is needed to confirm it.
MiRNAs are small non-coding RNAs that play a key role in tumor initiation, progression, and metastasis and have been closely related to disease diagnosis, patient prognosis and response to treatment.Citation12 Compared with other circulating markers, miRNAs can exist stably in blood, are not easily degraded, and the detection technology is more uniform and less restricted.Citation22 MiRNAs also act as an effective bridge connecting clinical research and basic research. The studies on the pathogenesis of miRNAs can provide new targets for disease prevention and treatment. Meanwhile, the abnormally expressed miRNAs under specific disease can provide a reliable reference for the study of disease pathogenesis. Therefore, after identifying circulating-HPV-DNA as a potential risk factor for NSCLC, this study sought to find changes in plasma miRNAs, hoping that it could provide clues for the early detection of circulating HPV-DNA-positive NSCLC. After screening miRNAs by using the TCGA database and RT-qPCR, only miR-183, miR-210, miR-182 and miR-144 were differentially expressed in circulating HPV-DNA-positive NSCLC patients. The ROC curve was used to evaluate their efficacy. When we used a single miRNA to differentiate HPV-DNA-positive NSCLC patients from healthy controls, miR-210 had the best diagnostic value. When we used two combined miRNAs, miR-210+ miR144 showed the best diagnostic performance, better than any single miRNA. When we analyzed the miRNA expression of circulating HPV-DNA-positive and negative NSCLC patients, we found that miR-210 alone or in combination with miR-144 had a high diagnostic value.
MiR-182 and miR-183 belong to the miR-183 family, and members of the miR-183 family have been shown to be upregulated in tumor tissues and plasma of patients with various malignancies, such as lung cancer.Citation23,Citation24 Overexpression of members of the miR-183 family have been confirmed to be associated with poor survival rate in patients with lung cancer.Citation25 And In NSCLC, serum miR-182 and miR-183 were reported to be able to serve as a high sensitive and specific diagnostic biomarker for early detection.Citation24 Moreover, miR-183 may be closely related to the metastasis of NSCLC.Citation26 So, miR-183 family may be considered potential novel biomarkers for the diagnosis and prognosis of lung cancer. Besides, miR-182 and miR-183 are upregulated in HPV-associated cancers and act as oncogenic miRNAs.Citation27 Recent study showed that high-risk HPV E7 upregulates miR-182 expression through the TGF-β/Smad4 pathway in progression of human cervical cancer.Citation28 In our study, miR-182 and miR-183 were significantly up-regulated in the plasma of HPV-DNA-positive patients. Such patients have a higher degree of malignancy, which may be due to the abnormal expression of miR-182 and miR-183. Further studies are needed to confirm their pathogenicity in circulating HPV-DNA-positive NSCLC.
MiR-210 has long been known as the “micromanager of the hypoxic pathway” and is involved in tumor growth, apoptosis and angiogenesis, which have carcinogenic effects.Citation29 It has been reported that miR-210 is upregulated in the tissues,Citation30 sputum,Citation31 and plasmaCitation24 of NSCLC patients. MiR-210 overexpression was associated with poor progression-free survival in NSCLC and negatively related to overall survival and disease-free survival.Citation32 Meanwhile, it has been reported that miR-210 could indicate the poor outcomes in HPV16-positive cervical cancer.Citation33 In our study, circulating-HPV-DNA-positive patients with higher disease severity have elevated plasma miR-210 levels, which is consistent with previous studies. The role of miR-144 has been widely studied in recent years. This miRNA affects the occurrence of various tumors. Studies have shown that miR-144 is downregulated in many cancers and acts as a potential tumor suppressor.Citation34 In NSCLC, the up-regulated miR-144 could inhibit tumor growth.Citation35 In this study, we found that miR-144 was also decreased in the plasma of circulating-HPV-DNA-positive NSCLC patients. The decrease in the inhibitory effect on cancers caused by the down-regulation of miR-144 may be a potential factor for the progression of circulating-HPV-DNA-positive NSCLC. Moreover, among the differentially expressed miRNAs screened by the TCGA database, the levels of miR-9-3, miR-451a, miR-486-1 and miR-486-2 were not significantly different between circulating-HPV-DNA-positive and negative NSCLC patients. This suggested that there may be some differences in the pathogenic mechanism between HPV-infected and non-HPV-infected NSCLC. Whether this difference is mediated by miRNAs requires further study to confirm.
Some studies indicated that the role of miRNAs in inhibiting or promoting tumors depends on the different tissues and cellular environments. The expression of the same miRNAs in different tumors may be completely opposite.Citation36 During the development of NSCLC, miR-182 can promote tumor cell proliferationCitation37 but in turn inhibit tumor metastasis.Citation38 MiR-183 has the ability to promote tumor cell proliferation, migration and invasion,Citation39,Citation40 while miR-144 mediates tumor cell apoptosis and inhibits tumor invasion.Citation41,Citation42 Therefore, the progression of cancers may be caused by the joint action of multiple miRNAs. With further researches, it was found that miRNAs also play an important role in regulating the response of HPV-related cancers to drugs. Studies have reported that elevated miR-182 and miR-210 levels promote the response of HPV-positive oropharyngeal and cervical cancer to therapies.Citation43,Citation44 In circulating HPV-DNA-positive NSCLC patients, the role of miRNAs in mediating therapy responses still need more studies to investigate. Our study mainly focused on the relationship among miRNAs, circulating-HPV-DNA and clinicopathological characteristics of NSCLC. However, considering the complexity of clinical practice, more studies are needed to confirm the correlation between miRNAs and the progression or prognosis of circulating-HPV-DNA-positive NSCLC, as well as the biological function of these miRNAs in circulating HPV-DNA-positive NSCLC.
It has been reported that miRNAs enter the plasma in approximately two ways:Citation45,Citation46 1) secreted by damaged broken cells or 2) transported by microvesicles, exosomes, etc. And circulating miRNAs are relatively stable, which provides possibilities for their experimental use. Although circulating miRNAs are reliable biomarkers for the diagnosis of cancer,Citation47 the following limitations may still exist in this study: (1) different detection methods may produce different results, (2) some miRNA functions are variable, and (3) small sample size may lead to selection bias.
Overall, this study reveals the potential risk of circulating HPV in NSCLC patients, and evaluates the value of plasma miR-182, miR-183, miR-210 and miR-144 in the identification of circulating- HPV-DNA-positive NSCLC patients. This study may provide new ideas for the research on the pathogenic mechanism of circulating-HPV-DNA and new clues for the estimation of the condition and prognosis of NSCLC patients.
Materials and methods
Samples collection and clinical information
All blood samples were obtained from Affiliated Hospital of Jiangsu University (2017 to 2019). Among the cases, 52 were healthy controls from the physical examination center of the hospital who did not have a tumor diagnosis of or a history of HPV infection and 100 were patients with NSCLC. All patients were newly diagnosed based on histopathology. No patients received chemotherapy or radiotherapy before the samples were collected. The NSCLC patients included 41 with squamous cell carcinoma, 59 with adenocarcinoma, 43 males and 57 females, aged 28 to 81 years old, with an average age of 63 years. According to the pathological reports, 33 were poorly differentiated and 67 were moderately or highly differentiated. The patients had various tumor diameters. Fifty-seven cases were over 3 cm, and 43 cases were less than 3 cm. According to the TNM staging standard established by the International Union Against Cancer (UICC) in 2017, there were 14 cases in stage I, 30 cases in stage II, 25 cases in stage III, and 31 cases in stage IV. The control group included 25 males and 27 females with an average age of 61.45 years. Venous blood samples were collected from each participant into an EDTA tube. Two hundred microliters of whole blood samples were dispensed into EP tubes and stored at 4°C. The remaining samples were separated into plasma and cellular components by centrifugation at 1600 rcf for 10 min. The supernatants were collected in RNase-free EP tubes, and the samples were centrifuged again at 16,000 rcf for 10 min to obtain the plasma, which was stored at −80°C until use.
Tissue and blood samples of the validation cohort were also collected. The validation cohorts included 32 patients and fulfil the same eligibility criteria (Supplementary Table S1).
The study was approved by the Ethics Committee of the Affiliated Hospital of Jiangsu University, and all subjects received informed consent before collection.
DNA extraction and HPV type detection
DNA was extracted from whole blood and tissues using the DNA rapid extraction kit (Sangon, Shanghai, China). All steps followed the manufacturer’s instructions. Then HPV-DNA-positive blood samples were screened by HPV universal primers, and then HPV16- and HPV18-specific primers were used for nested-PCR amplification to identify the subtype. The primers were designed in reference to the studies by Kleter et alCitation48 and Pao et alCitation49 and submitted to Hong Xun Biotech Co (Suzhou, China) for synthesis. The specific primer information is shown in . The HPV universal primers PCR conditions were as follows: pre-denaturation at 94°C for 10 min, denaturation at 94°C for 45 s, annealing at 50°C for 45 s, and extension at 72°C for 60 s for 30 cycles, and finally extension at 72°C for 7 min. Next, 10 µl of the final PCR product was loaded onto a 2% agarose gel. PCR conditions for the HPV16- and HPV18-specific primers were as follows: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 40 s, annealing at 55°C for 60 s, and extension at 72°C for 60 s for 30 cycles, and finally extension at 72°C for 10 min. Then, 10 µl of the final PCR product was loaded onto a 1% agarose gel, stained with ethidium bromide, and visualized under ultraviolet illumination. DNA preparations from SiHa cells (containing HPV16) and HeLa cells (containing HPV18) were used as positive controls. Furthermore, after the PCR amplification was completed, the PCR product was sent to Hong Xun Biotech Co for sequencing analysis, and the sequencing results were compared through using the BLAST website to confirm the HPV type.
Table 6. HPV universal primers, HPV16 and HPV18 nested-PCR primers
Differential miRNA screening
In this study miRNA expression data for lung adenocarcinoma and squamous cell carcinoma were obtained from the TCGA public database (http://cancergenome.nih.gov/), which included 567 samples of lung adenocarcinoma (including 59 paracancerous samples) and 523 samples of lung squamous cell carcinoma (including 49 paracancerous samples). Data preprocessing was first performed, low abundance data were filtered out, and the data were normalized. Using the SAM software, miRNAs that met the criteria of P< .05, FDR<0.01, and |log2FC|>4 were screened out.
RNA extraction, reverse transcription and quantitative PCR
Total RNA containing small RNA was extracted from 1 ml of plasma using a miRNA Purification Kit (CoWin, Shanghai, China) according to the manufacturer’s protocol. The specific steps were as follows: 200 μl of plasma was taken and 1 mL of TRIzol reagent was added, and the mixture was shaken vigorously for 30 s and then allowed to stand for 5 min to fully lyse. Total RNA was extracted by adding 200 μL of chloroform, precipitated with absolute ethanol, and then RWT and RW2 reagents were added to elute the total RNA in the center of the adsorption column and finally resuspend it in 30 μL of DEPC water. The concentration and purity of sample RNA were determined by a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, USA). Reverse transcription was performed using the miRNA cDNA Synthesis Kit (CoWin, Shanghai, China) via the poly-A tail method and oligo(dT)-universal tag primer to finally synthesize the first strand cDNA. Each real-time PCR mixture (20 μl) contained 10 μl of 2× miRNA qPCR mix (ROX) (CoWin), 0.8 μl of primer (10 μM) and 2 μl of cDNA. Real-time PCR was performed with the miRNA-specific forward primers (sequences as shown in ) and the sequence complementary to the poly(T) adapter as the reverse primer (5ʹ-GCGAGCACAGAATTAATACGAC-3ʹ) in a CFX96 TouchTM System (Bio-Rad). The PCR was carried out as follows: initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min, and then a dissociation curve analysis was conducted to confirm the specificity. U6 was used as a control. To reduce errors, the experiments were performed in triplicate for every independent RNA sample.
Table 7. The sequence of miRNA-specific forward primer
Statistical analysis
The expression levels of the miRNAs were calculated using the ΔCt method, where ΔCt = Cttarget – Ctreference, with a smaller ΔCt value indicating higher expression. The RT-qPCR data were analyzed with nonparametric statistical methods by SPSS 24.0 software, and comparisons between groups were performed with the Kruskal-Wallis H test. Then, the corresponding graphs were generated using GraphPad Prism 5.0. Fisher’s exact test was used to analyze the correlation between tissue HPV and blood HPV in the validation cohort. The relationship between the circulating HPV-DNA and clinical-pathologic parameters was calculated by a Pearson Chi-Square or a Fisher’s Exact Test. The diagnostic value of miRNAs was analyzed by receiver operator characteristic curves (ROC curves) using MedCalc software. Youden Index = Sensitivity + Specificity −1 was used to determine the best cutoff value. P< .05 (bilateral) was considered statistically significant, and P< .01 was considered significant.
Disclosure of potential conflicts of interest
The authors have no conflicts of interest to report for this work.
Supplemental Material
Download MS Word (15.5 KB)Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Xu S, Ma H, Bo Y, Shao M. The oncogenic role of CB2 in the progression of non-small-cell lung cancer. Biomed Pharmacother. 2019;117:109080. doi:10.1016/j.biopha.2019.109080.
- Xiong WM, Xu QP, Li X, Xiao RD, Cai L, He F. The association between human papillomavirus infection and lung cancer: a system review and meta-analysis. Oncotarget. 2017;8(56):96419–96432. doi:10.18632/oncotarget.21682.
- Clément-Duchêne C, Stock S, Xu X, Chang ET, Gomez SL, West DW, Wakelee HA, Gould MK. Survival among never-smokers with lung cancer in the cancer care outcomes research and surveillance study. Ann Am Thorac Soc. 2016;13(1):58–66. doi:10.1513/AnnalsATS.201504-241OC.
- de Freitas AC, Gurgel AP, de Lima EG, de França São Marcos B, Do Amaral CM. Human papillomavirus and lung cancinogenesis: an overview. J Cancer Res Clin Oncol. 2016;142(12):2415–2427. doi:10.1007/s00432-016-2197-1.
- Yuan S, Qiu Y, Xu Y, Wang H. Human papillomavirus infection and female infertility: a systematic review and meta-analysis. Reprod Biomed Online. 2020;40(2):229–237. doi:10.1016/j.rbmo.2019.10.019.
- Zhang R, Chen L, Cui YD, Li G. The association between human papillomavirus infection and smoking, age, gender in lung cancer patients: a meta-analysis. Iran J Public Health. 2019;48(1):1–8.
- Salman NA, Davies G, Majidy F, Shakir F, Akinrinade H, Perumal D, Ashrafi GH. Association of high risk human papillomavirus and breast cancer: a UK based study. Sci Rep. 2017;7:43591. doi:10.1038/srep43591.
- Coppock JD, Lee JH. mTOR, metabolism, and the immune response in HPV-positive head and neck squamous cell cancer. World J Otorhinolaryngol Head Neck Surg. 2016;2(2):76–83. doi:10.1016/j.wjorl.2016.05.010.
- Jensen KK, Grønhøj C, Jensen DH, von Buchwald C. Circulating human papillomavirus DNA as a surveillance tool in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Clin Otolaryngol. 2018;43(5):1242–1249. doi:10.1111/coa.13136.
- Pornthanakasem W, Shotelersuk K, Termrungruanglert W, Voravud N, Niruthisard S, Mutirangura A. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Cancer. 2001;1:2. doi:10.1186/1471-2407-1-2.
- Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67(1):129–139. doi:10.1007/s13105-010-0050-6.
- Santos JMO, Gil da Costa RM, Medeiros R. Dysregulation of cellular microRNAs by human oncogenic viruses - implications for tumorigenesis. Biochim Biophys Acta Gene Regul Mech. 2018;1861(2):95–105. doi:10.1016/j.bbagrm.2018.01.017.
- Zheng ZM, Wang X. Regulation of cellular miRNA expression by human papillomaviruses. Biochim Biophys Acta. 2011;1809(11–12):668–677. doi:10.1016/j.bbagrm.2011.05.005.
- Lu J, Han B. Liquid biopsy promotes non-small cell lung cancer precision therapy. Technol Cancer Res Treat. 2018;17:1533033818801809. doi:10.1177/1533033818801809.
- Paiva I, Gil da Costa RM, Ribeiro J, Sousa H, Bastos MM, Faustino-Rocha A, Lopes C, Oliveira PA, Medeiros R. MicroRNA-21 expression and susceptibility to HPV-induced carcinogenesis - role of microenvironment in K14-HPV16 mice model. Life Sci. 2015;128:8–14. doi:10.1016/j.lfs.2015.02.020.
- Araújo R, Santos JMO, Fernandes M, Dias F, Sousa H, Ribeiro J, Bastos MMSM, Oliveira PA, Carmo D, Casaca F, et al. Expression profile of microRNA-146a along HPV-induced multistep carcinogenesis: a study in HPV16 transgenic mice. J Cancer Res Clin Oncol. 2018;144(2):241–248. doi:10.1007/s00432-017-2549-5.
- Syrjänen KJ. Condylomatous changes in neoplastic bronchial epithelium. Report of a case. Respiration. 1979;38(5):299–304. doi:10.1159/000194095.
- Shikova E, Ivanova Z, Alexandrova D, Shindov M, Lekov A. Human papillomavirus prevalence in lung carcinomas in Bulgaria. Microbiol Immunol. 2017;61(10):427–432. doi:10.1111/1348-0421.12535.
- Silva EM, Mariano VS, Pastrez PRA, Pinto MC, Nunes EM, Sichero L, Villa LL, Scapulatempo-Neto C, Syrjanen KJ, Longatto-Filho A. Human papillomavirus is notassociated to non-small cell lung cancer: data from a prospective cross-sectional study. Infect Agents Cancer. 2019;14:18. doi:10.1186/s13027-019-0235-8.
- Ferreira LL, Biasoli ÉR, Bernabé DG, Nunes CM, Miyahara GI. Plasma HPV DNA is detectable in oral leukoplakia patients. Pathol Res Pract. 2017;213(7):759–765. doi:10.1016/j.prp.2017.04.005.
- Chiou HL, Wu MF, Liaw YC, Cheng YW, Wong RH, Chen CY, Lee H. The presence of human papillomavirus type 16/18 DNA in blood circulation may act as a risk marker of lung cancer in Taiwan. Cancer. 2003;97(6):1558–1563. doi:10.1002/cncr.11191.
- Wu L, Wang J, Zhu D, Zhang S, Zhou X, Zhu W, Zhu J, He X. Circulating epstein-barr virus microRNA profile reveals novel biomarker for nasopharyngeal carcinoma diagnosis. Cancer Biomark. 2020;27(3):365–375. doi:10.3233/CBM-190160.
- Zhang QH, Sun HM, Zheng RZ, Li YC, Zhang Q, Cheng P, Tang ZH, Huang F. Meta-analysis of microRNA-183 family expression in human cancer studies comparing cancer tissues with noncancerous tissues. Gene. 2013;527(1):26–32. doi:10.1016/j.gene.2013.06.006.
- Zhu W, Zhou K, Zha Y, Chen D, He J, Ma H, Liu X, Le H, Zhang Y. Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS One. 2016;11(4):e0153046. doi:10.1371/journal.pone.0153046.
- Zhu W, Liu X, He J, Chen D, Hunag Y, Zhang YK. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer. 2011;11:393. doi:10.1186/1471-2407-11-393.
- Lin Q, Mao W, Shu Y, Lin F, Liu S, Shen H, Gao W, Li S, Shen D. A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol. 2012;138(1):85–93. doi:10.1007/s00432-011-1068-z.
- Tang T, Wong HK, Gu W, Yu MY, To KF, Wang CC, Wong YF, Cheung TH, Chung TK, Choy KW. MicroRNA-182 plays an onco-miRNA role in cervical cancer. Gynecol Oncol. 2013;129(1):199–208. doi:10.1016/j.ygyno.2012.12.043.
- Chen J, Deng Y, Ao L, Song Y, Xu Y, Wang CC, Choy KW, Tony Chung KH, Du Q, Sui Y, et al. The high-risk HPV oncogene E7 upregulates miR-182 expression through the TGF-β/Smad pathway in cervical cancer. Cancer Lett. 2019;460:75–85. doi:10.1016/j.canlet.2019.06.015.
- Huang X, Le QT, Giaccia AJ. MiR-210–micromanager of the hypoxia pathway. Trends Mol Med. 2010;16(5):230–237. doi:10.1016/j.molmed.2010.03.004.
- Eilertsen M, Andersen S, Al-Saad S, Richardsen E, Stenvold H, Hald SM, Al-Shibli K, Donnem T, Busund LT, Bremnes RM. Positive prognostic impact of miR-210 in non-small cell lung cancer. Lung Cancer. 2014;83(2):272–278. doi:10.1016/j.lungcan.2013.11.005.
- Roa WH, Kim JO, Razzak R, Du H, Guo L, Singh R, Gazala S, Ghosh S, Wong E, Joy AA, et al. Sputum microRNA profiling: a novel approach for the early detection of non-small cell lung cancer. Clin Invest Med. 2012;35(5):E271. doi:10.25011/cim.v35i5.18700.
- He RQ, Cen WL, Cen JM, Cen WN, Li JY, Li MW, Gan TQ, Hu XH, Chen G. Clinical significance of miR-210 and its prospective signaling pathways in non-small cell lung cancer: evidence from gene expression omnibus and the cancer genome atlas data mining with 2763 samples and validation via real-time quantitative PCR. Cell Physiol Biochem. 2018;46(3):925–952. doi:10.1159/000488823.
- Nilsen A, Jonsson M, Aarnes EK, Kristensen GB, Lyng H. Reference microRNAs for RT-qPCR assays in cervical cancer patients and their application to studies of HPV16 and hypoxia biomarkers. Transl Oncol. 2019;12(3):576–584. doi:10.1016/j.tranon.2018.12.010.
- Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7(10):e48086. doi:10.1371/journal.pone.0048086.
- Huang Y, Ni R, Wang J, Liu Y. Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed Pharmacother. 2019;109:1851–1859. doi:10.1016/j.biopha.2018.09.151.
- Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81(3):388–396. doi:10.1016/j.lungcan.2013.05.007.
- Chen G, Yu L, Dong H, Liu Z, Sun Y. MiR-182 enhances radioresistance in non-small cell lung cancer cells by regulating FOXO3. Clin Exp Pharmacol Physiol. 2019;46(2):137–143. doi:10.1111/1440-1681.
- Li Y, Zhang H, Li Y, Zhao C, Fan Y, Liu J, Li X, Liu H, Chen J. MiR-182 inhibits the epithelial to mesenchymal transition and metastasis of lung cancer cells by targeting the Met gene. Mol Carcinog. 2018;57(1):125–136. doi:10.1002/mc.22741.
- Zhang L, Quan H, Wang S, Li X, Che X. MiR-183 promotes growth of non-small cell lung cancer cells through FoxO1 inhibition. Tumour Biol. 2015;36(10):8121–8126. doi:10.1007/s13277-015-3550-8.
- Zhu C, Deng X, Wu J, Zhang J, Yang H, Fu S, Zhang Y, Han Y, Zou Y, Chen Z, et al. MicroRNA-183 promotes migration and invasion of CD133(+)/CD326(+) lung adenocarcinoma initiating cells via PTPN4 inhibition. Tumour Biol. 2016;37(8):11289–11297. doi:10.1007/s13277-016-4955-8.
- Zha W, Cao L, Shen Y, Huang M. Roles of Mir-144-ZFX pathway in growth regulation of non-small-cell lung cancer. PLoS One. 2013;8(9):e74175. doi:10.1371/journal.pone.0074175.
- Gao F, Wang T, Zhang Z, Wang R, Guo Y, Liu J. Regulation of activating protein-4-associated metastases of non-small cell lung cancer cells by miR-144. Tumour Biol. 2015. doi:10.1007/s13277-015-3866-4.
- Liu Q, Ma L, Jones T, Palomero L, Pujana MA, Martinez-Ruiz H, Ha PK, Murnane J, Cuartas I, Seoane J, et al. Subjugation of TGF β signaling by human papilloma virus in head and neck squamous cell carcinoma shifts DNA repair from homologous recombination to alternative end joining. Clin Cancer Res. 2018;24(23):6001–6014. doi:10.1158/1078-0432.
- Zhu Y, Huang Y, Liu M, Yan Q, Zhao W, Yang P, Gao Q, Wei J, Zhao W, Ma L. Epigallocatechin gallate inhibits cell growth and regulates miRNA expression in cervical carcinoma cell lines infected with different high-risk human papillomavirus subtypes. Exp Ther Med. 2019;17(3):1742–1748. doi:10.3892/etm.2018.7131.
- Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol. 2013;228(8):1713–1719. doi:10.1002/jcp.24344.
- Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi:10.1016/j.tcb.2011.12.001.
- Qu H, Xu W, Huang Y, Yang S. Circulating miRNAs: promising biomarkers of human cancer. Asian Pac J Cancer Prev. 2011;12(5):1117–1125.
- Kleter B, van Doorn LJ, Ter Schegget J, Schrauwen L, van Krimpen K, Burger M, Ter Harmsel B, Quint W. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153(6):1731–1739. doi:10.1016/S0002-9440(10)65688-X.
- Pao CC, Kao SM, Tang GC, Lee K, Si J, Ruan S. Prevalence of human papillomavirus DNA sequences in an area with very high incidence of cervical carcinoma. Br J Cancer. 1994;70(4):694–696. doi:10.1038/bjc.1994.375.
