ABSTRACT
Double homeobox A pseudogene 8 (DUXAP8) belongs to long non-coding RNAs (lncRNAs), which has been proven to promote the biological processes of multiple human cancers. Triple-negative breast cancer (TNBC) is the leading cause of cancer-related death in women worldwide. However, the specific role of lncRNA DUXAP8 and its underlying mechanism in TNBC remains to be unclear. We detected the expression of DUXAP8 in TNBC cells through qRT-PCR analysis. The effects of DUXAP8 silencing on TNBC cell proliferation and apoptosis were identified using CCK-8 assay, EdU assay, flow cytometry analysis and TUNEL assay. The downstream microRNA (miRNA) and messenger RNA (mRNA) of DUXAP8 were searched out through bioinformatics analysis and mechanism experiments. Rescue assays were conducted to verify the involvement of suppressor APC domain containing 2 (SAPCD2) in DUXAP8-mediated TNBC cell proliferation and apoptosis. DUXAP8 was highly expressed in TNBC cells compared to that in normal breast cells. Knockdown of DUXAP8 inhibited TNBC cell proliferation and accelerated cell apoptosis. DUXAP8 interacted with miR-29a-3p and thus enhanced the expression of SAPCD2. Moreover, YY1 transcription factor could bind to DUXAP8 promoter to activate the transcription of DUXAP8. YY1-induced transcriptional activation of DUXAP8 promotes TNBC cell growth through miR-29a-3p/SAPCD2 axis.
Introduction
Triple-negative breast cancer (TNBC) is one of the most common cancers and the second leading cause of women cancer-related death worldwide.Citation1 Therapeutic methods for TNBC patients usually include surgery, chemotherapy, and radiotherapy.Citation1,Citation2 However, the prognosis of patients in the advanced stage is still poor.Citation3 Thus, investigation of molecular mechanism associated with the tumor growth and metastasis of triple-negative breast cancer is of great significance to improve the therapeutic effectiveness.
Long non-coding RNAs (lncRNAs) can act as potential regulators in various biological processes, including cell proliferation, apoptosis, invasion, and migration.Citation4–6 LncRNAs have been identified to affect cell functions in different manners, including transcriptional regulation, mRNA post-transcriptional processing, and microRNAs (miRNAs) sponges.Citation7–9 Dysregulation of lncRNAs can promote the development of multiple cancers.Citation10 It has been verified that lncRNAs facilitate the expression of homologous mRNAs by sponging miRNAs to promote the biological process of cancers.Citation9,Citation11,Citation12 LncRNA DUXAP8 has been proven to be up-regulated in glioma, bladder cancer, and pancreatic carcinoma.Citation13–16 However, it is undefined that whether DUXAP8 acts as an oncogene in TNBC. The current study was aimed at exploring the functions and relevant mechanism of DUXAP8 in TNBC.
Transcription factors could bind to the promoter of lncRNAs, thereby enhancing the expression level of lnRNAs.Citation17–19 Here, we also tried to explore the upstream regulator for DUXAP8 in TNBC. Competing endogenous RNA (ceRNA) refers to a lncRNA-miRNA-mRNA pathway, among which lncRNA can affect the expression of mRNA through sponging miRNA.Citation20 In our study, we also investigated whether DUXAP8 can exert functions through serving as a ceRNA in TNBC.
Materials and methods
Cell culture
Normal human mammary gland cell line (MCF-10A) and human breast cancer cell lines (MDA-MB-231, MDA-MB-468, MDA-MB-436) were bought from the Chinese Academy of Sciences (Shanghai, China). Cell lines were maintained in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Sigma-Aldrich, Milan, Italy) at 37°C in 5% CO2.
Cell transfection
For the knockdown of DUXAP8, YY1, SAPCD2, MDA-MB-231 and MDA-MB-468 cells were transfected with shRNAs against DUXAP8 (sh-DUXAP8#1#2), YY1 (sh-YY1#1#2), SAPCD2 (sh-SAPCD2#1#2). shRNAs that cannot target them were used as the negative controls (sh-NC). To overexpress SAPCD2, pcDNA3.1/SAPCD2 and the empty pcDNA3.1 vector were constructed. The miR-29a-3p mimics and NC mimics were synthesized to upregulate miR-29a-3p. All plasmids and vectors used for transfections were synthesized by GenePharma (Shanghai, China) and transfected into cells with Lipofectamine 2000 (Invitrogen).
qRT-PCR
Total RNA was extracted from cells by using TRIzol reagent (Invitrogen). First-strand cDNA was generated using a FastKing RT Kit (YiHui Biotechnology, Shanghai, China). qRT-PCR was completed using SYBR Green Real-Time PCR Kit (Takara, Tokyo, Japan). Fold changes of RNA expression were calculated with 2−∆∆Ct method. GAPDH and U6 were used as the internal controls.
Bioinformatics analysis
Through the UCSC database (http://genome.ucsc.edu/), we predicted that YY1 is a potential upstream regulator for DUXAP8. The DNA motif of YY1 and its binding sites on DUXAP8 promoter were obtained from JASPAR (http://jaspar.genereg.net/). The miRNAs that potentially bind with DUXAP8 as well as mRNAs that were potentially the downstream target of miR-29a-3p were predicted from starBase (http://starbase.sysu.edu.cn/).
CCK-8 assay
MDA-MB-231 and MDA-MB-468 cells were inoculated in 96-well plates (1 × 103 cells per well). Then, cell viability was examined at specific time points by adding 10 µL of CCK-8 solution to each well. After 4 h incubation, a microplate reader purchased from Bio-Tek Instruments (Hopkinton, MA, USA) was used to monitor the absorbance at 450 nm.
EdU incorporation assay
An EdU assay kit (Ribobio, Guangzhou, China) was used to evaluate cell proliferation ability. Transfected cells were put into plates and then were cultured with 50 μM EdU buffer for 2 h. Then, cells were fixed with 4% formaldehyde (Sigma-Aldrich) and permeabilized using 0.1% Triton X-100 for 20 min. After being dying of nuclei with Hoechst33342, EdU-positive cells were visualized via a fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry analysis
MDA-MB-231 and MDA-MB-468 cells were cultured in 6-well plates for 48 h, and rinsed with PBS (Solarbio, Beijing, China). After being fixed in 70% pre-cooled ethanol (Sigma-Aldrich) at 4°C, double-staining was conducted using propidium iodide and Annexin V-fluorescein isothiocyanate. Cell apoptosis rate was measured via a flow cytometer.
Western blot
Cells were first lysed in RIPA buffer (Invitrogen). Protein concentrations were quantified by BCA Kit. Proteins were isolated utilizing 10% SDS-PAGE and then transferred to PVDF membranes (Millipore, Bedford, USA). After blocked with skim milk, membranes were incubated with primary antibodies, including anti-SAPCD2 (ab126342, Abcam, Cambridge, USA) and anti-GAPDH (ab8245, Abcam). Next, membranes were incubated with secondary antibody for 1 h. Protein bands were visualized on a chemiluminescence detection system.
TUNEL assay
TUNEL assays were performed in MDA-MB-231 and MDA-MB-468 cells by utilizing an Apoptosis Detection Kit (Ribobio). Cells were stained in DAPI (Sigma-Aldrich) or TUNEL (Thermo Fisher Scientific, Waltham, MA, USA). TUNEL-positive cells were counted in a randomly chosen field using a microplate (Olympus).
Subcellular fractionation assay
Nuclear/cytoplasmic fractionation PARIS Kit (Life Technologies, Carlsbad, CA, USA) was applied to gather nuclear and cytoplasmic fractions. Afterward, DUXAP8, GAPDH (cytoplasmic control) and U6 (nuclear control) in a cytoplasmic faction or nuclear fraction were detected by qRT-PCR.
Luciferase reporter assay
The wild-type (WT) and mutant (Mut) binding sites of miR-29a-3p in DUXAP8 or SAPCD2 3ʹ-UTR were sub-cloned into pmirGLO dual-luciferase vector to construct plasmids, named DUXAP8-WT/Mut, SAPCD2-WT/Mut, and then co-transfected into MDA-MB-231 and MDA-MB-468 cells with miR-29a-3p mimics or NC mimics, respectively.
To demonstrate the interaction between YY1 and DUXAP8 promoter, pGL3-DUXAP8 promoter was co-transfected into cells along with sh-YY1#1#2 or sh-NC using Lipofectamine 2000 (Invitrogen). The luciferase activity was measured by a Dual-Luciferase reporter assay system (Promega, USA).
RNA pull-down assay
Briefly, cell lysates were incubated with biotinylated RNAs including Bio-miR-29a-3p-WT, Bio-miR-29a-3p-Mut, Bio-SAPCD2-WT, Bio-SAPCD2-Mut and Bio-NC, respectively. Later, cell lysates were incubated with M-280 streptavidin magnetic bead (Sigma-Aldrich) for 48 h. Relative enrichment of miR-29a-3p or DUXAP8 was evaluated via qRT-PCR.
ChIP assay
ChIP assay was performed using an EZ-ChIP kit (Millipore). Immunoprecipitation was incubated with anti-YY1 (Millipore) or anti-IgG (negative control). Finally, qRT-PCR detected Precipitated chromatin DNA.
Statistical analysis
Statistical analysis was made using GraphPad Prism 5 software (Graph Pad, La Jolla, CA, USA). Experimental data obtained from three or more independent experiments were shown as mean ± SD. The differences between two or more groups were estimated via Student’s t-test or one-way ANOVA. P < .05 indicated data had statistical significance.
Results
DUXAP8 promotes TNBC cell proliferation and suppresses apoptosis
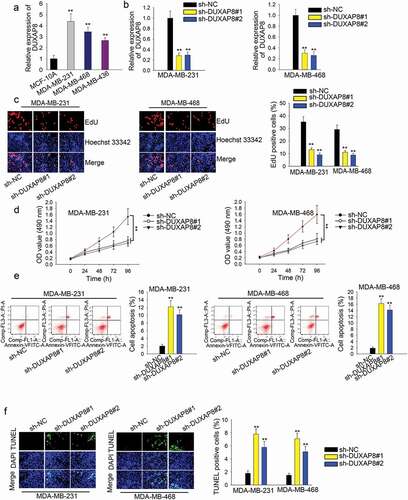
Firstly, we evaluated DUXAP8 expression in human TNBC cells with qRT-PCR. Intriguingly, DUXAP8 expression was higher in human TNBC cells than that in human breast cells (), n = 3, P < .01). The highest level was observed in MDA-MB-231 (4.38) and MDA-MB-468 cells (3.45) when the level in MCF-10A was set as 1.0. Then, we silenced DUXAP8 in two TNBC cells with specific shRNAs (), n = 3, P < .01). The level of DUXAP8 was decreased to 0.28 and 0.29 in MDA-MB-231 cells and reduced to 0.30 and 0.26 in MDA-MB-468 cells. The percent of EdU-positive cells in MDA-MB-231 cells was decreased from 35.10% to 13.10% and 9.10%, and that in MDA-MB-468 cells was decreased from 29.10% to 11.10% and 8.90% (), n = 3, P < .01). CCK-8 assay also indicted that the low OD value was lowered in DUXAP8-silenced cells (), n = 3, P < .01). Additionally, the apoptosis rate was increased remarkably after DUXAP8 was silenced (), n = 3, P < .01). According to the result of the TUNEL assay, the apoptosis rate in MDA-MB-231 cells was enhanced from 1.80% to 7.80% and 5.80%. The apoptosis rate in MDA-MB-468 cells was increased from 1.50% to 7.10% and 5.10% (), n = 3, P < .01).
Figure 1. DUXAP8 promotes TNBC cell proliferation and suppresses apoptosis
YY1 binds to DUXAP8 promoter and activates the transcription of DUXAP8
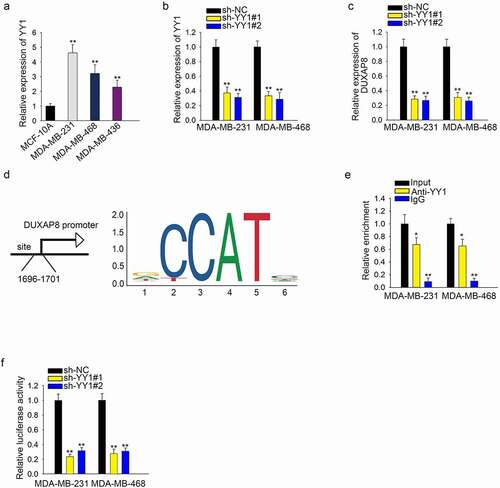
Here, we found that YY1 was highly expressed in three kinds of TNBC cell lines (), n = 3, P < .01). Knockdown of YY1 caused the downregulation of YY1 (), n = 3, P < .01). Then, we found that DUXAP8 expression was significantly decreased after YY1 silencing (), n = 3, P < .01). The binding sites of YY1 and DUXAP8 promoter were predicted via UCSC and JASPAR ()). ChIP assay showed that the enrichment of DUXAP8 promoter in the complexes conjugated with anti-YY1 showed that YY1 can bind with DUXAP8 promoter (), n = 3, P < .01). Then, the luciferase activity of vector containing DUAP8 promoter was decreased significantly after the knockdown of YY1 (), n = 3, P < .01).
Figure 2. YY1 induces upregulation of DUXAP8 by activating the transcription of DUXAP8
MiR-29a-3p can interact with DUXAP8 and suppress TNBC cell growth
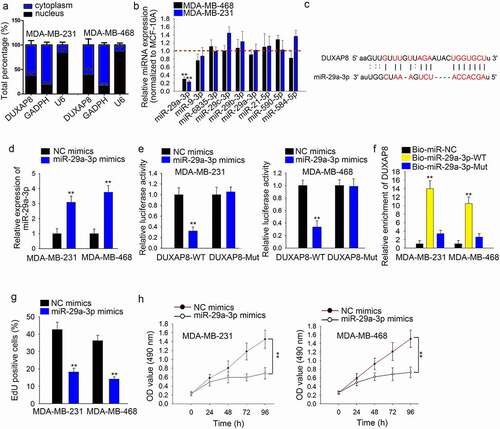
Through subcellular fractionation assay, we found that DUXAP8 was mainly located in the cytoplasm (), n = 3). Then, we used starBase to predict miRNAs that potentially bind with DUXAP8 and nine miRNAs were selected according to the high binding degree. The expression of these nine miRNAs in TNBC cells was detected by qRT-PCR. It was found that miR-29a-3p was expressed lower in two TNBC cells than other miRNAs (), n = 3, P < .0). We found the binding sites of DUXAP8 and miR-29a-3p ()). Afterward, we overexpressed miR-29a-3p in TNBC cells (), n = 3, P < .01). Through luciferase reporter assay, we identified that the luciferase activity in DUXAP8 wild-type reporter vector was decreased after miR-29a-3p overexpression (), n = 3, P < .01). RNA pull-down assay revealed that the enrichment of DUXAP8 was significantly high in the complexes conjugated with Bio-miR-29a-3p-WT (), n = 3, P < .01). Moreover, EdU results showed that overexpression of miR-29a-3p decreased TNBC cells proliferation (), n = 3, P < .01). The reduced OD value was also detected in cells with miR-29a-3p overexpression through CCK-8 assay (), n = 3, P < .01).
Figure 3. MiR-29a-3p can interact with DUXAP8 and suppresses TNBC cell growth
SAPCD2 promotes TNBC cell growth by DUXAP8/miR-29a-3p axis
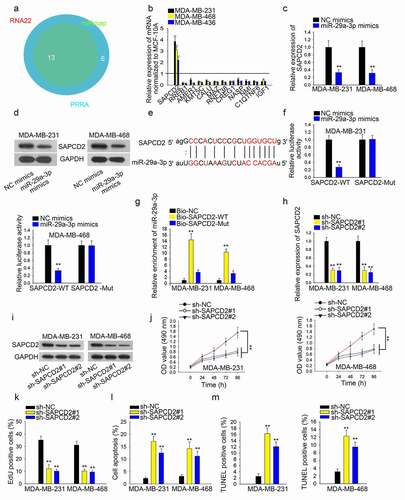
Subsequently, we predicted potential mRNAs that bind with miR-29a-3p from starBase with the condition of three programs ()). We selected four mRNAs according to the binding degree. qRT-PCR showed that SAPCD2 was expressed higher in TNBC cells than other mRNAs (, n = 3, P < .01). Then, overexpression of miR-29a-3p decreased both mRNA and protein levels of SAPCD2 (, n = 3, P < .01). Afterward, we predicted the binding sites of SAPCD2 and miR-29a-3p ()). Through luciferase reporter assay, we determined that miR-29a-3p could reduce the luciferase activity in SAPCD2-WT group rather than the SAPCD2-Mut group (), n = 3, P < .01). High enrichment of miR-29a-3p in Bio-SAPCD2-WT-conjugated complexes supported the above result (), n = 3, P < .01). Then, knockdown of SAPCD2 decreased the levels of SAPCD2 mRNA and protein in TBNC cells (, n = 3, P < .01). Moreover, CCK-8 and EdU assays results showed that inhibition of SAPCD2 impaired the proliferative ability of TNBC cells (, n = 3, P < .01). Flow cytometry analysis and TUNEL results manifested the positive effect of SAPCD2 knockdown on TBNC cell apoptosis (, n = 3, P < .01).
Figure 4. SAPCD2 promotes TNBC cell growth by DUXAP8/miR-29a-3p axis
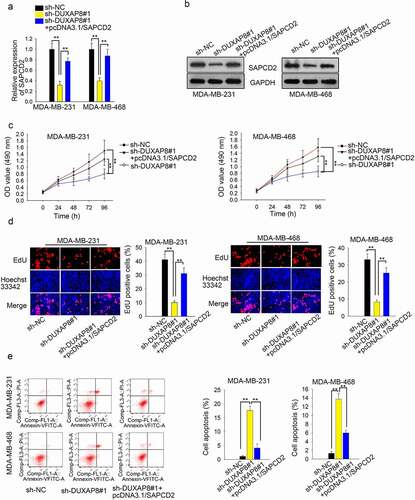
DUXAP8 promotes TNBC cell growth through upregulating SAPCD2
First of all, knockdown of DUXAP8 caused the decreased expression of SAPCD2, while the tendency was reversed after overexpression of SAPCD2 (), n = 3, P < .01). Results of western blot assay indicated that knockdown of DUXAP8 suppressed the level of SAPCD2 protein, but then rescued by overexpression of SAPCD2 (), n = 3). Afterward, results of CCK-8 assay and EdU assay showed that SAPCD2 induced the recovery of cell proliferation hampered by DUXAP8 silencing (, n = 3, P < .01). Finally, overexpression of SAPCD2 reversed the effect of DUXAP8 knockdown on cell apoptosis (), n = 3, P < .01).
Figure 5. DUXAP8 promotes TNBC cell growth through upregulating SAPCD2
Discussion
Long non-coding RNAs (lncRNAs) have been reported as biological regulators in various human cancers.Citation9 Dysregulation of lncRNAs is involved in the initiation and development of human cancers.Citation6,Citation21,Citation22 Functions of some lncRNAs have been identified in breast cancers, including triple-negative breast cancer.Citation23,Citation24 LncRNA DUXAP8 has been detected to be a dysregulated lncRNA in colorectal cancer and glioma.Citation13,Citation25 In the present study, we also determined that DUXAP8 was highly expressed in TNBC cells compared to normal breast cells. Importantly, knockdown of DUXAP8 suppressed TNBC cell proliferation and accelerated apoptosis. Hereto, we determined that DUXAP8 exerted an oncogenic role in TNBC.
Upregulation of lncRNAs may be induced by their upstream transcription factors. YY1 is known as a transcription activator for lncRNA,Citation26,Citation27 which is overexpressed in several human cancers and is correlated with poor prognosis.Citation28 However, it is unknown whether YY1 regulated DUXAP8 transcription till now. Therefore, we predicted and proved the binding of YY1 to DUXAP8 promoter. Importantly, YY1 positively regulated DUXAP8 expression through activating DUXAP8 transcription. Thus, we confirmed that YY1-induced upregulation of DUXAP8 in TNBC cells.
Competing endogenous RNA (ceRNA) has been proven to be correlated with tumorigenesis in many studies.Citation20 In our study, we identified the cytoplasmic localization of DUXAP8 in TNBC cells. MiR-29a-3p has been unveiled to involve in the ceRNA network by interacting with lncRNAs.Citation29 Similarly, we predicted and demonstrated that miR-29a-3p could bind with DUXAP8 in TNBC cells. The anti-tumor role of miR-29a-3p has been elucidated in hepatocellular carcinoma,Citation30 papillary thyroid carcinoma,Citation31 gastric cancer.Citation32 Consistently, we defined the tumor-suppressive role of miR-29a-3p in TNBC due to its suppressive effect on cell growth. These data indicated that DUXAP8 could bind with miR-29a-3p to regulate TNBC cell growth. Subsequently, we also verified that SAPCD2 was the downstream target of miR-29a-3p. According to previous reports, SAPCD2 possesses oncogenic potentials in colorectal carcinoma,Citation33 breast cancer.Citation34 In our current study, we determined that SAPCD2 silencing led to the growth inhibition of TNBC cells. Moreover, SAPCD2 can participate in the ceRNA pathway.Citation35 Here, we also determined the DUXAP8/miR-29a-3p/SAPCD2 ceRNA pathway in TNBC.
In conclusion, DUXAP8 is transcriptionally activated by YY1 and promotes TNBC cell proliferation but hampers apoptosis through modulating the miR-29a-3p/SAPCD2 axis. Our study revealed a novel ceRNA pathway in TNBC cell growth, which may help to unveil novel therapeutic target for TNBC patients.
However, lncRNAs usually regulate various biological functions through multiple downstream targets.Citation36–38 According to previous studies, lncRNAs can exert functions through regulating multiple downstream targets.Citation39,Citation40Citation41 In the current study, we only reported one downstream target of DUXAP8 in TNBC, which is a limitation of our current study. To enrich our research findings, we will explore multiple targets of DUXAP8 in the future study. Additionally, lack of clinical data is also a limitation of this study. Therefore, we will analyze the clinical significance of the ceRNA pathway in the next study.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We appreciate the supports of all our experimenter.
Additional information
Funding
References
- Mouh FZ, Mzibri ME, Slaoui M, Amrani M. Recent progress in triple negative breast cancer research. Asian Pac J Cancer Prev: APJCP. 2016;17:1595–1608. doi:10.7314/APJCP.2016.17.4.1595.
- Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer–deciphering the heterogeneity. Clin Cancer Res. 2014;20:782–790. doi:10.1158/1078-0432.CCR-13-0583.
- Sharma P. Biology and management of patients with triple-negative breast cancer. oncologist. 2016;21:1050–1062. doi:10.1634/theoncologist.2016-0067.
- Li X, Wu Z, Fu X, Han W. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21. doi:10.1016/j.mrrev.2014.04.002.
- Duguang L, Jin H, Xiaowei Q, Peng X, Xiaodong W, Zhennan L, Jianjun Q, Jie Y. The involvement of lncRNAs in the development and progression of pancreatic cancer. Cancer Biol Ther. 2017;18:927–936. doi:10.1080/15384047.2017.1385682.
- Huo X, Han S, Wu G, Latchoumanin O, Zhou G, Hebbard L, George J, Qiao L. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16:165. doi:10.1186/s12943-017-0734-4.
- Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK, Diehl JA, Howe PH. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115. doi:10.1038/ncb3595.
- Tang XJ, Wang W, Hann SS. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie. 2019;163:58–72. doi:10.1016/j.biochi.2019.05.010.
- Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi:10.1111/cas.13342.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi:10.1158/0008-5472.CAN-16-2634.
- Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99:494–501. doi:10.1002/cpt.355.
- Li L, Wang M, Mei Z, Cao W, Yang Y, Wang Y, Wen A. lncRNAs HIF1A-AS2 facilitates the up-regulation of HIF-1alpha by sponging to miR-153-3p, whereby promoting angiogenesis in HUVECs in hypoxia. Biomed Pharmacother = Biomedecine & Pharmacotherapie. 2017;96:165–172. doi:10.1016/j.biopha.2017.09.113.
- Zhao X, Hao S, Wang M, Xing D, Wang C. Knockdown of pseudogene DUXAP8 expression in glioma suppresses tumor cell proliferation. Oncol Lett. 2019;17:3511–3516. doi:10.3892/ol.2019.9994.
- Lian Y, Yang J, Lian Y, Xiao C, Hu X, Xu H. DUXAP8, a pseudogene derived lncRNA, promotes growth of pancreatic carcinoma cells by epigenetically silencing CDKN1A and KLF2. Cancer Commun (London, England). 2018;38:64. doi:10.1186/s40880-018-0333-9.
- Lin MG, Hong YK, Zhang Y, Lin BB, He XJ. Mechanism of lncRNA DUXAP8 in promoting proliferation of bladder cancer cells by regulating PTEN. Eur Rev Med Pharmacol Sci. 2018;22:3370–3377. doi:10.26355/eurrev_201806_15158.
- Jiang B, Hailong S, Yuan J, Zhao H, Xia W, Zha Z, Bin W, Liu Z. Identification of oncogenic long noncoding RNA SNHG12 and DUXAP8 in human bladder cancer through a comprehensive profiling analysis. Biomed Pharmacother = Biomedecine & Pharmacotherapie. 2018;108:500–507. doi:10.1016/j.biopha.2018.09.025.
- Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao S, et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. 2015;32:693–706. doi:10.1016/j.devcel.2015.01.028.
- Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi:10.1016/j.molcel.2013.07.017.
- Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi:10.1186/s12943-017-0680-1.
- Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi:10.3748/wjg.v21.i41.11680.
- Marin-Bejar O, Mas AM, Gonzalez J, Martinez D, Athie A, Morales X, Galduroz M, Raimondi I, Grossi E, Guo S, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017;18:202. doi:10.1186/s13059-017-1331-y.
- Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol. 2014;6:181–191. doi:10.1093/jmcb/mju013.
- Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi:10.1038/ncb3295.
- Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res: MCR. 2015;13:330–338. doi:10.1158/1541-7786.MCR-14-0251.
- Du C, Wang HX, Chen P, Chen CH. STAT3-induced upregulation of lncRNA DUXAP8 functions as ceRNA for miR-577 to promote the migration and invasion in colorectal cancer through the regulation of RAB14. Eur Rev Med Pharmacol Sci. 2019;23:6105–6118. doi:10.26355/eurrev_201907_18424.
- Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X, Xu S, Pang D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res: CR. 2019;38:418. doi:10.1186/s13046-019-1421-7.
- Yan Z, Yang Q, Xue M, Wang S, Hong W. YY1-induced lncRNA ZFPM2-AS1 facilitates cell proliferation and invasion in small cell lung cancer via upregulating of TRAF4. Cancer Cell Int. 2020;20:108.
- Zhou H, Li J, Zhang Z, Ye R, Shao N, Cheang T, Wang S. RING1 and YY1 binding protein suppresses breast cancer growth and metastasis. Int J Oncol. 2016;49:2442–2452. doi:10.3892/ijo.2016.3718.
- He H, Wang N, Yi X, Tang C, Wang D. Long non-coding RNA H19 regulates E2F1 expression by competitively sponging endogenous miR-29a-3p in clear cell renal cell carcinoma. Cell Biosci. 2017;7:65. doi:10.1186/s13578-017-0193-z.
- Wang X, Liu S, Cao L, Zhang T, Yue D, Wang L, Ping Y, He Q, Zhang C, Wang M, et al. miR-29a-3p suppresses cell proliferation and migration by downregulating IGF1R in hepatocellular carcinoma. Oncotarget. 2017;8:86592–86603. doi:10.18632/oncotarget.21246.
- Ma Y, Sun Y. miR-29a-3p inhibits growth, proliferation, and invasion of papillary thyroid carcinoma by suppressing NF-κB signaling via direct targeting of OTUB2. Cancer Manag Res. 2019;11:13–23. doi:10.2147/CMAR.S184781.
- Bai F, Jiu M, You Y, Feng Y, Xin R, Liu X, Mo L, Nie Y. miR‑29a‑3p represses proliferation and metastasis of gastric cancer cells via attenuating HAS3 levels. Mol Med Rep. 2018;17:8145–8152. doi:10.3892/mmr.2018.8896.
- Luo Y, Wang L, Ran W, Li G, Xiao Y, Wang X, Zhao H, Xing X. Overexpression of SAPCD2 correlates with proliferation and invasion of colorectal carcinoma cells. Cancer Cell Int. 2020;20:43.
- Zhang Y, Liu JL, Wang J. SAPCD2 promotes invasiveness and migration ability of breast cancer cells via YAP/TAZ. Eur Rev Med Pharmacol Sci. 2020;24:3786–3794. doi:10.26355/eurrev_202004_20844.
- Jia X, Niu P, Xie C, Liu H. Long noncoding RNA PXN-AS1-L promotes the malignancy of nasopharyngeal carcinoma cells via upregulation of SAPCD2. Cancer Medicine. 2019;8:4278–4291. doi:10.1002/cam4.2227.
- Wang X, Lai Q, He J, Li Q, Ding J, Lan Z, Gu C, Yan Q, Fang Y, Zhao X, et al. LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci. 2019;16:51–59. doi:10.7150/ijms.27359.
- Su W, Feng S, Chen X, Yang X, Mao R, Guo C, Wang Z, Thomas DG, Lin J, Reddy RM, et al. Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 2018;78:3207–3219. doi:10.1158/0008-5472.CAN-18-0222.
- Li Z, Hong S, Liu Z. LncRNA LINC00641 predicts prognosis and inhibits bladder cancer progression through miR-197-3p/KLF10/PTEN/PI3K/AKT cascade. Biochem Biophys Res Commun. 2018;503:1825–1829. doi:10.1016/j.bbrc.2018.07.120.
- Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29:e95.
- Zhang E, Han L, Yin D, He X, Hong L, Si X, Qiu M, Xu T, De W, Xu L, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101. doi:10.1093/nar/gkw1247.
- Tang R, Chen J, Tang M, Liao Z, Zhou L, Jiang J, Hu Y, Liao Q, Xiong W, Tang Y, et al. LncRNA SLCO4A1-AS1 predicts poor prognosis and promotes proliferation and metastasis via the EGFR/MAPK pathway in colorectal cancer. Int J Biol Sci. 2019;15:2885–2896. doi:10.7150/ijbs.38041.
