ABSTRACT
In recent years, FGD5 antisense RNA 1 (FGD5-AS1) was confirmed to be the long non-coding RNAs (lncRNAs) that could accelerate the development of multiple cancers. Nevertheless, specific biological functions and latent mechanism of FGD5-AS1 were not yet clear in pancreatic cancer (PC). This research was aimed to search the functions of FGD5-AS1 on the PC progression. The expression of FGD5-AS1 in PC cells was tested by using RT-qPCR assay. Colony formation assay, EdU assay, flow cytometry assay and transwell assay as well as western blot were adopted to test the cell abilities of proliferation, apoptosis and migration, separately. Furthermore, RIP experiment and pull down assay were applied for validating the correlation FGD5-AS1, miR-520a-3p and KIAA1522. As a result, the abnormal high expression of FGD5-AS1 was observed in PC cells. And cell proliferative and migratory abilities could be restrained via FGD5-AS1 depletion. Moreover, FGD5-AS1 was proven to combine with miR-520a-3p directly. It was also confirmed that KIAA1522 could be targeted by miR-520a-3p. Rescue assay results indicated that overexpressed KIAA1522 could reverse the repressive function of silencing FGD5-AS1 on PC progression. Taken together, FGD5-AS1 accelerated cell proliferation and migration via sponging miR-520a-3p and upregulating KIAA1522.
Introduction
Pancreatic cancer (PC) is a common gastrointestinal cancer in the world with high invasiveness.Citation1 It has been considered to be one of the most deadly malignant tumors since the current five-year survival rate of patients is far below 10%.Citation2 Due to limited medical conditions and technology, early PC is difficult to diagnose. Therefore, the majority of patients with PC had local metastasis when they were initially diagnosed, which greatly elevated the difficulty of treatment and declined the survival rate of patients.Citation3,Citation4 Accordingly, it is extremely necessary to explore a new and effective therapeutic target to improve the survival rate of patients.
Long non-coding RNAs (lncRNAs) have been identified as the RNAs which compose of more than 200 nucleotides in length without the capacity of coding protein.Citation5 Previously, lncRNAs were regarded as the transcriptional noise since they did not take part in encoding proteins, and it was accordingly considered that lncRNAs did not have the biological function.Citation6 Until in recent years, a flow of researches have suggested that various lncRNAs exert critical roles in biological process, such as cell proliferation, apoptosis and migration.Citation7 Even an increasing number of studies demonstrated that lncRNA could take part in modulating tumorigenesis and development of multiple cancers, and PC was included.Citation8 For instance, ZEB2-AS1 accelerated the proliferative and invasive capabilities of PC via regulation of the miR-204/HMGB1 axis.Citation9 The lncrna-TUG1/EZH2 axis was reporter to expedite cell growth and EMT progression of PC via sponging miR-382.Citation10 FGD5 antisense RNA 1 (FGD5-AS1) is an lncRNA and its carcinogenic nature has been discovered in several cancers. For instance, FGD5-AS1 facilitated cell migration and invasion of colorectal cancer by the upregulation of CDCA7 via sponging miR-302e.Citation11 Moreover, FGD5-AS1 accelerated cell growth of non-small cell lung cancer cell proliferation through sponging miR-107 to upregulate FGFRL1.Citation12 However, the molecule mechanism of FGD5-AS1 in PC was not clear.
MicroRNAs (miRNAs) have also been verified as the critical modulators in multiple cancers.Citation13 They are a kind of endogenous small noncoding RNA molecules with 19–26 nucleotides in length, and can negatively regulate their target gene expression.Citation14,Citation15 For example, miR-135b-5p could accelerate cell migration and invasion of PC via targeting NR3C2.Citation16 On the contrast, miR-9-5p could suppress cell proliferation and invasion of PC through targeting GOT1.Citation17 Nevertheless, the function of miR-520a-3p in PC remains to be investigated.
In our study, we investigated the function and the regulatory mechanism of FGD5-AS1 as well as its downstream genes miR-520a-3p and KIAA1522 in the progression of PC, which might provide a promising target for TNBC therapy.
Materials and methods
Cell lines
PC cell lines (Capan-1, BxPc3, PANC-1 and SW1990) purchased from ATCC (Manassas, VA, USA) and the normal human pancreatic duct epithelial cell line (HPDE-C7) purchased from Huzhen Biological Technology Co., Ltd (Shanghai, China) were selected for conducting our assays. Capan-1 cells were cultivated in Iscove’s Modified Dulbecco’s medium (GIBCO, Grand Island, NY, USA). BxPc3 and HPDE-C7 were cultivated in RPMI-1640 medium (GIBCO). PANC-1 cells were cultured in Dulbecco’s Modified Eagle’s medium (GIBCO). SW1990 cells were cultured in Leibovitz’s L-15 medium (GIBCO). All mediums were supplemented with 10% fetal bovine serum (FBS; GIBCO) and 1% p/s and deposited at 37°C with 5% CO2.
Cell transfection
PANC-1 and SW1990 cells were incubated in 6-well plates with plasmids utilizing Lipofectamine 3000 (Invitrogen) for 48 hours. The specific shRNAs targeting FGD5-AS1 or KIAA1522 and their negative control (NC) were devised and synthesized from GenePharma (Shanghai, China). Besides, the miR-520a-3p inhibitor/mimics and NC inhibitor/mimics were structured via Ribobio (Guangzhou, China). Bio-repeats were run in triplicate.
RT-qPCR
Total RNA of PANC-1 and SW1990 cells were subjected to isolation using TRIzol Reagent (Invitrogen, Carlsbad CA, USA). Then they were synthesized to complementary DNA by the utilization of PrimeScript™ RT Master Mix kit (TaKaRa BIO, Shiga, Japan). After that, gene expression was tested utilizing SYBR Green PCR Kit (Takara) on ABI Prism 7900HT sequence detector (Applied Biosystems, Foster City, CA, USA). In the end, we performed the 2−ΔΔCt method to calculate. GAPDH or U6 served as control. Bio-repeats were run in triplicate.
Colony formation
Clonogenic cells were seeded in the 6-well-plate at the density of 600 cells per well and cultivated for 14 days. After the cultivation was finished, cells were subjected to fixation by 4% paraformaldehyde, and then stained with 0.1% crystal violet. Ultimately, the colonies were calculated manually. Bio-repeats were run in triplicate.
EdU staining assay
Based on the protocols of the supplier, BeyoClick™ EdU Cell Proliferation Kit (Beyotime, Shanghai, China) was utilized to implement this assay. 1 × 104 cells were put in the 96-well-plate and treated for 2 hours by EdU kit at room temperature. Then DAPI stained cells for 5 minutes. Finally, cells were observed via fluorescence microscope (Olympus, Tokyo, Japan). Bio-repeats were run in triplicate.
Transwell migration assay
Cell migratory capability was measured utilizing a transwell chamber (8 μm pore size; Costar, Cambridge, MA, USA). We gather the transfected cells (2 × 104) and then put them in the upper chamber with a serum-free medium. Besides, the lower chamber was supplemented with a complete medium. Twenty-four hours later, the cells migrated to the lower surface were fixed and dyed by 4% paraformaldehyde and crystal violet separately. Finally, the optical microscope (Olympus) was utilized to observe the cells. Bio-repeats were run in triplicate.
Flow cytometry
In accordance with the protocols of the supplier, Annexin V-FITC/PI Apoptosis kit (BD Biosciences, San Jose, CA) was utilized to perform this assay. Cells were double-stained in Binding Buffer for 10 minutes and then observed via FACS Calibur flow cytometer (BD Biosciences). Bio-repeats were run in triplicate.
Western blot
The total cell protein specimens lysed in RIPA lysis buffer were gathered and isolated on 10% SDS-PAGE. Next they were transferred onto PVDF membranes. Following, 5% nonfat milk was applied for blockade the membranes which were then cultured for a whole night at 4°C with the primary antibodies against GAPDH (ab8245, Abcam) and Bax (ab32503, Abcam), Bcl2 (ab32124, Abcam), MMP2 (ab92536, Abcam), MMP9 (ab38898, Abcam) and KIAA1522 (ab122203, Abcam). After that, we cultured the membranes with the HRP-tagged secondary antibody for at least 2 hours. In the end, the signals were tested via ECL substrate (Pierce, Rockford, IL).
FISH
PANC-1 and SW1990 cells were subjected to fixation and then rinsed in PBS. Following, they were cultivated with the FGD5-AS1-specific FISH probe (Ribobio) in a hybridization buffer. After supplementing the DAPI solution, the Olympus fluorescence microscope was applied for analysis. Bio-repeats were run in triplicate.
Subcellular fractionation assay
PARIS™ Kit (Invitrogen) was utilized to perform this assay in accordance with the user guide. PANC-1 and SW1990 cells were treated in cell fractionation buffer for gathering cytoplasm. Following, they were subjected to centrifugation. The nucleus of cells was gathered through cell disruption buffer. In the end, RT-qPCR was applied for analyzing the content of FGD5-AS1. GAPDH and U6 served as control. Bio-repeats were run in triplicate.
RNA immunoprecipitation (RIP)
Cells were firstly treated in RIP lysis buffer and then Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was applied for carrying out this assay. Then the magnetic beads which bound with Ago2 antibody or control IgG antibody were treated for collecting the immunoprecipitates. And the RT-qPCR was conducted to analyze the RNA extraction. Bio-repeats were run in triplicate.
RNA pull down
The cell protein lysate obtained from RIPA lysis buffer was utilized to treat with Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA) for this assay in line with the user guide. Magnetic beads were then added to specimens for obtaining the RNA-protein mixture. Finally, RT-qPCR was applied for detecting RNA enrichment. Bio-repeats were run in triplicate.
Luciferase reporter assay
FGD5-AS1 or KIAA1522 fragments covering miR-520a-3p binding sites (wild-type and mutant) were inserted into the pmirGLO luciferase vector (Promega, Madison, WI). Following, cells were subjected to co-transfection with these vectors and miR-520a-3p mimics or NC mimics. Forty-eight hours later, the luciferase activity was measured by the Luciferase Reporter Assay System (Promega). Bio-repeats were run in triplicate.
Statistical analyses
The assays in our study were conducted in triplicate. GraphPad PRISM 6 (GraphPad, San Diego, CA, USA) was utilized for analysis and data was displayed as means ± SD. Besides, the difference of group was detected through Student’s t-test or one-way ANOVA. P < .05 served as the threshold of statistical significance.
Results
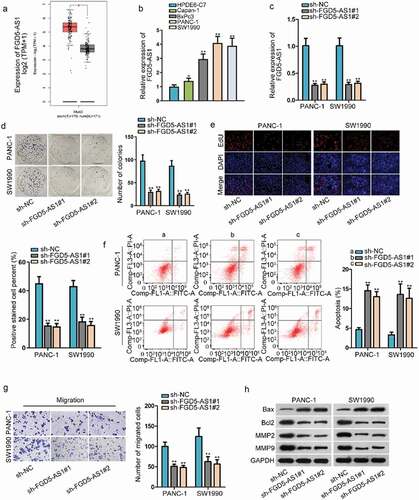
FGD5-AS1 expression is abnormal high in PC cells and plays carcinogenic effect
Increasing researches have confirmed that how lncRNAs exert their function depends on their expression level in cancer cells. Thus, we evaluated FGD5-AS1 expression in PAAD (pancreatic adenocarcinoma) tissues through TCGA database. After utilizing the GEPIA (http://gepia.cancer-pku.cn/), we found that FGD5-AS1 expression in PAAD tissues was obviously higher than that in normal tissues (). Then we further tested its expression in PC cell lines (Capan-1, BxPc3, PANC-1 and SW1990) through RT-qPCR. In accordance with the outcomes, we discovered an elevated expression of FGD5-AS1 in PC cells in comparison to normal human pancreatic duct epithelial cell line (HPDE-C7) (). Therefore, we initially suspected its carcinogenic nature in PC. Later, for the sake of silencing FGD5-AS1 expression, we transfected the specific shRNA targeting to FGD5-AS1 in PANC-1 and SW1990 cells which were selected for conducting the following experiments because of the higher FGD5-AS1 expression. Through observation of RT-qPCR, the interference efficiency of FGD5-AS1 displayed a desirable effect (). Then a chain of loss-of-functional assays was carried out for detecting FGD5-AS1 function on PC cells. It was indicated through colony formation assay that cell proliferation was restrained in sh-FGD5-AS1-tarnsfected PANC-1 and SW1990 cells since the number of colonies was declined in transfected cells after incubation of two weeks (). Then, we observed in EdU assay that the EdU positive cells were decreased after silencing FGD5-AS1 in cells, which further confirmed that cell proliferation could be weakened by FGD5-AS1 depletion (). Next, cell apoptosis was detected by flow cytometry assay and the results indicated that cell apoptosis rate was elevated when FGD5-AS1 was knocked down in cells (). Following, transwell assay was conducted to measure cell migratory capability and the results illustrated that the number of cells that succeeded in migrating to the lower chamber was declined in sh-FGD5-AS1-transfection groups, suggesting knockdown of FGD5-AS1 played the inhibitory role on cell migratory capability (). Finally, the western blot assay estimated protein levels of apoptosis-related proteins (Bax and Bcl2) and migration-related proteins (MMP2 and MMP9), and we discovered that the protein level of Bax was increased after FGD5-AS1 was inhibited in cells while the protein level of Bcl2, MMP2 and MMP9 was reduced (). Taken together, we proved that FGD5-AS1 expression was extremely high in PC cells and cell proliferative and migratory capabilities could be enhanced.
Figure 1. FGD5-AS1 expression is abnormal high in PC cells and plays carcinogenic effect
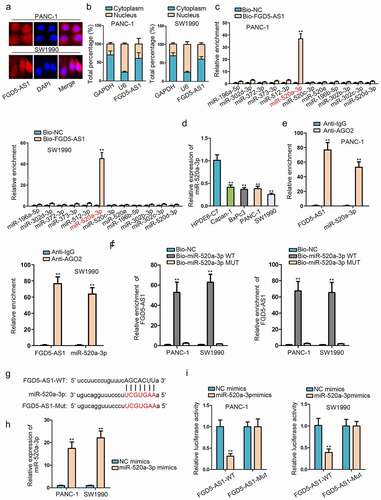
FGD5-AS1 acts as a sponge of miR-520a-3p
After validating the carcinogenic properties of FGD5-AS1, we began to explore the regulatory mechanism of FGD5-AS1 in PC cells. Recently, a competing endogenous RNA (ceRNA) network has been frequently discovered in human cancers, including PC.Citation9 Thus, we wondered to whether FGD5-AS1 held the potential to be a ceRNA. Cytoplasmic lncRNA is considered to have the possibility to exert the ceRNA function, so we performed the subcellular fractionation and FISH experiments to test the distribution of FGD5-AS1 in PC cells. On the basis of the results, we affirmed that FGD5-AS1 was a cytoplasmic lncRNA, signifying the ceRNA possibility of FGD5-AS1 (). Then we utilized starBase (http://starbase.sysu.edu.cn/index.php) to discover the miRNA that might combine with FGD5-AS1. With the strict condition (CLIP-Data ≥5, Degradome-Data ≥2), we found 50 miRNAs had the binding possibility with FGD5-AS1. Among them, three miRNAs (miR-142-3p, miR-302e, miR-520b) has been investigated with FGD5-AS1 in some cancers by others, thus we selected the remainder 12 miRNAs for keeping the novelty of our study. For screening, we utilized the biotinylated FGD5-AS1 probe to conduct an RNA pull down assay and the result indicated that miR-520a-3p enrichment was most abundant in the biotinylated FGD5-AS1 group (). Accordingly, miR-520a-3p was selected for the later experiments. Then, RT-qPCR detected the expression of miR-520a-3p in PC cells. As we expected, miR-520a-3p was lowly expressed in PC cells (). Following, a series of mechanism assay was conducted to verify the binding situation of miR-520a-3p and FGD5-AS1. RIP assay indicated that miR-520a-3p and FGD5-AS1 enriched in the AGO2 group, indicating that they existed in the RISC complex (). Then RNA pull down assay further proved that FGD5-AS1 was pulled down by biotinylated miR-520a-3p-WT, while biotinylated miR-520a-3p-Mut did not work on FGD5-AS1, which implied that FGD5-AS1 could bind to miR-520a-3p (). Furthermore, we obtained the binding sites of miR-520a-3p and FGD5-AS1 through starBase (). After we overexpressed miR-520a-3p by transfecting miR-520a-3p mimics into cells (), we performed luciferase reporter assay to validate the binding sites. The results indicated that the luciferase activity of FGD5-AS1-WT was decreased by overexpressed miR-520a-3p, while that of FGD5-AS1-Mut almost unchanged (). Overall, we got the conclusion that FGD5-AS1 could sponge miR-520a-3p by acting as a ceRNA in PC cells.
Figure 2. FGD5-AS1 acts as a sponge of miR-520a-3p
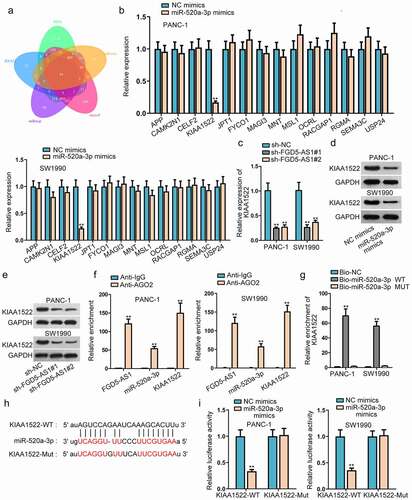
KIAA1522 is targeted by miR-520a-3p in PC cells
For the sake of further exploring the ceRNA network in PC cells, we began to investigate the downstream of miR-520a-3p. Through the prediction of PITA, RNA22, miRmap, microT and miRanda database under the strict condition (CLIP-Data ≥5), we found that there were 14 messenger RNAs (mRNAs) that might have the possibility of binding with miR-520a-3p (). Then we performed the RT-qPCR assay to screen out a suitable mRNA. In accordance with the experimental outcomes, we discovered that only KIAA1522 expression was declined when we overexpressed miR-520a-3p in PANC-1 and SW1990 cells, thus we selected KIAA1522 to conduct the further experiments (). Accordingly, the correlation between FGD5-AS1 and miR-520a-3p was detected through RT-qPCR. The results validated that the expression level of KIAA1522 were decreased in sh-FGD5-AS1-transfected cells, which illustrated that KIAA1522 was positively associated with FGD5-AS1 (). Western blot assay further verified that KIAA1522 protein level was declined via miR-520a-3p upregulation or FGD5-AS1 depletion (, e). Next, RIP assay was conducted and we discovered that FGD5-AS1, miR-520a-3p and KIAA1522 were enriched in Ago2 group, suggesting they coexisted in the RISC complex (). RNA pull down assay further validated that biotinylated miR-520a-3p-WT obviously pulled down KIAA1522 (). Moreover, through searching starBase, we discovered the binding sites of KIAA1522 and miR-520a-3p (). Then luciferase reporter assay was carried out to validate the binding of them and we observed that upregulation of miR-520a-3p restrained the luciferase activity of KIAA1522-WT, while it could not change the luciferase activity of KIAA1522-Mut, which further proved the combination of KIAA1522 and miR-520a-3p (). In a word, KIAA1522 was the downstream target gene of miR-520a-3p in PC.
Figure 3. KIAA1522 is targeted by miR-520a-3p in PC cells
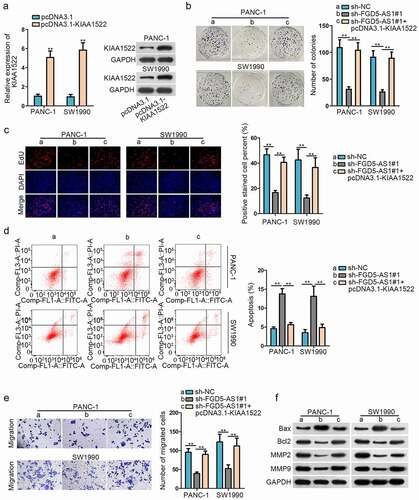
FGD5-AS1 expedites cell proliferation and migration of PC via upregulation of KIAA1522
In the end, we wondered whether FGD5-AS1 regulates the proliferation of PC through KIAA1522. Firstly, KIAA1522 was overexpressed in PANC-1 and SW1990 cells through transfection with pcDNA3.1-KIAA1522 and RT-qPCR result indicated that the expression of KIAA1522 was effectively elevated by that (). Western blot assay further proved this result (). Then colony formation assay and EdU assay displayed that the inhibited cell proliferation caused by silenced FGD5-AS1 could be recovered by overexpressed KIAA1522 (, c). In the contrary, cell apoptosis rate elevated by FGD5-AS1 depletion was restrained again by KIAA1522 overexpression (). Then, transwell assay demonstrated that cell migratory capability was hampered by FGD5-AS1 depletion, but then overexpressed KIAA1522 counteracted this effect (). Finally, we observed from western blot assay that protein levels of Bax was increased in sh-FGD5-AS1-transfected cells, but then reduced by co-transfection with pcDNA3.1-KIAA1522, while the overexpression of KIAA1522 offset the inhibitory function of silencing FGD5-AS1 on the protein level of Bcl2, MMP2 and MMP9 (). Overall, we concluded that FGD5-AS1 expedited the progression of PC through upregulating KIAA1522.
Figure 4. FGD5-AS1 expedites cell proliferation and migration of PC via upregulation of KIAA1522
Discussion
PC is a malignant tumor with high mortality rate, but no effective treatment has been developed. Therefore, it is urgent to find effective therapeutic targets. In recent years, an increasing number of researches have illustrated that there are various lncRNAs that are singularly expressed in human cancers and the aberrant expression has been regarded as the crucial index for lncRNA to exert a regulatory role in cancer. For example, LINC00261 was verified to have a low expression in colon cancer cells and it could suppress this cancer progression through sponging miR-324-3p and inactivating the Wnt/β-catenin pathway.Citation18 It was also reported that SNHG14 was highly expressed in cervical cancer cells and facilitated cell growth through the regulation of miR-206/YWHAZ.Citation19 That it can be seen that how lncRNAs exert the function in cancers depend on their expression in cancer cells. LncRNA FGD5-AS1 has been confirmed to be highly expressed in colorectal cancer cellsCitation11 and non-small cell lung cancer cellsCitation12 and exert the carcinogenic effect. Thus, we suspected whether FGD5-AS1 exerted the similar function in PC. Through the prediction of bioinformatics tools and RT-qPCR of cancer cells, FGD5-AS1 was discovered to be highly expressed in PAAD tissues and PC cells in contrast of the normal cells and tissues. Then a chain of loss-of-functional assays was applied for evaluation of FGD5-AS1’s biological function. As we expected, inhibition of FGD5-AS1 could restrain cell proliferative and migratory capability but accelerate cell apoptosis. Accordingly, we proved that FGD5-AS1 served as the oncogene in PC cells.
In recent years, more and more existing literatures have clarified that plenty of lncRNAs can act as a ceRNA to boost the progression of assorted cancers, such as cervical cancer,Citation20 hepatocellular carcinoma,Citation21 gastric cancerCitation22 and PC.Citation9 CeRNA network refers to that lncRNA can regulate the mRNA expression through acting as a sponge of miRNA at the post-transcriptional level, so as to exert the regulatory function in cancers.Citation23,Citation24 Therefore, we wondered whether FGD5-AS1 mediated PC progression through acting as a ceRNA. In our study, we firstly determined that the distribution of FGD5-AS1 was mostly in the cytoplasm, suggesting the regulation at the post-transcriptional level. Accordingly, we searched the bioinformatics tools and conducted mechanism assays, and the results verified that miR-520a-3p could be sponged by FGD5-AS1. Also, miR-520a-3p was lowly expression in PC cells. Previously, reports indicated the repressing function of miR-520a-3p in breast cancer through CCND1 and CD44.Citation25 Besides, miR-520a-3p expression was low in non-small cell lung cancer cells and restrained cell proliferation in it.Citation26 Moreover, the low expression and repressive function of miR-520a-3p was also proven in papillary thyroid carcinoma.Citation27
KIAA1522 has been verified to be closely associated with different kinds of cancers. For instance, KIAA1522 upregulation could accelerate tumorigenicity of esophageal cancer cells via enhancing the ERK activity.Citation28 KIAA1522 was confirmed to be targeted by miR-125b-5p and played the carcinogenic effect in breast cancer.Citation29 Furthermore, overexpression of KIAA1522 was verified to be utilized as an independent biomarker for predication of bad survival of patients with non-small cell lung cancer.Citation30 In our research, KIAA1522 served as the target gene of miR-520a-3p, was discovered through bioinformatics tools. Similar as those previous researches, KIAA1522 played the oncogene function since the rescue assays demonstrated that overexpression of KIAA1522 could reverse the repressive function of FGD5-AS1 depletion on PC progression.
In a word, our study validated that FGD5-AS1 expedited the progression of PC via sponging miR-520a-3p to upregulate KIAA1522 expression. This discovery implied that the ceRNA network of FGD5-AS1 existed in PC, which might provide a promising target for TNBC therapy.
Acknowledgments
We sincerely appreciate all lab members.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ilic M, Ilic I. Epidemiology of pancreatic cancer. World Journal of Gastroenterology. 2016;22(44):9694–9705. doi:10.3748/wjg.v22.i44.9694.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442.
- Ansari D, Tingstedt B, Andersson B, Holmquist F, Sturesson C, Williamsson C, Sasor A, Borg D, Bauden M, Andersson R. Pancreatic cancer: yesterday, today and tomorrow. Future Oncology (London, England). 2016;12(16):1929–1946. doi:10.2217/fon-2016-0010.
- Goral V. Pancreatic cancer: pathogenesis and diagnosis. Asian Pacific Journal of Cancer Prevention: APJCP. 2015;16(14):5619–5624. doi:10.7314/APJCP.2015.16.14.5619.
- Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol. 2017;1008:283–323.
- Chen X, Sun Y, Cai R, Wang G, Shu X, Pang W. Long noncoding RNA: multiple players in gene expression. BMB Rep. 2018;51(6):280–289. doi:10.5483/BMBRep.2018.51.6.025.
- Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014. doi:10.4161/21541272.2014.944014.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634.
- Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y, Zhang G. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int J Biol Macromol. 2018;116:545–551. doi:10.1016/j.ijbiomac.2018.05.044.
- Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The Lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMT phenotype formation through sponging Mir-382. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2017;42(6):2145–2158. doi:10.1159/000479990.
- Li D, Jiang X, Zhang X, Cao G, Wang D, Chen Z. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. In Vitro Cell Dev Biol Anim. 2019;55(8):577–585. doi:10.1007/s11626-019-00376-x.
- Fan Y, Li H, Yu Z, Dong W, Cui X, Ma J, Li S. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Biosci Rep. 2020;40(1):BSR20193309.
- Qadir MI, Faheem A. miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27(3):197–204. doi:10.1615/CritRevEukaryotGeneExpr.2017019494.
- Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi:10.1016/j.critrevonc.2015.10.003.
- Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods in Molecular Biology (Clifton, NJ). 2017;1509:1–10.
- Zhang Z, Che X, Yang N, Bai Z, Wu Y, Zhao L, Pei H. miR-135b-5p promotes migration, invasion and EMT of pancreatic cancer cells by targeting NR3C2. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2017;96:1341–1348. doi:10.1016/j.biopha.2017.11.074.
- Wang J, Wang B, Ren H, Chen W. miR-9-5p inhibits pancreatic cancer cell proliferation, invasion and glutamine metabolism by targeting GOT1. Biochem Biophys Res Commun. 2019;509(1):241–248. doi:10.1016/j.bbrc.2018.12.114.
- Yan D, Liu W, Liu Y, Luo M. LINC00261 suppresses human colon cancer progression via sponging miR-324-3p and inactivating the Wnt/β-catenin pathway. J Cell Physiol. 2019;234(12):22648–22656. doi:10.1002/jcp.28831.
- Ji N, Wang Y, Bao G, Yan J, Ji S. LncRNA SNHG14 promotes the progression of cervical cancer by regulating miR-206/YWHAZ. Pathol Res Pract. 2019;215(4):668–675. doi:10.1016/j.prp.2018.12.026.
- Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29(6):e95. doi:10.3802/jgo.2018.29.e95.
- Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi:10.1186/s12943-017-0680-1.
- Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, Zhao Q, Wu D, Gong W, Du M. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87. doi:10.1186/s12943-018-0829-6.
- Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334.
- Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. TheScientificWorldJournal. 2014;2014:896206. doi:10.1155/2014/896206.
- Li J, Wei J, Mei Z, Yin Y, Li Y, Lu M, Jin S. Suppressing role of miR-520a-3p in breast cancer through CCND1 and CD44. Am J Transl Res. 2017;9(1):146–154.
- Lv X, Li CY, Han P, Xu XY. MicroRNA-520a-3p inhibits cell growth and metastasis of non-small cell lung cancer through PI3K/AKT/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(8):2321–2327. doi:10.26355/eurrev_201804_14822.
- Bi CL, Zhang YQ, Li B, Guo M, Fu YL. MicroRNA-520a-3p suppresses epithelial-mesenchymal transition, invasion, and migration of papillary thyroid carcinoma cells via the JAK1-mediated JAK/STAT signaling pathway. J Cell Physiol. 2019;234:4054–4067. doi:10.1002/jcp.27199.
- Xie ZH, Yu J, Shang L, Zhu YQ, Hao JJ, Cai Y, Xu X, Zhang Y, Wang MR. KIAA1522 overexpression promotes tumorigenicity and metastasis of esophageal cancer cells through potentiating the ERK activity. Onco Targets Ther. 2017;10:3743–3754. doi:10.2147/OTT.S142610.
- Li Y, Wang Y, Fan H, Zhang Z, Li N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. 2018;504(1):277–282. doi:10.1016/j.bbrc.2018.08.172.
- Liu YZ, Yang H, Cao J, Jiang YY, Hao JJ, Xu X, Cai Y, Wang MR. KIAA1522 is a novel prognostic biomarker in patients with non-small cell lung cancer. Sci Rep. 2016;6(1):24786. doi:10.1038/srep24786.
