ABSTRACT
Emerging documents revealed that E2 enzyme family has been implicated in regulating the progression of numerous human cancers. Ubiquitin-conjugating enzyme E2 J1 (UBE2J1), a member of E2 enzyme family, has been reported to participate in the biological process of medulloblastoma, while little is known about its functionality in endometrial cancer (EC). Gene expression at the mRNA and protein levels were identified using RT-qPCR and western blot analysis, separately. The alteration on cell proliferation, adhesion, migration, invasion, and epithelial-mesenchymal transition (EMT) process was determined through 5-Ethynyl-2ʹ-deoxyuridine, cell adhesion, wound healing and transwell assays as well as western blot analysis. The role of UBE2J1 in xenograft tumor in mice was determined. Luciferase reporter and chromatin immunoprecipitation assays were conducted to reveal the undering mechanism of UBE2J1. Our results indicated that UBE2J1 displayed high level in EC tissues and cells and predicted poor prognosis of EC patients. In addition, UBE2J1 depletion inhibited cell proliferation, adhesion, motion, EMT process invitro, and repressed tumor growth invivo. Rescue assays manifested that ethyl 2-amino-6-chloro-4-(1-cyano-2-ethoxy-2-oxoethyl)-4H-chromene-3-carboxylate treatment reversed the effects of UBE2J1 on PI3K/AKT pathway activation and malignant phenotypes of EC cells. Finally, zinc finger X-chromosomal protein (zfx), with high expression in EC tissues, was verified to activate UBE2J1 transcription by binding to UBE2J1 promoter. In conclusion, all facts signified that zfx-induced upregulation of UBE2J1 accelerated the progression of EC via regulating the PI3K/AKT signaling pathway, which suggested that UBE2J1 might be of great significance in probing into the underlying therapeutic strategies of EC.
KEYWORDS:
Introduction
Endometrial carcinoma (EC), as a kind of gynecologic cancer, ranks the fifth in cancer-associated malignancies in females. EC has been widely concerned on account of its ever-increasing incidence.Citation1 Known factors including diets, diabetes, female hormone, and genetic factors were reported to be closely associated with the initiation and progression of EC.Citation2–4 Clinical outcomes revealed that hysterectomy, radiotherapy and lymphadenectomy significantly reduced the mortality of patients with early EC, while the survival rates of patients with advanced metastatic EC remain quite unsatisfactory.Citation5 The 5-year survival rate of EC patients at stage I is 96%, while the 5-year survival rate of patients at stage IV is only 17%. Therefore, elucidating EC pathogenesis and understanding the underlying mechanisms of EC development have become the first issue we need to address.
E2 enzyme family consists of approximately 40 ubiquitin E2 family genes and many of which are involved in human diseases.Citation6 Different E2 enzymes affect different kinds of ubiquitin linkages.Citation7,Citation8 Ubiquitin-conjugating enzyme E2 J1 (UBE2J1), also known as Ubc6, a member of the E2 ubiquitin-conjugating enzyme family, is located on the cytoplasmic surface of the endoplasmic reticulum.Citation9 Currently, it has been reported that UBE2J1 is engaged in the progression of protein degradation in several investigations. For example, UBE2J1 plays a critical role in degrading misfolded proteins in endoplasmic reticulum.Citation10 The c-IAP1/TRAF2 complex translocated to Ubc6-containing section and TRAF2 ubiquitination was caused by TNF-α.Citation11 Additionally, it has been reported that UBE2J1 is implicated in the progression of human carcinomas. A literature has pointed out that UBE2J1 participates in the progression of human medulloblastoma.Citation12 UBE2J1 is identified to be upregulated in prostatic cancer (PCa) and it serves as a prognosis biomarker for PCa patients.Citation13 However, the role and potential mechanism of UBE2J1 in EC remain completely unknown.
It is well-recognized that PI3K/AKT often displays activation status during cancer initiation and development. In addition, the PI3K/AKT signaling pathway is associated with plenty of cancer cell activities.Citation14,Citation15 For instance, a study has manifested that lncRNA KCNQ1OT1 promotes the proliferative and migratory abilities of colorectal cancer cells through modulating the PI3K/AKT pathway.Citation16 Another document proposed that lncRNA LINC01305 deficiency suppresses EMT process via the PI3K/Akt signaling in lung cancer.Citation17 Moreover, lncRNA LINP1 exerts the oncogene role in EC development with the regulation of the PI3K/AKT signaling pathway.Citation18 All these fully certified the crucial function of PI3K/AKT signaling pathway in cancers, including EC. It has been reported that Ubc9, another protein of ubiquitin-conjugating enzyme family, can activate PI3K/AKT.Citation19,Citation20 However, whether UBE2J1 modulates the PI3K/AKT signaling pathway to exert certain function in EC has not been elucidated.
Our research aimed to investigate biological role and underlying molecular mechanism of UBE2J1 in EC. Our findings demonstrated that zfx-induced upregulation of UBE2J1 elicits the cancerogenic function via regulating PI3K/AKT, which may provide further insights and implications for EC therapy.
Results
UBE2JI is highly expressed in EC tissues and cells, and predicts adverse prognosis
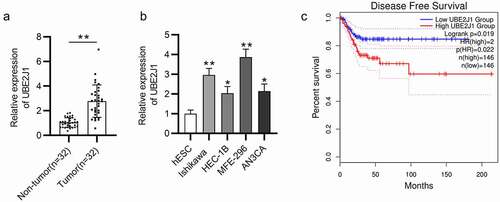
First, UBE2J1 expression status was detected in EC tumor tissues by RT-qPCR analysis and the results elucidated that UBE2J1 expression level was significantly elevated in EC tissues (n = 32) relative to the adjacent non-tumor endometrial tissues (n = 32) (). In addition, compared with the corresponding normal endometrial cells (hESC), UBE2J1 expression was up-regulated in EC cells (Ishikawa, HEC-1B, MFE-296, and AN3CA), especially in Ishikawa and MFE-296 cells(). Hence, these two cell lines were selected for further experiments. Importantly, as exhibited in , data from Kaplan-Meier Plotter website (https://kmplot.com/) shown that patients with higher UBE2J1 level have shorter disease-free survival (DFS) time while patients with low UBE2J1 expression present high OS rate. To conclude, UBE2J1 displayed high level in EC tissues and cells, and predicted unfavorable prognosis of EC patients.
Figure 1. UBE2JI is highly expressed in EC tissues and cells and predicts poor prognosis. (a) RT-qPCR analysis was performed to examine UBE2J1 expression in EC tumor tissues (n = 32) and adjacent non-tumor endometrial tissues (n = 32). (b) UBE2J1 expression was detected in normal endometrial cells (hESC) and in EC cells (Ishikawa, HEC-1B, RL95-2 and AN3CA) by RT-qPCR. (c) The correlation between UBE2J1 level and disease-free survival time of EC patients. *P < .05, **P < .01.
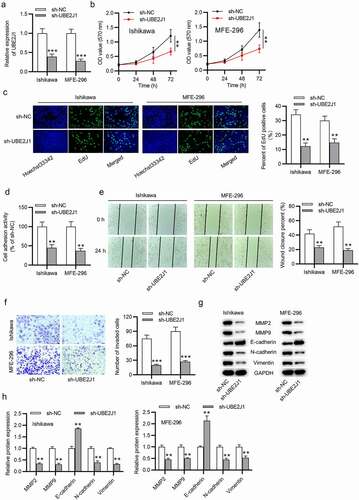
UBE2J1 depletion suppresses EC cell proliferation, adhesion, and motion
To investigate the biological role of UBE2J1 in EC cells, we conducted a serious of functional experiments. UBE2J1 level was effectively knocked down by transfecting sh-UBE2J1 into Ishikawa and MFE-296 cells (). UBE2J1-depletion induced decreased viability of Ishikawa and MFE-296 cells (). By performing EdU assay, we observed that number of EdU-positive cells was reduced by UBE2J1 downregulation (). In addition, cell adhesion assay suggested that EC cell adhesion activity of Ishikawa and MFE-296 cells was weakened by UBE2J1 silencing (). Next, we aimed to investigate the effects of UBE2J1 on EC cell motion. The result of wound healing assay revealed that UBE2J1 knockdown decreased the number of migrated Ishikawa and MFE-296 cells (). Similarly, we found that the invasive capacity of Ishikawa and MFE-296 cells was suppressed by transfection with sh-UBE2J1 through transwell assay (). In subsequent western blot analysis, in comparison with the sh-NC group, low MMP2, MMP9, N-cadherin, and Vimentin protein levels as well as the high E-cadherin protein level were observed in UBE2J1-silenced Ishikawa and MFE-296 cells (). These findings verified that knockdown of UBE2J1 resulted in a suppressive effect on ECcell malignant phenotypes.
Figure 2. UBE2J1 depletion suppresses EC cell growth. (a) The interference efficiency of sh-NC or sh-UBE2J1 in Ishikawa and MFE-296 cells was determined utilizing RT-qPCR analysis. (b) MTT assay was adopted to examine the effect of sh-NC or sh-UBE2J1 on Ishikawa and MFE-296 cell viability. (c) Examination for cell proliferation was conducted by EdU assay in UBE2J1-silenced Ishikawa and MFE-296 cells. (d) Cell adhesion activity was examined in Ishikawa and MFE-296 cells upon UBE2J1 knockdown. (e-f) The effects of sh-NC or sh-UBE2J1 on Ishikawa and MFE-296 cell migration and invasion were tested via wound healing assay and transwell assay, separately. (g-h) Western blot analysis was used to test MMP2, MMP9, E-cadherin, N-cadherin, and Vimentin protein levels. **P < .01, ***P < .001.
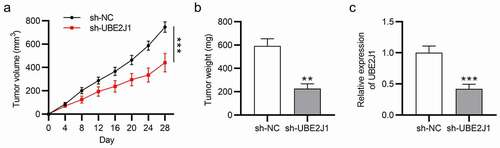
UBE2J1 facilitates EC tumor growth in vivo
Next, we aimed to probe into the effect of UBE2J1 on EC tumor growth in vivo. Nude mice were subcutaneously injected with Ishikawa cells pre-transfected with sh-NC or sh-UBE2J1. Four weeks later, tumor volume and weight were measured after excising the tumors from those nude mice. Compared with the sh-NC group, the final tumor volume and weight were reduced in sh-UBE2J1 group, indicating that UBE2J1 knockdown limited tumor growth and tumor weight invivo (). In addition, with the implementation of RT-qPCR analysis, we found the down-regulated UBE2J1 level in sh-UBE2J1 group compared with sh-NC group (). These data signified that UBE2J1 deficiency inhibited EC tumor growth invivo.
UBE2J1 activates the PI3K/AKT pathway to facilitate EC cell growth
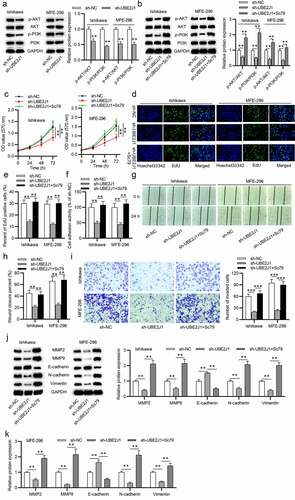
Recently, many researchers have certified that the activation of PI3K/AKT signaling pathway promotes the proliferative and invasive capacities of EC cells.Citation21,Citation22 Ubc9, a member of the E2 ubiquitin-binding enzyme family, has been reported to activate PI3K/AKT.Citation19,Citation20 Considering UBE2J1 is a member of the E2 ubiquitin-binding enzyme family, we speculated that UBE2J1 might also activate the PI3K/AKT pathway to facilitate EC cell growth. As shown in , p-AKT and p-PI3K protein levels were apparently decreased in response to UBE2J1 knockdown. Moreover, Sc79 (the activator of AKT) treatment regained phosphorylation level of PI3K and AKT resulting from UBE2J1 knockdown, suggesting that UBE2J1 exerts significant effect on the activation of PI3K/AKT pathway (). To further validated that UBE2J1 mediates cell malignant phenotype via PI3K/AKT signaling, Ishikawa and MFE-296 cells were treated with Sc79. According to MTT and EdU assays, UBE2J1 deficiency-repressed cell viability and proliferation was counteracted by Sc79 treatment (). Additionally, the inhibitive effects of UBE2J1 knockdown on cell adhesion was reversed by Sc79 treatment (). The suppressive influence of UBE2J1 deficiency on migratory and invasive abilities was also retrieved by treatment of Sc79 (). Additionally, Sc79 treatment countervailed the effects of UBE2J1 depletion on MMP2, MMP9, E-cadherin, N-cadherin, and Vimentin protein levels (). Conclusively, these results suggested that UBE2J1 accelerated EC cell growth via the activation of the PI3K/AKT pathway.
Figure 4. UBE2J1 activates the PI3K/AKT pathway to facilitate EC cell growth. (a) The expression levels of p-AKT and p-PI3K were detected by western blot analysis in sh-NC and sh-UBE2J1 groups. (b) p-AKT and p-PI3K expression was examined in sh-NC, sh-UBE2J1 and sh-UBE2J1+ Sc79 groups. (c-e) In UBE2J1-silenced EC cells, the viability and proliferation were respectively detected by MTT and EdU assays after Sc79 treatment. (f) Cell adhesion assay was utilized to detect cell adhesion activity after sh-UBE2J1 transfection and Sc79 treatment. (g-i) The migration and invasion in cells were estimated using wound healing and transwell assays with indicated transfection and treatment. (j-k) The expression of EMT-related proteins was detected using western blot analysis. **P < .01.
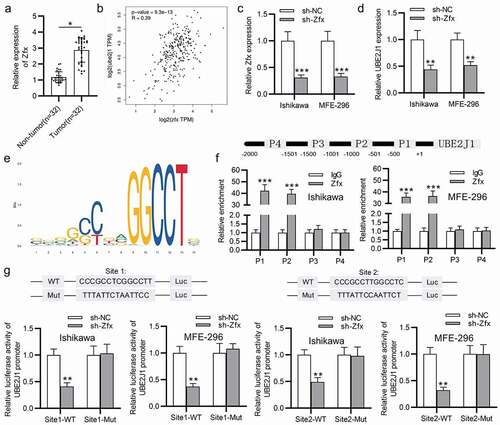
Zfx binds to UBE2J1 promoter
We then investigate the upstream mechanism of UBE2J1 upregulation in EC tissues and cells. Increasing researches have manifested that transcription factors (TFs) can activate gene transcription to upregulate mRNAs in EC.Citation23–25 Moreover, UBE2J1 was proposed to be transcriptionally activated by zfx in hepatocellular carcinoma.Citation12 In our study, zfx expression was found to be upregulated in EC tissues (), and data from GEPIA (http://gepia2.cancer-pku.cn/) website indicated that UBE2J1 is positively correlated with zfx in EC tissues (). Therefore, we conjectured that zfx might be implicated in the transcriptional activation of UBE2J1 expression in EC cells. First, zfx expression was knocked down with the transfection of sh-zfx into Ishikawa and MFE-296 cells (). Data from RT-qPCR analysis delineated that UBE2J1 expression was decreased by sh-zfx (). Based on UCSC (http://genome.ucsc.edu/) and JASPAR (http://jaspar.genereg.net/), fourteen putative binding sites between Zfx and UBE2J1 promoter were predicted (supplementary table 1), and the binding motif was shown in . The top 4 with higher binding score were selected for further exploration. The ChIP assay demonstrated that zfx was enriched in P1 and P2 group, implying that zfx bind with UBE2J1 promoter at P1 and P2 region (). Furthermore, luciferase reporter assay showed that knockdown of zfx suppressed luciferase activities of UBE2J1 promoter of site1/2-Wt, while site1/2-Mut displayed no differentiated expression in EC cells (), suggesting that zfx binds with site1 and site2 of UBE2J1 promoter. Overall, zfx bound with UBE2J1 promoter to activate the transcription of UBE2J1.
Figure 5. Zfx binds to UBE2J1 promoter. (a) Zfx expression in EC tissues and adjacent normal endometrial tissues was tested via RT-qPCR. (b) The correlation between zfx and UBE2J1. (c) Knockdown efficiency of sh-zfx. (d) UBE2J1 expression in zfx-silenced cells. (e) The binding motif between zfx and UBE2J1 promoter. (f) The binding capacity between zfx and UBE2J1 promoter was determined using ChIP assay. (g) The binding capacity between zfx and UBE2J1 promoter was assessed using luciferase reporter assay. *P < .05, **P < .01. ***P < .001.
Discussion
E2 family members are pivotal to the formation of ubiquitin chains, and UBE2J1, as a member of E2 family, participates in the progression of protein degradation.Citation10,Citation11,Citation26,Citation27 Recently, it has been indicated that UBE2J1 is involved in the development of medulloblastoma,Citation12 and it functions as a prognostic biomarker for PCa patients.Citation13 Nevertheless, identification of the function and molecule mechanism of UBE2J1 in EC has not been conducted yet. The results in our current study reported that UBE2J1 was highly expressed in EC tissues and cells. Besides, highly expressed UBE2J1 predicted adverse prognosis of EC patients. Functional experiments manifested that UBE2J1 silencing caused a suppression on EC cell growth via inhibiting cell viability, proliferation, adhesion, migration, invasion, and EMT process. In vivo experiments demonstrated that UBE2J1 knockdown reduced the tumor volume and weight of mice that were implanted with EC cells. These results implied the oncogenic function of UBE2J1 in EC.
A myriad of reports disclosed that the PI3K/AKT signaling pathway participates in various processes of human cancers such as cell proliferation, migration, angiogenesis, and lymphangiogenesis.Citation28–30 For example, lncRNA HOXB-AS3 exacerbates proliferative, and migratory abilities of lung cancer cells with the activation of PI3K/AKT pathway.Citation31 LncRNA TCL6-mediated miR-106a-5p promotes the invasive properties of hepatocellular carcinoma cells via modulating the PI3K/AKT signaling pathway.Citation32 Through the PI3K/Akt signaling pathway, lncRNA HULC knockdown inhibits angiogenesis in human gliomas.Citation33 It is revealed by some documents that EC cell proliferation and invasion can be facilitated by activating the PI3K/AKT pathway.Citation34,Citation35 In recent research, Ubc9, has been documented to activate the PI3K/AKT pathway.Citation19 Nevertheless, whether UBE2J1 can activate the PI3K/AKT signaling in EC is unknown. Our research depicted that down-regulated UBE2J1 inactivate the PI3K/AKT signaling in EC cells. In addition, restoration assays illuminated that Sc79 treatment reversed UBE2J1 silencing-mediated inhibition in PI3K/AKT signaling and EC cell growth.
Zinc finger X-chromosomal protein is localized on the X chromosome and it possesses an acidic transcriptional activation domain.Citation36 Nowadays, zfx has generated great concern due to its association with stem cell and human tumor development. It is reported that zfx exerts a pivotal function in regulating the self-renewal of embryos and hematopoietic stem cells.Citation37 zfx contributes to the malignancy of laryngeal squamous cell carcinoma,Citation38 hepatocellular carcinoma,Citation39 gastric cancer,Citation40 bladder cancer,Citation41 and pancreatic cancer.Citation42 It is noteworthy that zfx can function as a TF to activate the transcription of downstream genes in diverse human carcinomas.Citation43 Specifically, zfx was reported to transcriptionally activate UBE2J1 in hepatocellular carcinoma.Citation12 Likewise, our study reported that zfx expression was upregulated in EC tissues and it was in positive correlation with UBE2J1 in EC tissues. In addition, zfx was validated to combine with UBE2J1 promoter region to activate the transcription of UBE2J1.
Based on our current investigation, it is reasonable to summarize that Zfx-mediated upregulation of UBE2J1 accelerated EC progression through activating the PI3K/AKT pathway, which may provide new directions for EC treatment.
Materials and methods
Specimens
The Zhangjiagang First People’s Hospital (Jiangsu, China) provided EC tissue samples (n = 32) and corresponding adjacent normal tissue samples (n = 32) from patients diagnosed with EC. The patients did not receive any chemotherapy or radiation before surgery. After the patients underwent hysterectomy, their tissue samples were collected and snap-stored in liquid nitrogen at −80°C for subsequent experiments. Each patient signed a written informed consent and understood its content. This study was approved by the Ethics Committee of Zhangjiagang First People’s Hospital (Jiangsu, China).
Cell culture and transfection
Human normal endometrium (NE) cell line (hESC) and EC cell lines (Ishikawa, HEC-1B, MFE-296 and AN3CA) were purchased from American Type Culture Collection. All cells were incubated in RPMI-1640 medium which was produced by Invitrogen in USA. RPMI-1640 medium containing 1/10 FBS and 100 U/mL penicillin/streptomycin (HyClone, USA) was used in a humidified environment at 37°C with 5% CO2. The activator of PI3K/AKT signaling pathway (Sc79, 8 µg/mL) was purchased from Sigma Aldrich (St Louis, MI). For cell transfection, shRNA of UBE2J1 (sh-UBE2J1) and zfx (sh-zfx) were adopted to knock down UBE2J1 and zfx levels with sh-NC as the negative control. UBE2J1 overexpressing plasmid (pcDNA3.1/UBE2J1) and its negative control (pcDNA3.1) were designed and generated by RiboBio (Guangzhou, China). Cell transfection was conducted for 48 h using Lipofectamine 3000 transfection reagent (Life Technologies, USA) according to the supplier’s guidelines.
RT-qPCR analysis
TRIzol reagent (Takara, China) was used for extracting total RNA from EC tissues and cells. The cDNA reverse-transcription was accomplished using a Reverse Transcription Kit (Takara, Dalian, China). SYBR Premix Ex Taq (Takara) was applied for RT-qPCR analysis on an Applied Biosystems 7500 Real-Time PCR System. Glyceraldehyde-3phosphate dehydrogenase (GAPDH) functioned as the internal reference for mRNA. The primers for RT-qPCR are as follows:
UBE2J1: forward, 5ʹ-CACCATGGAGACCCGCTACAACCTG-3ʹ; reverse, 5ʹ-TCACAAACCAGCATTATAACTCAAAGTC-3ʹ.
Zfx: forward, 5ʹ-GGCAGTCCACAGCAAGAAC-3ʹ; reverse, 5ʹ-TTGGTATCCGAGAAAGTCAGAAG-3ʹ.
GAPDH: forward, 5ʹ-CCC ATG TTC GTC ATG GGT GT-3ʹ; reverse, 5ʹ-CCG TTC AGC TCA GGG ATG AC-3ʹ.
Data were calculated through the 2−ΔΔCt method.
Western blot analysis
Ishikawa and MFE-296 cells were first lysed by cell lysis buffer, RIPA (Beyotime, Shanghai, China), following which the proteins were subjected to 10% SDS-PAGE. Then, the proteins were moved onto the PVDF membranes in a conventional way. Subsequently, the membranes were cultivated with the primary antibodies against MMP2 (Abcam, ab92536), MMP9 (Abcam, ab38898), E-cadherin (Abcam, ab1416), N-cadherin (Abcam, ab76011), Vimentin (Abcam, ab92547), and GAPDH (Abcam, ab8245) for overnight at 4°C. The next day, the membranes were cultured with the secondary antibodies for 2 h. Imaging Analysis System (Odyssey Infrared, USA) was used for visualization of the protein bands at last.
MTT assay
MTT assay was conducted for the detection of cell viability according to the instructions of manufacturer. The 96-well plates were used to ensconce the cells (3 × 103), which were cultured in RPMI-1640 medium. After transfection at 0, 24, 48, and 72 h, MTT was added into each well to cultivate the cells for 4 h. Then, dimethylsulfoxide was supplemented into the wells and the optical density (OD) was examined at 570 nm.
EdU (5-Ethynyl-2ʹ-deoxyuridine) assay
Transfected cells were inoculated in 96-well plates and incubated with RPMI-1640 for 24 h. Subsequently, 4% paraformaldehyde was used to immobilize the cells which were then permeabilized by 0.5% Triton X-100 and dyed with Apollo dye reaction liquid. Afterward, cell nuclei was stained with 1× Hoechst33342. EdU-positive cells were observed from three randomly selected fields under a microscope.
Transwell assay
Transfected Ishikawa and MFE-296 cells were inoculated into the upper chamber which was coated with Matrigel. Medium (200 μL) without serum was added to the upper chamber while the lower chamber was supplemented with 600 μL medium including 10% FBS. After cell incubation for 48 h, formaldehyde (4%) was implemented to immobilize the cells and 0.1% of crystal violet was utilized for cell staining. Finally, the invaded cells were visualized through an inverted fluorescence microscope.
Wound healing assay
Cells were seeded into the 6-well plates and cultured with 2 ml culture media. When the confluence was up to about 85%, the pipette tips were utilized to generate a scratch in the monolayers. Then, cultivating the cells in 37°C with 5% CO2 was performed, following which an inverted microscope was applied to photograph the scratch width at 0 and 24 h.
Cell adhesion assay
Cell adhesion assay was performed as required by the manufacturer as previously described.Citation44 The 96 well-plate that was blocked with 1% bovine serum albumin (BSA; Sigma-Aldrich) was coated by fibronectin (100 mg/ml; BD Biosciences, USA) at 37°C for 2 h. Afterward, 3 × 104 cells were plated in the plates and cultivated for 2 h. PBS was used to discard the non-adherent cells and 4% PFA (Solarbio, Beijing, China) was applied for cell immobilization. Finally, a microscope (Olympus, Japan) was utilized to observe and count the cells.
Animal study
All invivo experiments were performed as instructed by the regulations of the Use Committee for Animal Care of Zhangjiagang First People’s Hospital (Jiangsu, China). BALB/c female nude mice (4-6-week-old, n = 10) were purchased from Vital River Co., Ltd (Beijing, China) and housed under specific pathogen-free condition. The transfected Ishikawa cells (4 × 106) were subcutaneously injected into each mouse. Tumor volume was examined every 4 days based on the calculating formula: Volume = 0.5× length×width2. The mice were euthanized at day 28, and the tumors were excised for further experiments.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was conducted to verify the interaction between zfx and UBE2J1 promoter. Briefly, a commercial kit (Upstate Biotechnology) was employed to perform ChIP assay as per the product manuals. EC cells were cultivated at 37°C with 5% CO2. Then, the anti-zfx was used for chromatin immunoprecipitation while anti-IgG functioned as the negative control. RT-qPCR analysis was performed to examine the level of target fragment which were purified by elution buffer.
Luciferase reporter assay
Luciferase reporter assay was carried out to detect the combination between zfx and UBE2J1 promoter. Briefly, the wild type or mutant sequences supplemented with site1/2 in UBE2J1 promoter region was separately synthesized and subcloned into the pGL3 luciferase vector (Promega, USA) to form site1/2-WT and site1/2-Mut. Then, Ishikawa or MFE-296 cells were co-transfected with sh-zfx or its negative control (sh-NC) and site1/2-WT and site1/2-Mut, separately. Finally, the luciferase activity of UBE2J1 promoter was detected utilizing the luciferase reporter assay system (Promega, USA).
Statistical analysis
Experimental data was exhibited as the mean ± standard deviation. The variance was compared with Student’s t-tests and ANOVA for two groups and multiple groups, respectively. Statistical analysis was realized using SPSS version 17.0 (Chicago, USA) software. Each experiment was carried out at least three times. Pearson’s correlation analysis was used for correlation analyses. P < .05 suggested a statistically difference.
Disclosure of potential conflicts of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Supplemental Material
Download Comma-Separated Values File (987 B)Supplemental data
Supplemental data for this article can be accessed on the publisher’s website.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Lee JY, Kim JW, Lee TS, Zang R, Chen X, Yang J, Wang K-L, Sugiyama T. Difference in practice patterns in the management of endometrial cancer: a survey of the members of 4 East Asian gynecologic oncology groups. Int j gynecol cancer. 2017 Nov;27(9):1888–1894. doi: 10.1097/igc.0000000000001078. PubMed PMID: 28953133; eng.
- Dashti SG, Chau R, Ouakrim DA, Buchanan DD, Clendenning M, Young JP, Winship IM, Arnold J, Ahnen DJ, Haile RW, et al. Female hormonal factors and the risk of endometrial cancer in lynch syndrome. Jama. 2015 July 7;314(1):61–71. doi: 10.1001/jama.2015.6789. PubMed PMID: 26151267; PubMed Central PMCID: PMCPmc4688894. eng.
- Smits A, Lopes A, Das N, Bekkers R, Galaal K. The impact of BMI on quality of life in obese endometrial cancer survivors: does size matter? Gynecol Oncol. 2014 Jan;132(1):137–141. doi: 10.1016/j.ygyno.2013.11.018. PubMed PMID: 24262880; eng.
- Tseng CH. Metformin and endometrial cancer risk in Chinese women with type 2 diabetes mellitus in Taiwan. Gynecol Oncol. 2015 July;138(1):147–153. doi: 10.1016/j.ygyno.2015.03.059. PubMed PMID: 25913129; eng.
- Mell LK, Carmona R, Gulaya S, Lu T, Wu J, Saenz CC, Vaida F. Cause-specific effects of radiotherapy and lymphadenectomy in stage I-II endometrial cancer: a population-based study. J Natl Cancer Inst. 2013 Nov 6;105(21):1656–1666. doi: 10.1093/jnci/djt279. PubMed PMID: 24123960; PubMed Central PMCID: PMCPmc6281051. eng.
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009 Nov;10(11):755–764. doi: 10.1038/nrm2780. PubMed PMID: 19851334; PubMed Central PMCID: PMCPmc3107738. eng.
- David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010 Mar 19;285(12):8595–8604. doi: 10.1074/jbc.M109.089003. PubMed PMID: 20061386; PubMed Central PMCID: PMCPmc2838281. eng.
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81(1):203–229. doi: 10.1146/annurev-biochem-060310-170328. PubMed PMID: 22524316; eng
- Lester D, Farquharson C, Russell G, Houston B. Identification of a family of noncanonical ubiquitin-conjugating enzymes structurally related to yeast UBC6. Biochem Biophys Res Commun. 2000 Mar 16;269(2):474–480. doi: 10.1006/bbrc.2000.2302. PubMed PMID: 10708578; eng.
- Lenk U, Yu H, Walter J, Gelman MS, Hartmann E, Kopito RR, Sommer T. A role for mammalian Ubc6 homologues in ER-associated protein degradation. J Cell Sci. 2002 July 15;115(Pt14): 3007–3014. PubMed PMID: 12082160; eng.
- Wu CJ, Conze DB, Li X, Ying S-X, Hanover JA, Ashwell JD. TNF-alpha induced c-IAP1/TRAF2 complex translocation to a Ubc6-containing compartment and TRAF2 ubiquitination. EMBO J. 2005 May 18;24(10):1886–1898. doi: 10.1038/sj.emboj.7600649. PubMed PMID: 15861135; PubMed Central PMCID: PMCPmc1142588. eng.
- Palmer CJ, Galan-Caridad JM, Weisberg SP, Lei L, Esquilin JM, Croft GF, Wainwright B, Canoll P, Owens DM, Reizis B, et al. Zfx facilitates tumorigenesis caused by activation of the Hedgehog pathway. Cancer Res. 2014 Oct 15;74(20):5914–5924. doi: 10.1158/0008-5472.Can-14-0834. PubMed PMID: 25164012; PubMed Central PMCID: PMCPMC4199880. eng.
- Chen Z, Hu H. Identification of prognosis biomarkers of prostatic cancer in a cohort of 498 patients from TCGA. Curr Probl Cancer. 2019 Dec;43(6):100503. doi: 10.1016/j.currproblcancer.2019.100503. PubMed PMID: 31563279; eng.
- Hagihara T, Kondo J, Endo H, Ohue M, Sakai Y, Inoue M. Hydrodynamic stress stimulates growth of cell clusters via the ANXA1/PI3K/AKT axis in colorectal cancer. Sci Rep. 2019 Dec 27;9(1):20027. doi: 10.1038/s41598-019-56739-7. PubMed PMID: 31882967; PubMed Central PMCID: PMCPMC6934682 Development in Kyoto University, which is sponsored by KBBM, Inc. The other authors declare no competing interests. eng.
- Si X, Xu F, Xu F, Wei M, Ge Y, Chenge S. CADM1 inhibits ovarian cancer cell proliferation and migration by potentially regulating the PI3K/Akt/mTOR pathway. Biomed pharmacother = Biomedecine pharmacotherapie. 2020 Mar;123:109717. doi: 10.1016/j.biopha.2019.109717. PubMed PMID: 31865146; eng.
- Duan Q, Cai L, Zheng K, Cui C, Huang R, Zheng Z, Xie L, Wu C, Yu X, Yu J, et al. lncRNA KCNQ1OT1 knockdown inhibits colorectal cancer cell proliferation, migration and invasiveness via the PI3K/AKT pathway. Oncol Lett. 2020 July;20(1):601–610. doi: 10.3892/ol.2020.11619. PubMed PMID: 32565985; PubMed Central PMCID: PMCPMC7286112. eng.
- Yan F, Liu SW, Li XY, Li CC, Wu Y. Silencing LncRNA LINC01305 inhibits epithelial mesenchymal transition in lung cancer cells by regulating TNXB-mediated PI3K/Akt signaling pathway. J Biol Regul Homeost Agents. 2020 Mar-Apr;34(2):499–508. doi: 10.23812/20-73-a-33. PubMed PMID: 32549529; eng.
- Zhang XH, Li M, Kang YJ, Xie Y-Q, Cao Y-X. Long non-coding RNA LINP1 functions as an oncogene in endometrial cancer progression by regulating the PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2019 Aug;23(16):6830–6838. doi: 10.26355/eurrev_201908_18722. PubMed PMID: 31486482; eng.
- Dong M, Pang X, Xu Y, Wen F, Zhang Y. Ubiquitin-conjugating enzyme 9 promotes epithelial ovarian cancer cell proliferation in vitro. Int J Mol Sci. 2013 May 24;14(6):11061–11071. doi: 10.3390/ijms140611061. PubMed PMID: 23708104; PubMed Central PMCID: PMCPMC3709718. eng.
- Lin CH, Liu SY, Lee EH. SUMO modification of Akt regulates global SUMOylation and substrate SUMOylation specificity through Akt phosphorylation of Ubc9 and SUMO1. Oncogene. 2016 Feb 4;35(5):595–607. doi: 10.1038/onc.2015.115. PubMed PMID: 25867063; eng.
- Expression of Concern: upregulation of long non-coding RNA OGFRP1 facilitates endometrial cancer by regulating miR-124-3p/SIRT1 axis and by activating PI3K/AKT/GSK-3b pathway. Artif Cells, Nanomed Biotechnol. 2020 Dec;48(1):747. doi: 10.1080/21691401.2020.1752024. PubMed PMID: 32285700; eng.
- Xie P, Wang X, Kong M, Bai X, Jiang T. TRAF4 promotes endometrial cancer cell growth and migration by activation of PI3K/AKT/Oct4 signaling. Exp Mol Pathol. 2019 June;108:9–16. doi: 10.1016/j.yexmp.2019.03.003. PubMed PMID: 30853613; eng.
- Spoelstra NS, Manning NG, Higashi Y, Darling D, Singh M, Shroyer KR, Broaddus RR, Horwitz KB, Richer JK. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006 Apr 1;66( 7):3893–3902. doi: 10.1158/0008-5472.Can-05-2881. PubMed PMID: 16585218; eng.
- Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang P, Chan LKY, Zhang T, Yu J, Kwong J, et al. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013 July 18;32(29):3432–3442. doi: 10.1038/onc.2012.360. PubMed PMID: 22907428; eng.
- Stone DM, Murone M, Luoh S, Ye W, Armanini MP, Gurney A, Phillips H, Brush J, Goddard A, de Sauvage FJ, et al. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J Cell Sci. 1999 Dec;112 (Pt 23): 4437–4448. PubMed PMID: 10564661; eng.
- Lemus L, Goder V. Regulation of endoplasmic reticulum-associated protein degradation (ERAD) by ubiquitin. Cells. 2014 Aug 5;3(3):824–847. doi: 10.3390/cells3030824. PubMed PMID: 25100021; PubMed Central PMCID: PMCPmc4197631. eng.
- Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci U S A. 2011 Feb 1;108(5):2034–2039. doi: 10.1073/pnas.1016229108. PubMed PMID: 21245296; PubMed Central PMCID: PMCPmc3033308. eng.
- Wen X, Li H, Sun H, Zeng A, Lin R, Zhao J, Zhang Z. MiR-455-3p reduces apoptosis and alleviates degeneration of chondrocyte through regulating PI3K/AKT pathway. Life Sci. 2020 Apr 25:117718. doi: 10.1016/j.lfs.2020.117718. PubMed PMID: 32343998; eng. 253
- Liu HT, Ma RR, Lv BB, Zhang H, Shi D-B, Guo X-Y, Zhang G-H, Gao P. LncRNA-HNF1A-AS1 functions as a competing endogenous RNA to activate PI3K/AKT signalling pathway by sponging miR-30b-3p in gastric cancer. Br J Cancer. 2020 Apr 27. doi: 10.1038/s41416-020-0836-4. PubMed PMID: 32336754; eng. 122 1825–1836
- Liu HT, Ma RR, Lv BB, Zhang H, Shi D-B, Guo X-Y, Zhang G-H, Gao P. LncRNA-HNF1A-AS1 functions as a competing endogenous RNA to activate PI3K/AKT signalling pathway by sponging miR-30b-3p in gastric cancer. Br J Cancer. 2020 June;122(12):1825–1836. doi: 10.1038/s41416-020-0836-4. PubMed PMID: 32336754; PubMed Central PMCID: PMCPMC7283217. eng.
- Jiang W, Kai J, Li D, Wei Z, Wang Y, Wang W. lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J Cell Physiol. 2020 Oct;235(10):7194–7203. doi: 10.1002/jcp.29618. PubMed PMID: 32039488; eng.
- Luo LH, Jin M, Wang LQ, Xu G-J, Lin Z-Y, Yu -D-D, Yang S-L, Ran R-Z, Wu G, Zhang T, et al. Long noncoding RNA TCL6 binds to miR-106a-5p to regulate hepatocellular carcinoma cells through PI3K/AKT signaling pathway. J Cell Physiol. 2020 Sept;235(9):6154–6166. doi: 10.1002/jcp.29544. PubMed PMID: 32020591; eng.
- Zhu Y, Zhang X, Qi L, Cai Y, Yang P, Xuan G, Jiang Y. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016 Mar 22;7(12):14429–14440. doi: 10.18632/oncotarget.7418. PubMed PMID: 26894862; PubMed Central PMCID: PMCPMC4924726. eng.
- Wang Q, Yan SP, Chu DX, Xie Y, Wang C-F, Zhang J-Y, Li W-C, Guo R-X. Silencing of long non-coding RNA RP1-93H18.6 acts as a tumor suppressor in cervical cancer through the Blockade of the PI3K/Akt Axis. Mol Ther Nucleic Acids. 2020 Mar 6;19:304–317. doi: 10.1016/j.omtn.2019.10.041. PubMed PMID: 31877407; PubMed Central PMCID: PMCPMC6938856. eng.
- Bjeije H, Soltani BM, Behmanesh M, Zali MR. YWHAE long non-coding RNA competes with miR-323a-3p and miR-532-5p through activating K-Ras/Erk1/2 and PI3K/Akt signaling pathways in HCT116 cells. Hum Mol Genet. 2019 Oct 1;28(19):3219–3231. doi: 10.1093/hmg/ddz146. PubMed PMID: 31238337; eng.
- Iovanna J, Dusetti N. Speeding towards individualized treatment for pancreatic cancer by taking an alternative road. Cancer Lett. 2017 Dec 1;410:63–67. doi: 10.1016/j.canlet.2017.09.016. PubMed PMID: 28947138; eng.
- Weng H, Wang X, Li M, Wu X, Wang Z, Wu W, Zhang Z, Zhang Y, Zhao S, Liu S, et al. Zinc finger X-chromosomal protein (ZFX) is a significant prognostic indicator and promotes cellular malignant potential in gallbladder cancer. Cancer Biol Ther. 2015;16(10):1462–1470. doi: 10.1080/15384047.2015.1070994. PubMed PMID: 26230915; PubMed Central PMCID: PMCPMC4846125. eng.
- Fang J, Yu Z, Lian M, Ma H, Tai J, Zhang L, Han D. Knockdown of zinc finger protein, X-linked (ZFX) inhibits cell proliferation and induces apoptosis in human laryngeal squamous cell carcinoma. Mol Cell Biochem. 2012 Jan; 360(1–2): 301–307. doi: 10.1007/s11010-011-1069-x. PubMed PMID: 22009483; eng.
- Lai KP, Chen J, He M, Ching AKK, Lau C, Lai PBS, To K-F, Wong N. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014 Oct 15;135(8):1790–1799. doi: 10.1002/ijc.28819. PubMed PMID: 24585547; eng.
- Wu S, Lao XY, Sun TT, Ren -L-L, Kong X, Wang J-L, Wang Y-C, Du W, Yu Y-N, Weng Y-R, et al. Knockdown of ZFX inhibits gastric cancer cell growth in vitro and in vivo via downregulating the ERK-MAPK pathway. Cancer Lett. 2013 Sept 1;337(2):293–300. doi: 10.1016/j.canlet.2013.04.003. PubMed PMID: 23587796; eng.
- Liu TY, Gong W, Tan ZJ, Lu W, Wu X-S, Weng H, Ding Q, Shu Y-J, Bao R-F, Cao Y, et al. Baicalein inhibits progression of gallbladder cancer cells by downregulating ZFX. PloS one. 2015;10(1):e0114851. doi: 10.1371/journal.pone.0114851. PubMed PMID: 25617627; PubMed Central PMCID: PMCPMC4305301. eng.
- Song X, Zhu M, Zhang F, Zhang F, Zhang Y, Hu Y, Jiang L, Hao Y, Chen S, Zhu Q, et al. ZFX promotes proliferation and metastasis of pancreatic cancer cells via the MAPK pathway. Cell physiol biochem. 2018;48(1):274–284. doi: 10.1159/000491727. PubMed PMID: 30007968; eng.
- Rhie SK, Yao L, Luo Z, Witt H, Schreiner S, Guo Y, Perez AA, Farnham PJ. ZFX acts as a transcriptional activator in multiple types of human tumors by binding downstream of transcription start sites at the majority of CpG island promoters. Genome Res. 2018 Feb 2;28(3):310–320. doi: 10.1101/gr.228809.117. PubMed PMID: 29429977; PubMed Central PMCID: PMCPMC5848610. eng.
- Han L, Wang W, Ding W, Zhang L. MiR-9 is involved in TGF-β1-induced lung cancer cell invasion and adhesion by targeting SOX7. J Cell Mol Med. 2017 Sept;21(9):2000–2008. doi: 10.1111/jcmm.13120. PubMed PMID: 28266181; PubMed Central PMCID: PMCPMC5571535. eng.