ABSTRACT
In China, gastric cancer (GC) ranks first in the incidence of all malignant tumors. With high recurrence and distant metastasis, GC has caused considerable mortalities. LncRNA long intergenic non-protein-coding RNA 668 (LINC00668) has been reported to be upregulated in GC cells and predict poor prognosis of GC patients. However, the mechanism of LINC00668 has not been fully investigated in GC. This study aimed to investigate the role of LINC00668 in GC. We found that LINC00668 level was upregulated in GC tissue and cells and predicted poor prognosis. Functionally, LINC00668 knockdown suppressed GC cell migration and invasion. Additionally, LINC00668 knockdown inhibited epithelial to mesenchymal transition (EMT) process. PKN2 exerts similar effects with LINC00668 in GC cells. LINC00668 knockdown suppressed tumor growth and metastasis in vivo. Mechanistically, HuR was predicted to bind with LINC00668 and protein kinase N2 (PKN2). RNA pull-down assays validated the binding between HuR and LINC00668 (or PKN2). Moreover, either silencing of LINC00668 or HuR could decrease PKN2 mRNA stability or reduce PKN2 mRNA and protein levels. Furthermore, PKN2 expression was positively correlated with LINC00668 expression and HuR expression in GC tissues, and HuR expression was positively associated with LINC00668 expression in GC tissues. Finally, rescue assays confirmed that the suppressive effect of LINC00668 silencing on cell migration, invasion, and EMT process was reversed by PKN2 overexpression or HuR upregulation. In conclusion, LINC00668 cooperated with HuR-dependent upregulation of PKN2 to facilitate gastric cancer metastasis, which may provide a potential novel insight for GC treatment.
KEYWORDS:
Introduction
Gastric cancer (GC), a common malignant tumor, ranks third in all cancer-related deaths.Citation1,Citation2 In 2018, about 1,000,000 newly diagnosed GC cases and 780,000 mortalities were predicted worldwide.Citation3 It has been noted that the incidence rate of GC is prominently increased in Eastern Asia.Citation3 Known factors including Helicobacter pylori (Hp) infection, smoking, and genetic influence are closely related to the initiation or progression of GC.Citation4 However, the pathogenesis of GC remains unknown. Consequently, identifying potential molecular mechanism of GC development is essential to improve the situation of GC patients.
Long non-coding RNA (lncRNA), a category of noncoding transcripts, comprise more than 200 nucleotides and ack protein-coding capacity .Citation5,Citation6 Emerging evidence has revealed that lncRNAs are implicated in multiple cellular processes including cell proliferation, cell apoptosis, cell migration, and invasion in tumors.Citation6,Citation7 For example, lncRNA GOLGA2P10 protects tumor cells from apoptosis induced by ER stress via regulating Bcl-2 family members.Citation8 LncRNA EPIC1 promotes cell proliferation and migration during the progression of glioma.Citation9 Mechanistically, lncRNAs are capable of regulating gene expression by many ways including chromatin modification, transcriptional regulation, and post-transcriptional modulation.Citation10 In detail, certain lncRNAs serve as scaffolds or molecular signals in the nucleus.Citation11,Citation12 Additionally, some lncRNAs interact with specific RNA binding proteins (RBPs) to enhance mRNA stability in the cytoplasm.Citation13,Citation14 Recently, long intergenic non-protein coding RNA 668 (LINC00668) has been reported to regulate several tumors including breast cancer, lung adenocarcinoma, and glioma.Citation15–17 Moreover, LINC00668 is upregulated in GC cells and predicts poor prognosis of GC patients.Citation18 However, the mechanism of LINC00668 in GC remains to be further explored.
Protein kinase N2 (PKN2), also known as PAK2 or PRK2, a member of the atypical protein kinase C subfamily, which is known for modulating cell migration.Citation19 Additionally, PKN2 has been reported to regulate multiple tumors. For instance, PKN2 hinders M2 phenotype polarization of tumor-associated macrophages by targeting the DUSP6-Erk1/2 signaling.Citation20 Moreover, PKN2 was identified to be upregulated in triple-negative breast cancer and promote cell proliferation.Citation21 However, the role of PKN2 in GC is still unclear.
In the current exploration, we attempted to investigate the biological function and molecular mechanism of LINC00668 in GC cells. This discovery possibly provides a potential novel insight for GC treatment.
Results
Upregulated LINC00668 facilitated GC cell migration, invasion, and epithelial to mesenchymal transition (EMT) process and predicted unfavorable prognosis in GC patients
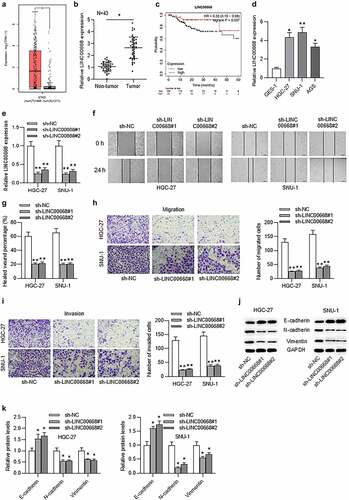
According to GEPIA (http://gepia.cancer-pku.cn/), the level of LINC00668 in stomach adenocarcinoma (STAD) tissues (n = 408) was significantly higher than that in normal tissues (n = 211) ()). Similarly, the result of RT-qPCR analysis showed that LINC0668 level was upregulated in GC tissues compared with adjacent normal tissues ()). Data from the Kaplan–Meier Plotter website (kmplot.com/analysis/) showed that GC patients with upregulated LINC00668 level had shorter overall survival time in 5 years ()). We then assessed the expression status of LINC00668 in GC cells and normal gastric epithelial cells and found higher level of LINC00668 in GC cells than in normal gastric epithelial cells ()). Considering that tumor cell migration and invasion are strongly associated with tumor progression, we evaluated the effect of LINC00668 on cell migration and invasion. First, LINC0068 level was knocked down by transfection with sh-lncRNA#1/2 in HGC-27 and SNU-1 cells ()). Then we demonstrated using a wound healing assay that cell migration ability was inhibited by LINC00668 knockdown (). In addition, trans-well assay showed that the migration or invasion of HGC-27 and SNU-1 cells was suppressed by LINC00668 knockdown (). It has been revealed that EMT process may induce tumor cell migration and invasion.Citation22,Citation23 Therefore, we evaluated EMT-related protein levels including E-cadherin, N-cadherin, and vimentin by western blot. The results depicted that knockdown of LINC00668 triggered a significant increase of E-cadherin and a significant decrease of N-cadherin and vimentin ().
Figure 1. Upregulated LINC00668 facilitated GC cell migration, invasion, EMT process, and predicted unfavorable prognosis in GC patients.
Upregulated PKN2 promoted GC cell migration, invasion, and EMT process and indicated poor prognosis in GC patients
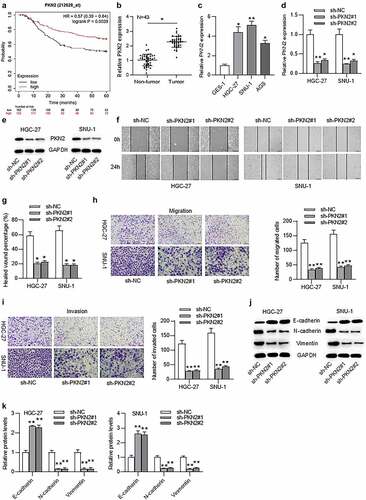
PKN2, an oncogene, has been demonstrated to promote the growth of several tumor types.Citation20,Citation21 Additionally, PKN2 promotes tumor metastasis in oral squamous cell carcinoma progression.Citation24 Findings in suggested that LINC00668- mediated proliferation and migration in GC cells. We thus speculated that PKN2 may be involved in the LINC00668-mediated proliferation and migration in gastric cancer progression. According to the Kaplan–Meier Plotter website (kmplot.com/analysis/), high level of PKN2 predicted poor prognosis of GC patients ()). Additionally, the result of RT-qPCR indicated that PKN2 mRNA level was higher in GC tissues compared to in adjacent normal tissues and higher in GC cells compared to in normal gastric epithelial cells (). To assess the effect of PKN2 on cell migration, invasion and EMT process, we knocked down PKN2 level in HGC-27 and SNU-1 cells by transfection with sh-PKN2#1/2 (). Furthermore, the wound healing and trans-well assays demonstrated that cell migration and invasion were suppressed by PKN2 silencing in HGC-27 and SNU-1 cells ()). Moreover, western blot analysis indicated that PKN2 silencing induced the increase of E-cadherin level and the reduction of N-cadherin and vimentin levels ().
Figure 2. Upregulated PKN2 drove cell migration, invasion, EMT process, and indicated poor prognosis in GC patients.
LINC00668 promoted GC growth and lung metastasis in vivo
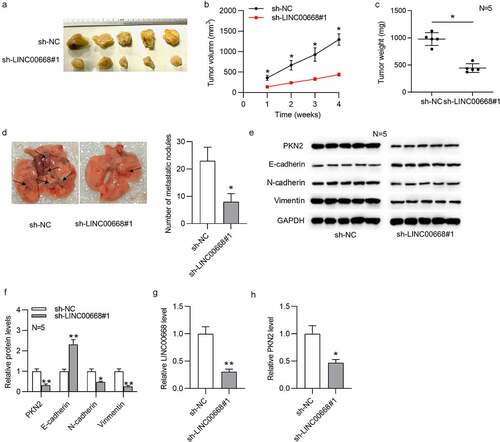
To further explore the role of LINC00668 in vivo, tumor xenograft assays were designed and carried out. As shown in ), the knockdown of LINC00668 significantly repressed tumor size, tumor volume, and tumor weight. Additionally, LINC00668 knockdown significantly reduced the lung metastatic nodules ()). Moreover, based on the result of western blot, LINC00668 knockdown induced a significant increase of E-cadherin level and a significant decrease of PKN2, N-cadherin, and vimentin levels in xenograft tumors of nude mice ()). RT-qPCR analysis indicated that both LINC00668 and PKN2 levels were downregulated in sh-LINC00668#1 group ()).
Figure 3. LINC00668 promoted GC growth and metastasis in vivo.
LINC00668 and PKN2 bound with HuR
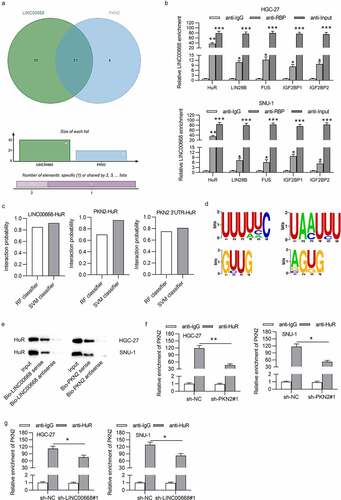
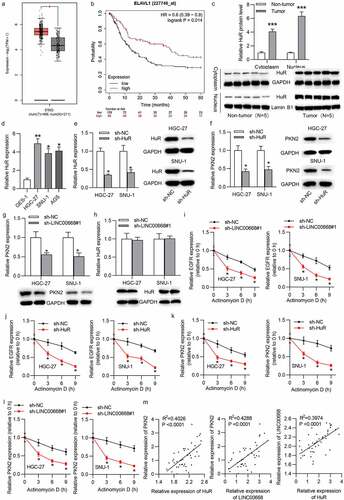
Since both LINC00668 and PKN2 levels were upregulated in GC cells and both LINC00668 and PKN2 promoted proliferation and migration in GC cells, we intended to explore the pattern by which LINC00668 regulated PKN2 expression in GC cells. Emerging studies proposed that lncRNA could recruit RBPs to stabilize mRNA, thereby upregulating target protein levels.Citation25,Citation26 We hypothesized that LINC00668 exerted regulation on PKN2 expression in such a manner. Through starBase, 42 RBPs were predicted to harbor binding motifs on LINC00668 and 21 RBPs were predicted to possess binding motifs on PKN2. The overlapped 13 RBPs shown by Venn diagram were chosen for further exploration ()). The RIP assay demonstrated that LINC00668 was significantly enriched in HuR-conjugated beads compared to in normal IgG in HGC-27 and SNU-1 cells (Supplementary Fig. 1A, B; )), suggesting the interaction of LINC00668 and HuR protein. Moreover, we proposed RNA–Protein Interaction Prediction (RPISeq) (http://pridb.gdcb.iastate.edu/RPISeq/#), a family of classifiers for predicting RNA–protein interactions using only sequence information. Two variants of RPISeq were shown: RPISeq-SVM, which uses a support vector machine (SVM) classifier, and RPISeq-RF, which uses a Random Forest classifier. The scores of SVM and RF classifiers both more than 0.5 indicated the high binding potential of molecules. Data from RPISeq website suggested that all the score of RF classifier and SVM classifier are no less than 0.7 ()), implying the high binding potential between HuR and LINC00668 (or PKN2 3ʹUTR). Additionally, the motifs of HuR on LINC00668 and PKN2 3ʹUTR are shown in ). RNA pull-down assays depicted that HuR was enriched in Bio-LINC00668 sense group or Bio-PKN2 sense group ()), indicating the binding between LINC00662 and HuR protein as well as binding between PKN2 and HuR protein. Furthermore, RIP assay indicated that the interaction of “HuR-PKN2” was suppressed under PKN2 downregulation or LINC00668 silencing ()), which further identified the interaction of HuR and PKN2.
Figure 4. LINC00668 and PKN2 bound with HuR
LINC00668 cooperated with HuR to stabilize PKN2
We then investigated HuR expression status in GC. Data from GEPIA showed that HuR was upregulated in STAD tissues (n = 408) compared with normal tissues (n = 211), and upregulated HuR predicted poor prognosis of GC patients ()). Western blot analysis indicated the higher protein levels of HuR in GC tissues compared to in adjacent non-tumor tissues. Additionally, HuR accounts for higher nuclear portion than cytoplasmic portion in GC tissues ()). Similarly, RT-qPCR verified that HuR was upregulated in GC cells ()). Then, we knocked down HuR level by transfecting sh-HuR into HGC-27 and SNU-1 cells. RT-qPCR and western blot results showed that mRNA and protein levels of HuR were reduced under transfection with sh-HuR. ()). Subsequently, we assessed the effect of LINC00668 and HuR on PKN2. As shown in ), either knockdown of LINC00668 or HuR significantly reduced PKN2 mRNA and protein levels. Moreover, RT-qPCR and western blot results depicted that LINC00668 depletion had no influence on HuR mRNA and protein levels ()), indicating the binding of “LINC00668-HuR”. It has been demonstrated that EGFR is a target of HuR and HuR mediates mRNA stability of EGFR.Citation27 We found that LINC00668 knockdown or HuR silencing reduced EGFR mRNA half-life in GC cells ()). The results served as the positive control for mRNA stability mediated by HuR in GC cells. Additionally, downregulation of HuR or LINC00668 significantly decreased PKN2 mRNA half-life ()). Based on GEPIA database, there is a positive correlation between PKN2 and HuR expression in stomach adenocarcinoma tissues (Supplementary Fig. 1C). Furthermore, Pearson correlation analysis illustrated that PKN2 expression was positively correlated with HuR expression and LINC00668 expression in GC tissues. There was a positive correlation between HuR expression and LINC00668 expression in GC tissues ()).
Figure 5. LINC00668 cooperated with HuR to stabilize PKN2
LINC00668-facilitated GC malignancy by upregulation of PKN2 at HuR-dependent way
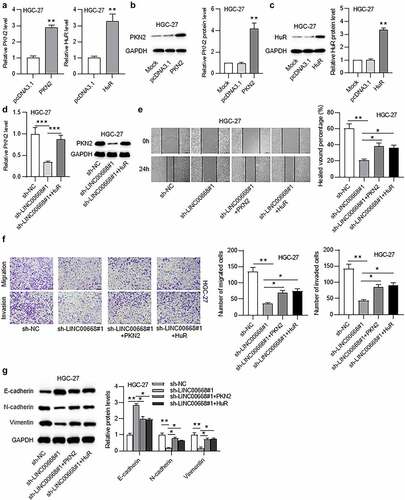
To confirm whether LINC00668 promoted GC malignancy by interacting with HuR and targeting PKN2, rescue assays were conducted in HGC-27 cells. First, mRNA and protein levels of PKN2 in HGC-27 cells were overexpressed by transfection with pcDNA3.1/PKN2,and mRNA and protein levels of HuR in HGC-27 cells were elevated under transfection with pcDNA3.1/HuR ()). Moreover, RT-qPCR and western blot results suggested that HuR overexpression rescued the decreased mRNA and protein levels of PKN2 under LINC00668 silencing in HGC-27 cells ()). Additionally, PKN2 overexpression or HuR upregulation reversed the decreased cell migration and invasion under LINC00668 knockdown ()). Furthermore, the increase of E-cadherin level and the decrease of N-cadherin and vimentin levels under LINC00668 knockdown were reversed by PKN2 overexpression or HuR upregulation ()).
Figure 6. LINC00668 facilitated GC malignancy by upregulation of PKN2 at HuR-dependent way.
Discussion
The progression of GC is extremely complicated, and tumor metastasis plays an essential role during GC progression.Citation28 Additionally, EMT process is strongly associated with tumor metastasis.Citation29 Several markers like E-cadherin, N-cadherin, and vimentin were widely used to evaluate EMT process.Citation30 Previously, a multitude of lncRNAs including XIST, HNRNPKP2, and UCA1 have been proven to regulate tumor metastasis in GC progression.Citation31–33 Recently, LINC00668 has been revealed to promote proliferation in GC cells and predict poor prognosis of GC patients.Citation18 Similarly, in our study, we found that LINC00668 level was upregulated in GC tissues and predicted poor prognosis of GC patients. Moreover, LINC00668 expression was significantly upregulated in GC cells. Additionally, LINC00668 can modulate the tumor metastasis of breast cancer and non-small-cell lung cancer via EMT signaling.Citation34,Citation35 In GC, LINC00668 has been confirmed to be an oncogene by promoting proliferation in vitro and in vivo.Citation18 In our exploration, knockdown of LINC00668 inhibited cell migration, invasion, and EMT process. Furthermore, knockdown of LINC00668 significantly suppressed tumor growth and tumor metastasis in xenograft tumors of mice.
PKN2, a member of the atypical protein kinase C subfamily, is also reported to regulate tumor cell proliferation and migration and tumor metastasis. PKN2 leads to increased colony formation, invasion, and migration in both smoke-exposed cells and head and neck cancer cell lines.Citation36 The expression of PKN2 in colon cancer cells suppresses tumor-associated M2 macrophage polarization and tumor growth.Citation20 In our study, PKN2 upregulation has been identified in GC tissues and cells, which indicated unfavorable prognosis of GC patients. Additionally, knockdown of PKN2 attenuated cell migration, invasion, and EMT process.
In recent years, growing research proposed that lncRNAs cooperated with RBPs to maintain mRNA stability.Citation37 Moreover, HuR is widely reported as RBPs recruited by lncRNAs to stabilize target molecules. For instance, long intergenic noncoding RNA UFC1 cooperates with HuR (ELAVL1) to stabilize the β-catenin mRNA and increase protein levels of β-catenin in hepatocellular carcinoma cells.Citation38 Moreover, LINC00707 interacts with HuR to enhance VAV3/F11R mRNAs stability in GC cells.Citation39 In the present research, HuR was verified to bind with LINC0068 and targeted 3ʹUTR of PKN2. LINC00668 depletion or PKN2 knockdown reduced binding ability of “HuR-PKN2”. According to previous research, HuR, a member of Drosophila embryonic lethal abnormal vision (ELAV) family, is a key regulator of cell growth and differentiation.Citation40,Citation41 Yet, the dysregulation of HuR can induce tumor occurrence.Citation39,Citation41 In our study, HuR was upregulated in GC tissues and predicted poor prognosis of GC patients. Either LINC00668 silencing or HuR downregulation reduced PKN2 mRNA and protein levels. LINC00668 and HuR could not mutually exerted regulation on gene expression. Furthermore, either knockdown of LINC00668 or HuR decrease PKN2 mRNA stability. These findings suggested that LINC00668 cooperated with HuR to stabilize PKN2. Additionally, PKN2 expression was positively correlated with LINC00668 expression and HuR expression in GC tissues, and HuR expression was positively correlated with LINC00668 expression in GC tissues. In addition, HuR overexpression rescued the decreased mRNA and protein levels of PKN2 under LINC00668 silencing in HGC-27 cells. Rescue assays further confirmed that the suppressive effect of LINC00668 downregulation on cell migration, invasion, and EMT process was reversed by PKN2 overexpression or HuR upregulation. LINC00668 recruits HuR protein to promote mRNA stability of PKN2, further reducing E-cadherin and increasing N-cadherin and vimentin. These findings indicated that LINC00668 promoted GC malignancy via upregulation of PKN2 at a HuR-dependent way.
In conclusion, we verified the upregulation of LINC00668, HuR, and PKN2 in GC tissues and cells and confirmed that high level of LINC00668, HuR or PKN2 predicted poor prognosis of GC patients. Zhang et al have previously identified that LINC00668 is associated with PRC2 and epigenetically regulates cyclin-dependent protein kinase inhibitors, while our study first revealed that LINC00668 recruits HuR protein to promote the mRNA stability of PKN2. Zhang et al have previously found that LINC00668 promotes cell proliferation and cell cycle of SGC-7901 and BGC-823 cells, while our study revealed that LINC00668 facilitates migration, invasion, EMT process of HGC-27 and SNU-1 cells. Moreover, we innovatively revealed that LINC00668 promotes lung metastasis. LINC00668 cooperates with HuR to facilitate gastric cancer metastasis by upregulation of PKN2. Our research possibly provides a potential novel molecular insight for GC pathogenesis. However, the pathology of GC is complicated and other mechanism of LINC00668 remains to be explored in future.
Materials and methods
Tissue samples and cell lines
Totally 43 paired GC and adjacent normal tissues were obtained from 60 patients at Eastern Hepatobiliary Surgery Hospital (Shanghai, China). All patients have signed written informed consent, and the Ethics Committee of Eastern Hepatobiliary Surgery Hospital (Shanghai, China) has approved this research. The resected tissues were preserved at −80°C.
Human normal gastric epithelium cells (GES-1) and GC cells including HGC-27, SNU-1, and AGS were bought from the Chinese Academy of Sciences (Shanghai, China). Cells were incubated in the Dulbecco’s Modified Eagle Medium (DMEM; Gibco, USA) containing 10% fetal bovine serum (FBS; Gibco), 100 mg/ml streptomycin, and 100 U/ml penicillin (Invitrogen, USA) in a humid atmosphere at 37°C with 5% CO2.
Cell transfection
The short hairpin RNAs (shRNAs) including sh-LINC00668#1/2 (targeting LINC00668), sh-HuR, and sh-PKN2#1/2 with their negative controls were used to knock down gene levels. The full length of PKN2 was cloned into pcDNA3.1 vector to overexpress PKN2, and empty pcDNA3.1 was used as control. The full length of HuR was cloned into pcDNA3.1 vector to overexpress HuR, and empty pcDNA3.1 was regarded as control. All vectors were transfected into HGC-27 and SNU-1 cells through Lipofectamine 3000 (Invitrogen) for 48 h under manufacturer’s instructions. Vectors used above were obtained from GenePharma (Shanghai, China). Non-transfected HGC-27 cells were taken as mock group.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from patient tissues, cells, or xenograft tumor tissues using TRIzol reagent (Invitrogen) and then reverse-transcribed into complementary DNA with a Reverse Transcription Kit (Takara, Dalian, China). SYBR Premix Ex Taq (Takara) was applied for RT-qPCR analysis on an Applied Biosystems 7500 Real-Time PCR System. Relative expression of LINC00668, HuR, and PKN2 was calculated by the 2−ΔΔCT method with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control. The primers would be provided under requirements.
Protein samples from cytoplasm and nucleus
The GC tissue suspensions were prepared by Trypsin-EDTA (Gibco, Beijing, China) treatment. The proteins were extracted from cytoplasm and nucleus of GC tissues using a kit from Pierce (Rockford, IL) according to the manufacturer’s protocols.
Western blot
Protein samples were extracted from cells or xenograft tumor tissues using Radio Immunoprecipitation Assay lysis buffer (Beyotime, Beijing, China). The extracted protein samples were detected using a Bicinchoninic Acid kit and isolated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. Then, samples were moved onto polyvinylidene fluoride membrane and blocked with 5% nonfat milk for 1 h at room temperature. Next, samples were incubated with primary antibodies including E-cadherin (ab1416; 1:50; Abcam, Shanghai, China), PKN2 (ab138514; 1:1000), N-cadherin (ab18203; 1:5000), HuR (ab200342; 1:1000), vimentin (ab92547; 1:1000), Lamin B1 (ab16048; 1:1000), and GAPDH (ab128915; 1:10,000). GAPDH was cytoplasmic loading control marker and Lamin B1 was nuclear loading control marker. Subsequently, horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies were added to incubate for 2 h at darkness. Finally, protein levels were evaluated by a chemiluminescence detection system (Thermo Fisher Scientific, USA). Antibodies used above were obtained from Abcam (Cambridge, USA).
Trans-well assays
Migration and invasion status of GC cells transfected with vectors was assessed by trans-well assays using trans-well chambers (pore size of 8 μm; Corning). For cell migration, 200 μL of cell suspension containing 1 × 105 cells was added into the top chamber with serum‐free DMEM, and complete DMEM with 10% FBS was added into the bottom chamber. Forty-eight hours after incubation, the migrating cells in the bottom chamber were fixed with 4% paraformaldehyde (Beyotime, Shanghai) and stained with 0.1% crystal violet. Finally, five views in each well were randomly selected, photographed, and counted. For cell invasion, GC cells were operated similarly in the top chamber pre-coated with Matrigel (BD Biosciences, USA).
Wound healing assay
Transfected GC cells (3 × 104 cells/well) were seeded in 24-well plates with lineation overnight at 37°C, scratched, and then washed with phosphate-buffered saline for three times. Then, cells were cultured at 37°C for 24 h. The relative wound width was detected, photographed at 0 and 24 h using microscopy (Leica Germany), and analyzed by ImageJ software (National Institutes of Health, USA).
In vivo assays
Four-week-old BALB/c nude mice (n = 10, male) were used for xenograft assay. All animal operations were authorized by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital (Shanghai, China). Stably transfected HGC-27 cells (1 × 107 cells/ml, 100 µl) containing lentivirus which expressed sh-NC or sh-LINC00668 were subcutaneously injected into the posterior flank of nude mice (n = 5 mice/group). Tumor growth was tested and recorded every 7 days. Tumor volume was calculated (equation V (volume) = L (longitudinal diameter) × W (latitudinal diameter) 2/2). Twenty-eight days after injection, mice were euthanized and sacrificed. The xenograft tumors were resected and photographed.
Tumor metastasis in vivo
To detect tumor metastasis in vivo, the stably transfected HGC-27 cells (1 × 107 cells/ml, 100 µl) were injected into 10 mice via tail vein. Mice were sacrificed 8 weeks after injection, and then the lungs of mice were resected and photographed. The metastatic nodules on lungs of mice were counted.
Bioinformatics analysis
Gene prediction was performed using starBase website (http://starbase.sysu.edu.cn/). The motif represents a structural component of a protein with a specific spatial conformation and specific function. Through starBase database, 42 RBPs were predicted to harbor binding motifs on LINC00668 and 21 RBPs were predicted to possess binding motifs on PKN2 (condition: strict stringency of CLIP Data). As shown in Supplementary file 1, LINC00668 or PKN2 3ʹUTR harbors the site uuuuuc (in yellow), LINC00668 or PKN2 3ʹUTR harbors the site guug (in blue), LINC00668 or PKN2 3ʹUTR harbors the site agug (in green), and only PKN2 3ʹUTR harbors the site uaauuu (in red).
RNA immunoprecipitation assay (RIP) assay
According to the Manufacturer’s instructions, a Magna RIP TM RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) was used for RIP assays. In brief, GC cells were lysed in complete RIP lysis buffer, and then whole cell extract (100 µl) was incubated with magnetic beads conjugated with HuR antibody (ab200342), CSTF2T antibody (ab138486), FUS antibody (ab124923), IGF2BP1 antibody (ab184305), IGF2BP2 antibody (ab128175), LIN28B antibody (ab191881), U2AF2 antibody (ab37530), HNRNPA1 antibody (ab5832), HNRNPC antibody (ab133607), HNRNPM antibody (ab177957), RBFOX2 antibody (ab57154), SRSF1 antibody (ab254935), and TARDBP antibody (ab109535) at 4°C for 6 h. Anti-IgG was regarded as the negative control and input was regarded as the positive control. All antibodies were obtained from Abcam (Cambridge, USA). After washing the beads, the complexes were treated with the Proteinase K to remove proteins. Finally, the purified RNA levels were measured by RT-qPCR analysis. The gel figures are shown in Supplementary Fig. 1A-B.
RNA pull-down assays
LINC00668 sense or antisense as well as PKN2 sense or antisense were transcribed by using T7 RNA polymerase (Ambio Life, Shanghai, China), and then purified with the RNeasy Plus Mini Kit (Qiagen, Germany). RNase-free DNase I (Qiagen) was used to treat LINC00668 or PKN2. Subsequently, LINC00668 sense or antisense as well as PKN2 sense or antisense were biotin-labeled with the Biotin RNA Labeling Mix (Ambio Life). Afterward, a Pierce TM Magnetic RNA-Protein Pull-Down Kit (Pierce, Thermo, USA) was used for RNA pull-down assays under the manufacturer’s instructions. Finally, the enrichment of LINC00668 was calculated by RT-qPCR analysis.
RNA stability assays
HGC-27 and SNU-1 cells were treated with actinomycin D (1 µg/ml) after the transfection with shRNAs or negative control. Then, GC cells were collected at 0, 3, 6, and 9 h. Finally, RNA levels were measured by RT-qPCR analysis.
Statistical analysis
All the data were shown as the mean ± standard deviation. The SPSS 18.0 software (SPSS Inc) was applied for statistical analysis. Kaplan–Meier survival plots were generated on the webpage (kmplot.com/analysis/). The hazard ratio (HR), 95% confidence interval (CI), and log-rank p value were automatically calculated and shown on the webpage. A log-rank p value < .05 was considered as statistically significant. Comparison between two groups was carried out using a two‐tailed Student’s t‐test. Comparison among multiple groups was conducted using one‐way analysis of variance. p < .05 was regarded statistically significant.
Authors’ contributions
J-TL, WD,Q-XJ and F-LZ contributed to the acquisition, analysis, or interpretation of data for this Article. J-TL, WD and F-LZ drafted the manuscript and HD contributed to the manuscript revision. HD conceived and supervised the project.
Statement of conflict of interest
The authors declare that there are no competing interests in this study.
Supplemental Material
Download Zip (3.5 MB)Acknowledgments
Not applicable.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014 May;23(5):700–713. doi:10.1158/1055-9965.Epi–13–1057.
- Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017 Jul;39(7):1010428317714626. doi:10.1177/1010428317714626.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68(6):394–424. doi:10.3322/caac.21492.
- Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013 Jun;42(2):211–217. doi:10.1016/j.gtc.2013.01.002.
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013 Jun;10(6):925–933. doi:10.4161/rna.24604.
- Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017 Oct;108(10):1927–1933. doi:10.1111/cas.13342.
- Hombach S, Kretz M. 2016. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 937:3–17. doi:10.1007/978-3-319-42059-2_1.
- Wu MZ, Fu T, Chen JX, Lin YY, Yang JE, Zhuang SM. LncRNA GOLGA2P10 is induced by PERK/ATF4/CHOP signaling and protects tumor cells from ER stress-induced apoptosis by regulating Bcl-2 family members. Cell Death Dis. 2020 Apr 24;11(4):276. doi:10.1038/s41419-020-2469–1.
- Wang J, Yang S, Ji Q, Li Q, Zhou F, Li Y, Yuan F, Liu J, Tian Y, Zhao Y, et al. Long non-coding RNA EPIC1 promotes cell proliferation and motility and drug resistance in glioma. Molecular Therapy Oncolytics. 2020 Jun 26;17:130–137. doi:10.1016/j.omto.2020.03.011.
- Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016 Feb;14(1):42–54. doi:10.1016/j.gpb.2015.09.006.
- Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014 Aug 12;111(4):736–748. doi:10.1038/bjc.2014.383.
- Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016 Jul;6(7):784–801. doi:10.1158/2159-8290.Cd–15–0921.
- Cao L, Zhang P, Li J, Wu M. LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. eLife. 2017 Dec 4;6. doi:10.7554/eLife.30433.
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3ʹ UTRs via Alu elements. Nature. 2011 Feb 10;470(7333):284–288. doi:10.1038/nature09701.
- Qiu X, Dong J, Zhao Z, Li J, Cai X. 2019. LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle. Onco Targets Ther. 12:5615–5625. doi:10.2147/ott.S188933.
- Hu C, Jiang R, Cheng Z, Lu Y, Gu L, Li H, Li L, Gao Q, Chen M, Zhang X. 2019. Ophiopogonin-B Suppresses Epithelial-mesenchymal Transition in Human Lung Adenocarcinoma Cells via the Linc00668/miR-432-5p/EMT Axis. J Cancer. 10(13):2849–2856. doi:10.7150/jca.31338.
- Liu Z, Wang J, Tong H, Wang X, Zhang D, Fan Q. LINC00668 modulates SOCS5 expression through competitively sponging miR-518c-3p to facilitate glioma cell proliferation. Neurochem Res. 2020 Jul;45(7):1614–1625. doi:10.1007/s11064-020-02988–2.
- Zhang E, Yin D, Han L, He X, Si X, Chen W, Xia R, Xu T, Gu D, De W, et al. E2F1-induced upregulation of long noncoding RNA LINC00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of CKIs. Oncotarget. 2016 Apr 26;7(17):23212–23226. doi:10.18632/oncotarget.6745.
- Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O, Lange C, Giese K, Kaufmann J, Khan M, et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol. 2014 Dec 20;32(36):4141–4148. doi:10.1200/jco.2013.55.0376.
- Cheng Y, Zhu Y, Xu J, Yang M, Chen P, Xu W, Zhao J, Geng L, Gong S. PKN2 in colon cancer cells inhibits M2 phenotype polarization of tumor-associated macrophages via regulating DUSP6-Erk1/2 pathway. Mol Cancer. 2018 Jan 24;17(1):13. doi:10.1186/s12943-017-0747–z.
- Lin W, Huang J, Yuan Z, Feng S, Xie Y, Ma W. Protein kinase C inhibitor chelerythrine selectively inhibits proliferation of triple-negative breast cancer cells. Sci Rep. 2017 May 17;7(1):2022. doi:10.1038/s41598-017-02222–0.
- Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, Yamazoe T, Black T, Sahmoud A, Furth EE, et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev Cell. 2018 Jun 18;45(6):681–695.e4. doi:10.1016/j.devcel.2018.05.027.
- Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017 Jan;11(1):28–39. doi:10.1002/1878–0261.12017.
- Ai Y, Tang Z, Zou C, Wei H, Wu S, Huang D. circ_SEPT9, a newly identified circular RNA, promotes oral squamous cell carcinoma progression through miR-1225/PKN2 axis. J Cell Mol Med. 2020 Oct 14;24:13266–13277. doi:10.1111/jcmm.15943.
- Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016 Jan;17(1):106–116. doi:10.1093/bib/bbv031.
- Song J, Tian S, Yu L, Xing Y, Yang Q, Duan X, Dai Q. AC-caps: attention based capsule network for predicting RBP binding sites of LncRNA. Interdiscip Sci. 2020 Dec;12(4):414–423. doi:10.1007/s12539-020-00379–3.
- Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi J, Liu B, Sun S, Yang F, Wang L, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019 Feb 15;10(3):154. doi:10.1038/s41419-019-1331–9.
- Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu X, Ye Y, Zhang Z, Mills T, Feng J, et al. Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med. 2019 Aug 26;11(1):55. doi:10.1186/s13073-019-0663–5.
- Zhang X, Zhang P, Shao M, Zang X, Zhang J, Mao F, Qian H, Xu W. 2018. SALL4 activates TGF-beta/SMAD signaling pathway to induce EMT and promote gastric cancer metastasis. Cancer Manag Res. 10:4459–4470. doi:10.2147/cmar.S177373.
- Chen D, Zhou H, Liu G, Zhao Y, Cao G, Liu Q. SPOCK1 promotes the invasion and metastasis of gastric cancer through Slug-induced epithelial-mesenchymal transition. J Cell Mol Med. 2018 Feb;22(2):797–807. doi:10.1111/jcmm.13357.
- Zhang Q, Chen B, Liu P, Yang J. XIST promotes gastric cancer (GC) progression through TGF-beta1 via targeting miR-185. J Cell Biochem. 2018 Mar;119(3):2787–2796. doi:10.1002/jcb.26447.
- Zhang Y, Zhang Q, Zhang M, Yuan M, Wang Z, Zhang J, Zhou X, Zhang Y, Lin F, Na H, et al. DC - SIGNR by influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4 in gastric cancer liver metastasis. Mol Cancer. 2017 Apr 13;16(1):78. doi:10.1186/s12943-017-0639–2.
- Gong P, Qiao F, Wu H, Cui H, Li Y, Zheng Y, Zhou M, Fan H. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018 Nov 21;9(12):1158. doi:10.1038/s41419-018-1170–0.
- Derieux S, Svrcek M, Manela S, Lagorce-Pages C, Berger A, André T, Taieb J, Paye F, Voron T. Evaluation of the prognostic impact of pathologic response to preoperative chemotherapy using mandard’s tumor regression grade (TRG) in gastric adenocarcinoma. Digestive Liver Dis. 2020 Jan;52(1):107–114. doi:10.1016/j.dld.2019.07.010.
- Qian W, Zhu Y, Wu M, Guo Q, Wu Z, Lobie PE, Zhu T. 2020. Linc00668 promotes invasion and stem cell-like properties of breast cancer cells by interaction with SND1. Front Oncol. 10:88. doi:10.3389/fonc.2020.00088.
- Rajagopalan P, Nanjappa V, Patel K, Jain AP, Mangalaparthi KK, Patil AH, Nair B, Mathur PP, Keshava Prasad TS, Califano JA, et al. Role of protein kinase N2 (PKN2) in cigarette smoke-mediated oncogenic transformation of oral cells. J Cell Commun Signal. 2018 Dec;12(4):709–721.
- Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. Febs J. 2014 Feb;281(3):802–813. doi:10.1111/febs.12625.
- Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, Wu D, Liu L. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015 Feb;148(2):415–26.e18. doi:10.1053/j.gastro.2014.10.012.
- Xie M, Ma T, Xue J, Ma H, Sun M, Zhang Z, Liu M, Liu Y, Ju S, Wang Z, et al. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett. 2019 Feb 28;443:67–79. doi:10.1016/j.canlet.2018.11.032.
- Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip Rev RNA. 2020 May;11(3):e1581.
- Choi RS, Lai WYX, Lee LTC, Wong WLC, Pei XM, Tsang HF, Leung JJ, Cho WCS, Chu MKM, Wong EYL, et al. Current and future molecular diagnostics of gastric cancer. Expert Rev Mol Diagn. 2019 Oct;19(10):863–874. doi:10.1080/14737159.2019.1660645.
